Abstract
Recently, autophagy has been found to be strongly activated in colon cancer cells, but few studies have addressed the normal colon mucosa. The aim of this study was to characterize autophagy in normal human intestinal cells. We used the expression of LC3-II and BECN1 as well as SQSTM1 as markers of autophagy activity. Using the normal human intestinal epithelial crypt (HIEC) cell experimental model, we found that autophagy was much more active in undifferentiated cells than in differentiated cells. In the normal adult colonic mucosa, BECN1 was found in the proliferative epithelial cells of the lower part of the gland while SQSTM1 was predominantly found in the differentiated cells of the upper part of the gland and surface epithelium. Interestingly, the weak punctate pattern of SQSTM1 expression in the lower gland colocalized with BECN1-labeled autophagosomes. The usefulness of SQSTM1 as an active autophagy marker was confirmed in colon cancer specimens at the protein and transcript levels. In conclusion, our results show that autophagy is active in the colonic gland and is associated with the intestinal proliferative/undifferentiated and progenitor cell populations.
Keywords: BECN1, p62/SQSTM1, autophagy, colon, epithelium, cancer
Introduction
Macroautophagy (referred to hereafter as autophagy) is the major physiological process that regulates degradation and recycling of long-lived proteins, cytosolic components and damaged cellular organelles such as mitochondria, endoplasmic reticulum and peroxisomes, as well as eliminating intracellular pathogens.1 The mechanistic events of autophagy consist of the formation of a double-membrane vesicle, called an autophagosome, surrounding portions of the cytoplasm. This process is mediated by specific autophagic-related (ATG) genes first discovered in yeast, and many orthologs have subsequently been identified in mammals.2 Briefly, a complex formed by the ATG protein BECN1 initiates the nucleation of a double membrane, called the phagophore, around the cytoplasmic material targeted for autophagy. In a second step, microtubule-associated protein 1 light chain 3 (LC3)-I is processed to LC3-II allowing the formation and expansion of the autophagosome, which finally fuses with a lysosome for degradation of the contents.3-5 Autophagy is evaluated in situ and in vitro by monitoring for the expression of the ATG proteins LC3 and BECN1.4,6 Recently, a new marker of autophagy has been identified, SQSTM1/p62, a ubiquitin-associated protein. This protein interacts with ubiquinated proteins through the C-terminal UBA and can self-polymerize via an N-terminal PB1 domain to form aggregates containing these ubiquitinated proteins. These aggregates are sequestered in autophagosomes by interaction of SQSTM1 with LC3 via a specific LC3-recognition sequence, which targets SQSTM1 and associated ubiquitinated proteins for autophagic degradation.7,8 The impaired turnover of SQSTM1 by autophagy deficiency causes an accumulation of these aggregates and is associated with various diseases.9
Since the discovery of ATG genes, autophagy has been linked to numerous pathologies.10,11 For instance, common coding variants of the ATG16L1 gene are associated with increased susceptibility to Crohn disease affecting Paneth cell functions.12,13 In cancer, the Atg gene, Becn1, has been characterized as a tumor suppressor since Becn1+/− mice display increased proliferation and increased frequency of spontaneous malignancy and mammary neoplasia.14,15 Furthermore, the BECN1 gene is monoallelically deleted in most cases of sporadic breast, ovarian, and prostate cancer16,17 and its expression suppresses tumorigenicity of cancer cells.18
In contrast to the majority of human cancers, gastrointestinal cancers (esophageal, stomach and colon) exhibit high levels of autophagy.19-21 More precisely, according to Ahn et al., BECN1 is highly expressed in 95% of colorectal carcinomas, a higher proportion than observed for other gastrointestinal cancers.22 A study by Houri et al. using a cancerous model of intestinal cell differentiation showed a downregulation of autophagic degradation of N-linked-glycoproteins suggesting that perhaps autophagy is regulated in colonic gland differentiation.23 However, the prevalence of autophagy occurring in the normal colon has not been fully investigated.
The aim of this study was to characterize sites of autophagy in the normal colon mucosa and to validate the occurrence of autophagy in colon cancer using BECN1 and SQSTM1 protein levels as indicators for autophagic activity/flux. In this study, we report that autophagy occurs in the normal human colon gland and is associated with the proliferative and progenitor/stem cell populations. We also show that SQSTM1 is degraded by autophagy in the normal colonic gland as well in colon cancer specimens.
Results
Regulation of autophagy during intestinal epithelial cell differentiation
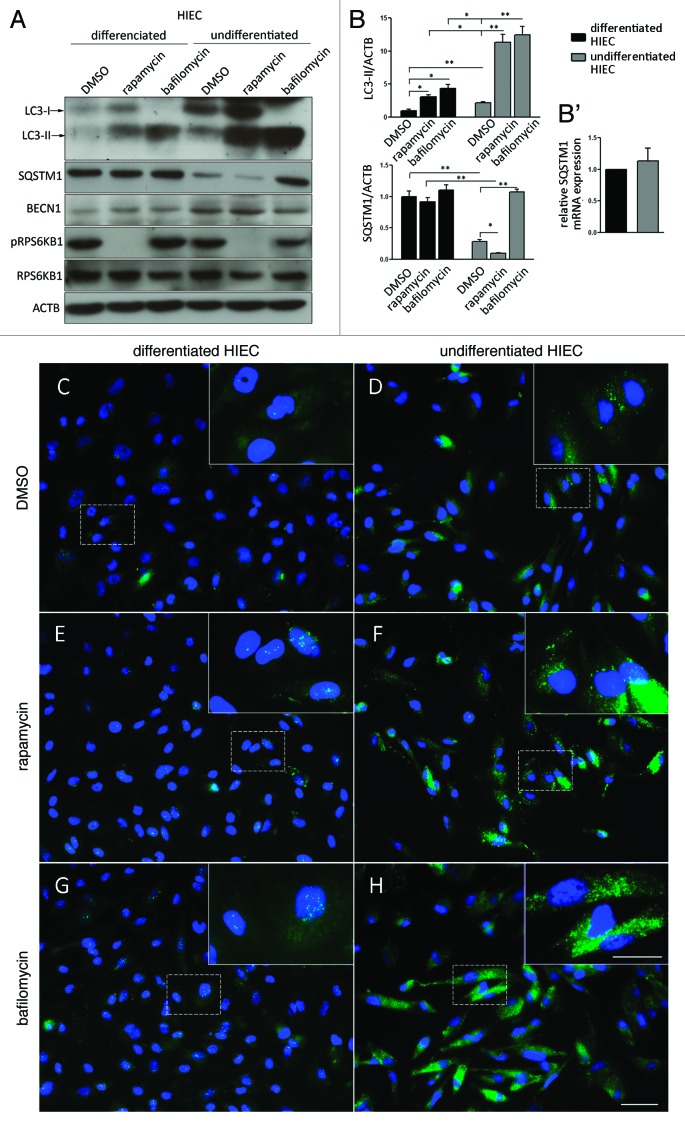
Autophagy was first analyzed as a function of intestinal differentiation. There are a limited number of normal experimental human intestinal cell models. Recently, we showed that undifferentiated human intestinal epithelial crypt (HIEC) cells can be induced to undertake a differentiation program after forced expression of the transcription factors CdX2, HNF1A and GATA4.24 Undifferentiated HIEC cells express the autophagic markers LC3-II, SQSTM1 and BECN1 (Fig. 1A and B) as well the differentiated HIEC. The high proportion of LC3-II and BECN1 and the lower proportion of SQSTM1 expression in undifferentiated cells compared with differentiated HIEC cells suggested that autophagy was highly active in these cells. No variation of SQSTM1 mRNA levels was observed between the two cell lines (Fig. 1B'). Treatment with the MTOR inhibitor rapamycin, a known stimulator of autophagy, significantly increased the amount of LC3-II (Fig. 1A and B) in both cell lines, indicating that while each cell population has different basal levels of autophagy, they are both sensitive to MTOR regulation. Addition of bafilomycin, which blocks the fusion of the autophagosome with the lysosome, also resulted in a significant increase of LC3-II in both cell types (Fig. 1A and B). A significantly higher proportion of LC3-II accumulated in undifferentiated compared with differentiated HIEC cells suggesting that autophagy is more active in proliferating HIEC cells. Expression of SQSTM1 was also monitored under the same conditions. Basal levels of SQSTM1 were significantly lower in undifferentiated HIEC cells than in differentiated cells. Moreover, SQSTM1 levels were lowered by rapamycin treatment and increased by bafilomycin only in undifferentiated HIEC cells despite the confirmed ability for both these treatments to regulate autophagy in differentiated and undifferentiated cells (Fig. 1A and B). Indirect immunofluorescence for LC3 confirmed that the increase in expression levels correlated with stronger overall fluorescence associated with both intense tiny puncta, the autophagosomes, as well as a more diffuse cytoplasmic staining in undifferentiated HIEC cells (Fig. 1C) than in differentiated HIEC cells (Fig. 1D). Treatment of differentiated and undifferentiated cells with rapamycin (Fig. 1E and F) and bafilomycin (Fig. 1G and H) revealed an observable increase in the abundance of bright puncta and in the overall cytoplasmic staining for both cell populations compared with the nontreated cells, but was consistently stronger in undifferentiated HIEC cells.
Figure 1. Downregulation of autophagy with intestinal differentiation in vitro. (A) Representative western blot analysis of LC3, BECN1, SQSTM1, phospho-RPS6KB1 and RPS6KB1 in differentiated and undifferentiated HIEC cells treated with DMSO, rapamycin and bafilomycin for 2 h. (B) Representative graph showing relative amounts of LC3 and SQSTM1 determined by optical densitometry. Data were normalized with ACTB/β-actin. Mean ± SEM, *p ≤ 0.05, **p ≤ 0.01, n = 3, paired t-test. (B') Quantitative RT-PCR of SQSTM1 mRNA in differentiated and undifferentiated HIEC cells. Mean ± SEM, n = 3, ns paired t-test. (C–H) Representative images of indirect immunofluroescence of LC3 on differentiated (C,E and G) and undifferentiated HIEC cells (D,F and H) treated with DMSO (C and D), rapamycin (E and F) and bafilomycin (G and H) for 2 h. Scale bars: 50 µm, insert: 25 µm.
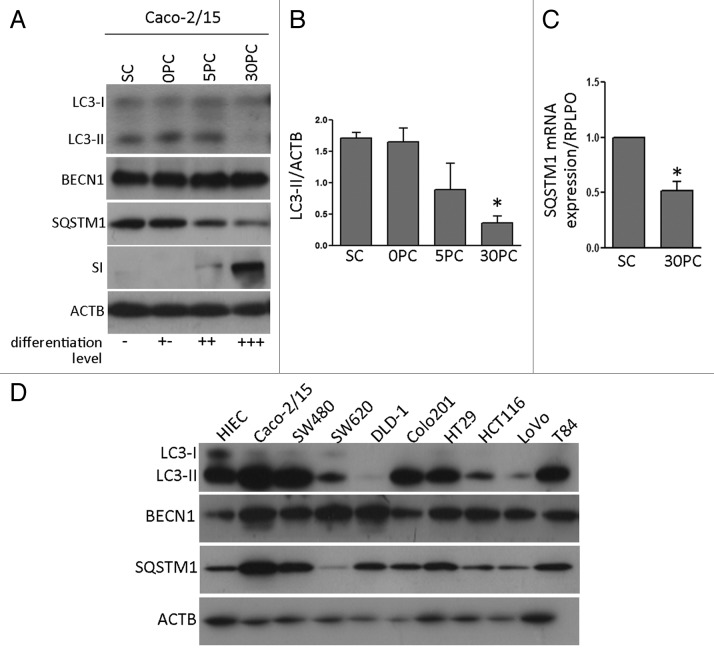
To confirm these observations, autophagy was monitored in Caco-2/15 cells, a cancerous cell model of intestinal differentiation (Fig. 2A and B). The levels of LC3-II decreased over the course of differentiation as monitored by sucrase-isomaltase expression. Unlike during differentiation in the normal cell model, BECN1 was not modulated through differentiation. Furthermore, SQSTM1 levels were decreased and this modulation was paralleled by a decrease in SQSTM1 mRNA expression in differentiated Caco-2/15 cells (Fig. 2C) suggesting that this was not due to autophagic degradation.
Figure 2. Autophagy in cancer cell models. (A) western blot analysis of LC3, BECN1, SQSTM1 and SI/sucrase in Caco-2/15 cells, proliferative at subconfluence (SC) and differentiating at postconfluent stages (PC). (B) Representative graph showing relative amounts of LC3 determined by optical densitometry. Data were normalized with ACTB. Mean ± SEM, *p ≤ 0.05, n = 3, paired t-test. (C) Quantitative RT-PCR of SQSTM1 mRNA in Caco-2/15 cells at sub-confluence (SC) and post-confluence (PC). Mean ± SEM, *p ≤ 0.05, n = 3, paired t-test. (D) Representative western blot analysis of LC3-II, BECN1 and SQSTM1 in 9 colon cancer cell lines for comparison with undifferentiated HIEC cells.
Expression of autophagic markers in various cancer cell models
Expression of LC3-II, BECN1 and SQSTM1 was analyzed in several commonly studied colon cancer cell lines and compared with normal HIEC cells. As shown in Figure 2D, each of these colon cancer cell lines expressed all three autophagic proteins although no correlation could be made between the levels of BECN1 and SQSTM1 protein, and autophagy, as measured by LC3-II. Interestingly, LC3-II was found to be lower in SW620, DLD-1, HCT116 and LoVo compared with HIEC. On the other hand, Caco-2/15, SW480, Colo201, HT29 and T84 exhibited higher or similar levels of LC3-II compared with HIEC. All cancerous cell lines expressed higher BECN1 levels than HIEC. Levels of SQSTM1 were higher or similar between the cancerous cell lines, except for SW620, which was lower.
Localization of BECN1 and SQSTM1 in normal colon tissue
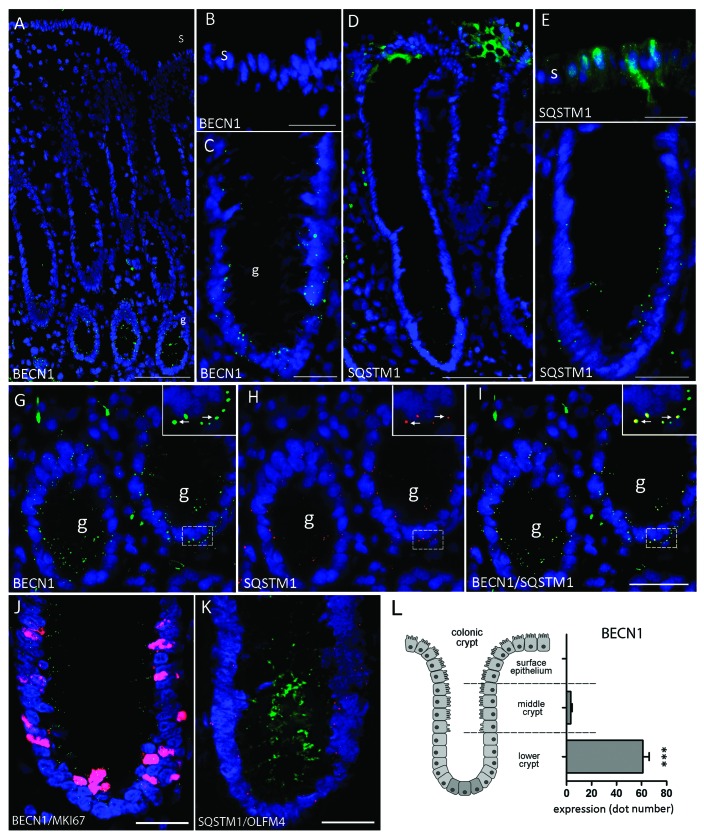
The distribution of BECN1 and SQSTM1 was investigated by indirect immunofluorescence in normal human adult colon specimens. In all 13 specimens studied (9 colon cancer resection margins and 4 noncancer resection margins), BECN1 staining was found to be concentrated in the undifferentiated epithelial cells located in the lower 1/3 of the gland, a region associated with the proliferative cell population (Fig. 3A and C). Positive staining was observed as bright puncta distributed throughout the cytoplasm, as expected for autophagosomes.25 BECN1 expression was not detected in the epithelium located in the upper two-thirds of the colonic gland or in the surface epithelium, regions containing the differentiated cell population (Fig. 3A and B).26 Consistent with its ubiquitous expression,27 using available antibodies that do not specifically recognize the autophagic LC3-II form, LC3 was detected in all epithelial cells as a diffuse staining and additionally as discrete puncta in the crypt region (Fig. S1).
Figure 3. Representative immunofluorescence staining for BECN1 and SQSTM1 in the normal adult colon. (A) Expression of BECN1 in the normal adult colon. (B) Surface epithelium was negative while (C) BECN1 expression showed strong staining in small puncta located in the lower part of the colonic gland (g). (D) Expression of SQSTM1 in the normal adult colon. (E) SQSTM1 was strongly detected in the upper third of the gland and in the surface (s) epithelium. (F) Weak expression was observed in the lower third of the gland (g). (G–I) Colocalization of BECN1 and SQSTM1 in the lower regions of the adult colonic gland. (G) Expression of BECN1 (green), (H) SQSTM1 (red) and (I) merged image showing the colocalization of both proteins (arrows). (J) Colocalization of BECN1 and MKI67 in the lower third of the gland. (K) Colocalization of SQSTM1 and OLFM4 in the lower third of the gland. (L) Estimation of BECN1 expression in the lower crypt, middle crypt and upper crypt/surface epithelium (n = 8; ***p < 0.001). Scale bars: (A) 100 µm; (B, C, E, F, J and K) 25 µm; (D and G–I) 50 µm.
To support these results, the distribution pattern of the newly described autophagic marker SQSTM17 was investigated. In the normal colonic mucosa, SQSTM1 was distributed relatively homogenously in the differentiated cells of the surface epithelium and upper gland according to a diffuse/granular staining pattern (Fig. 3D and E) while it was found as positive granules that appeared much less frequently, although still detectable as bright puncta, in the epithelial cells of the lower gland (Fig. 3F). Since the bright punctate staining of SQSTM1 was reminiscent of BECN1 staining in the proliferative region (Fig. 3C) colocalization of BECN1 and SQSTM1 was performed. As shown in high magnifications of the lower third of the colonic glands where BECN1 expression was concentrated (Fig. 3G–I), punctate staining of SQSTM1 was also detected, albeit more rarely (Fig. 3H). Importantly, these SQSTM1 dots colocalized with BECN1-positive autophagosomes (Fig. 2I, see arrows in insets) suggesting that SQSTM1 was targeted to autophagosomes in the normal tissue. The distribution of these two proteins strongly suggested that autophagy was predominantly associated with the actively growing cell population of the colonic gland, thus correlating with data obtained using the HIEC cell lines. Indeed, as confirmed in Figure 2J and K, BECN1 and SQSTM1 were found in the region associated with the proliferative marker Ki67 and the proliferative/ stem cell marker OLFM4, supporting the observation that autophagy was restricted to the proliferative cell region. Confinement of autophagy to the lower part of the crypt was also confirmed by quantitative analysis of BECN1 staining (Fig. 3L). Pertaining to OLFM4, it has to be pointed out that although reported as an intestinal stem cell marker by in situ hybridization,28 OLFM4 is also expressed throughout the crypt epithelium by immunohistochemistry29 consistent with the fact that it is a secreted protein.30
Localization of SQSTM1 in colon cancers
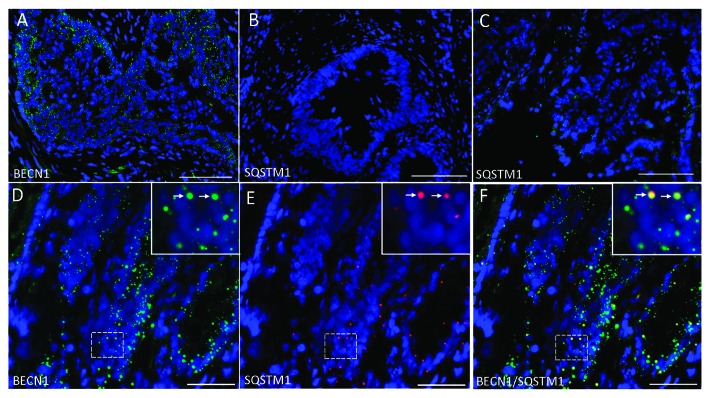
Previous studies have shown that BECN1 and LC3-II expression can be upregulated in colon carcinomas.19,22 As previously demonstrated, strong staining of BECN1 was found in colon cancer specimens (Fig. 4A and D). In the majority of the 12 colon cancer specimens studied (11 moderately differentiated, 1 poorly differentiated), SQSTM1 expression was consistently weaker than in the corresponding resection margin, varying from almost negative (Fig. 4B) to detectable (Fig. 4C and E). Positive staining was found as bright puncta distributed throughout the tissue and no longer appeared to be restricted to a specific region as found in normal tissue. Nevertheless, as observed in the normal colon, punctate staining of SQSTM1 colocalized with BECN1-positive autophagosomes (Fig. 4D–F, see arrows in insets) suggesting that SQSTM1 was also targeted to the autophagosomes in colon cancer.
Figure 4. Representative immunofluorescence staining for BECN1 and SQSTM1 in colon cancer. (A) Expression of BECN1 in colon cancer. (B) Expression of SQSTM1 in colon cancer showing no expression in some specimens or (C) weak expression in others. (D) Colocalization of BECN1 (green) and (E) SQSTM1 (red) and the (F) merged image showing colocalization of both proteins (arrows). Scale: (A–C) = 100 µm; (D–F) = 50 µm.
Colorectal cancer displays increased autophagy by SQSTM1
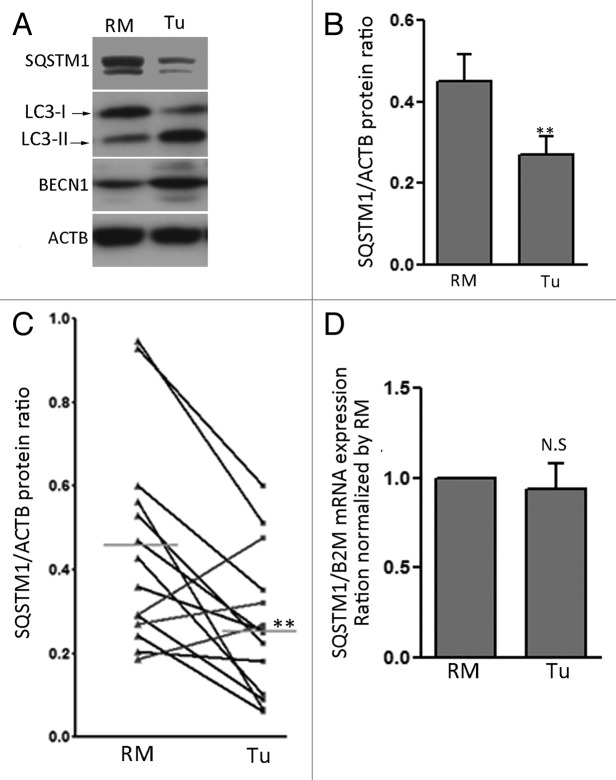
To confirm that increased degradation of SQSTM1 occurred in colon cancer tissue, protein and mRNA expression were analyzed in 14 colorectal cancers (6 stage 1–2, 8 stage 3–4; 12 moderately differentiated, 2 poorly differentiated) paired with their corresponding RMs. Levels of SQSTM1 protein were significantly decreased in cancer samples compared with their corresponding RMs (Fig. 5A and B). The decrease in SQSTM1 levels was seen in 11 of the 14 pairs, resulting in an overall statistically significant decrease of SQSTM1 in primary cancers compared with normal RMs (Fig. 5C). As expected,21,31 LC3-II (71%) and BECN1 (50%) protein levels were increased in tumors (Fig. 5A, data not shown). Interestingly, the decrease in SQSTM1 correlated with increased LC3-II levels (r = 0.547, p ≤ 0.05). Quantitative PCR analysis of the same 14 cancer pairs revealed that SQSTM1 mRNA levels were not affected (Fig. 5D), supporting the hypothesis that the observed decrease in SQSTM1 protein levels is a result of autophagic degradation.
Figure 5. SQSTM1 is degraded in colon cancer specimens. (A) Representative western blot analysis for SQSTM1, LC3-II, BECN1 and ACTB in patient-matched resection margins (RM) and corresponding primary tumors (Tu). Autophagy was strongly activated in Tu compared with RM. (B) Representative graph showing relative amounts of SQSTM1 in patient-matched resection margins and corresponding primary tumors determined by optical densitometry. Data were normalized with ACTB. Mean ± SEM, **p ≤ 0.01, n = 14, paired t-test. (C) Dot graph of individual SQSTM1/ACTB ratios showed a significant decrease in 10 (black) of the 14 paired samples analyzed. (D) Quantitative RT-PCR analyses of SQSTM1 mRNA levels in patient-matched resection margins and corresponding primary tumors. Data were normalized with B2M and RM values were set at 1. n = 14, paired t-test, N.S. = not significant.
Discussion
Autophagy is active in the majority of living cells of healthy human tissues. This process is crucial to mammalian embryonic development and differentiation (erythrocyte, lymphocyte and adipocyte) and as well as to adult cell homeostasis.32,33 In healthy differentiated adult cells, autophagy prevents aging by removing damaged proteins and organelles and actively participates in the clearance of protein aggregates that cause several neurodegenerative diseases, including Alzheimer and Parkinson.32,34
In this study, we used LC3-II, BECN1 and SQSTM1 as markers of autophagy activity4,6-8 in the normal human colonic mucosa and in HIEC cells as a normal experimental cell model. The results of the present study on HIEC cells and the human colon strongly suggest that autophagy was present and active in the normal human colon mucosa in a region known to contain the proliferative progenitor/stem cell populations,26,35 which are critical for the renewal of the colonic epithelium. An association between autophagy in Paneth cells of the small intestine and Crohn disease has been reported,12,13 making the restrictive distribution pattern of autophagy in the region of proliferative progenitor/stem cells of the colon quite interesting and suggests a unique function for autophagy in cell/tissue homeostasis. Interestingly, the reduction of autophagy flux as a function of differentiation in both cell models was consistently observed. Attempts to determine if this reduction correlated with proliferative status produced ambiguous results. Chemical induction of cell cycle arrest in Caco-2/15 cells caused a sharp increase in autophagy (data not shown) most likely due to their cancerous nature which, as a consequence to previous exposure to chemotherapeutic agents, may have adapted this response.36 HIEC cells, however, displayed reduced LC3-II levels when forced to quiescence after their maintenance at confluence (see Fig. S2). Because confluent HIEC cells also display significant decreases in all areas of general metabolism and cellular turnover (unpublished observations), it is difficult to interpret these results. Indeed, undifferentiated and nonproliferative cell population does not occur in the normal colonic epithelium in vivo.
One hypothesis for the restricted localization of this process in the colon could be that autophagy acts as a cytoprotective mechanism of highly proliferative cells.1 Moreover, autophagy could prevent the accumulation of DNA damage that would lead to the development of malignant epithelial cells that lead to cancer.37 Indeed, accumulating data in the literature indicates that colon cancer may originate from mutations or defects specifically affecting stem cells.35 In this way, active autophagy in the colonic gland could offer a protective “caretaker” mechanism to avert chromosomal instability, thereby acting as a tumor suppressor.37
Recently, it has been demonstrated by Mathew et al. that accumulation of SQSTM1 in autophagy-defective cells promotes tumorigenesis as a result of ROS accumulation and enhanced DNA damage through suppression of NFκB activation.8 Also, it has been demonstrated that SQSTM1 can regulate the WNT-CTNNB1/β-catenin pathway by sequestering ubiquinated proteins (e.g., CTNNB1) and prevent their degradation by limiting their access to the proteasome.38 More recently, SQSTM1 has been found to negatively regulate the WNT-CTNNB1 pathway under starvation conditions through the promotion of the DVL2-LC3 interaction, which consequently enhances DVL2 degradation through autophagy.39 Therefore, tight regulation of SQSTM1 levels by autophagy could be a natural mechanism that protects the progenitor cell from malignant transformation through regulation of the WNT-CTNNB1 pathway. Furthermore, the rapid elimination of differentiated cells by anoikis40 could also be considered as an essential mechanism to prevent malignant transformation of these cells resulting from an accumulation of SQSTM1. Future studies will be needed to confirm the precise role of autophagy in progenitor/stem cells and cancer.
Autophagy is regulated by several extracellular and intracellular signals.1 The molecular elements that ensure autophagy is restrained to the lower regions of the colonic gland remain largely unknown. A possible mechanism of autophagy regulation in these differentiated cells may occur via the regulation of the BECN1 protein level or activity by partners such as SH3GLB1/bif-1, UVRAG, AMBRA1, ATG14/BARKOR41 or BCL2.42 However, while the UVRAG gene is mutated in 20% of colon cancers, it has recently been shown that loss of UVRAG has no effect on autophagy initiation in colon cancer cells.43
Colorectal cancer is the second leading cause of death by cancer in the United States.44 Research for new therapeutic targets has been on the rise over the past few years and autophagy processes appear to be one of the most promising. The role of autophagy in cancer is controversial and has emerged as numerous studies have found that this process was lost in breast, ovarian, prostate, lung, liver and brain cancers, but was reported to be upregulated in colon cancer as evaluated by an increase in LC3 or BECN1 expression in a significant proportion of primary tumors as compared with normal adjacent tissue.19-22 Our data showing an increase in LC3-II and BECN1 expression and significant concomitant reduction of SQSTM1 at the protein level while the latter remains stable at the transcript level are in agreement with these studies. However, it has to be pointed out that other studies report opposite results, namely increased expression of SQSTM1 in colon tumor samples39,45 indicating that the role of autophagy in colon cancer may be complex. In a recent large study, Koukourakis et al. reported over- and underexpression of BECN1 in colon cancer, which were both associated with poor survival suggesting the existence of two distinct biological pathways linked with tumor aggressiveness.8,19,21,22,39,45-48
In conclusion, this study is the first to highlight that autophagy is physiologically active in the proliferative compartment of the normal colonic epithelium. As schematized in Figure 6, the localization of the autophagy marker BECN1 is restricted to the proliferative and progenitor/stem cells. A functional role for autophagy in this region is emphasized by the SQSTM1 expression pattern. SQSTM1 displays cytoplasmic staining in the surface epithelium, but in the proliferative region is weak, and more significantly, localizes in BECN1-positive structures, strongly suggesting regulation of SQSTM1 by autophagy. Our results thus suggest that autophagy may play an important function in intestinal proliferative/undifferentiated and progenitor/stem cell homeostasis. Future studies focusing on the targeting of autophagy for the treatment of colon cancer need to be cautious and should analyze the effect on normal colon beforehand, as this mechanism appears to be physiologically active.
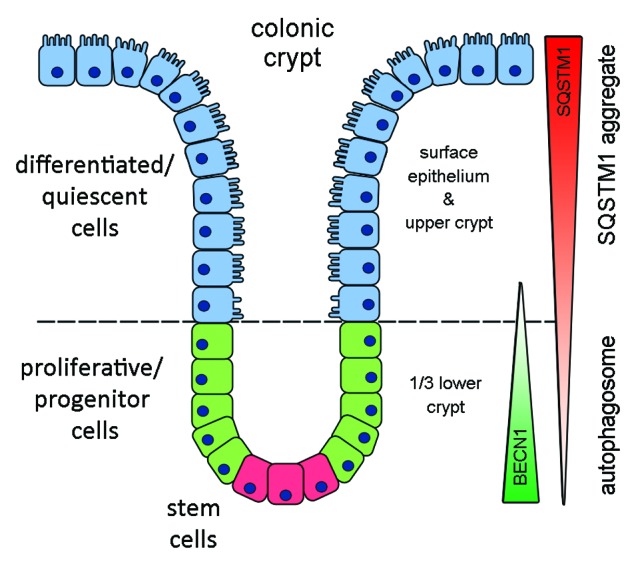
Figure 6. Schematic representation of BECN1 and SQSTM1 expression in normal intestinal cells. BECN1 was found predominantly expressed in the undifferentiated proliferative and progenitor/stem cells of the lower gland while SQSTM1 expression was weak in undifferentiated cells but stronger in differentiated cells where autophagy is minimal.
Materials and Methods
Human colon tissue samples
Adult human colon specimens were obtained from 14 patients between the ages of 49 and 86 undergoing surgical treatment for colon adenocarcinoma without neoadjuvant therapy. For each patient, samples from the primary tumor and from nondiseased areas (at least 10 cm from the lesion) corresponding to the resection margin were obtained. Diagnoses, staging and grading were performed by the pathologists of the Department of Pathology of the CHUS. Resection margins of colon specimens obtained from four patients undergoing surgery for pathologies other than colon cancer (bowel obstruction, diverticulitis, etc.) were also used in indirect immunofluorescence experiments. All tissues were obtained in accordance with protocols approved by the local Institutional Human Research Review Committee for the use of human material. The preparation and embedding of tissues for cryosectioning were performed as described previously.26
Antibodies and materials
Mouse primary antibodies used in this study were: anti-SQSTM1 (WB: 1:4000, IF 1:1000) (BD Transduction Laboratories, clone 3/P62 LCK LIGAND), anti-ACTB/β-actin (WB: 1/75000) (Chemicon International, C4), anti-MKI67 (IF: 1:1000) (Milipore, clone Ki-S5). Rabbit primary antibodies used in this study were: anti-BECN1 (WB: 1:2000, IF: 1:1000) (Abcam, ab16998), anti-LC3 (WB 1:1000, IF: 1:100) (Sigma-Aldrich, L8918), anti-RPS6KB1 (WB: 1:1000) (Cell Signaling, 9205), anti pRPS6KB1 (WB: 1:1000) (Cell Signaling, 9202), anti-OLFM4 (IF: 1:2000) (Imgenex, IMG-5983A), anti-CCND/cyclin D (WB:1:2000) (Santa Cruz Biotechnology, C-17). Secondary antibodies used were Alexa Fluor 488 or 594 goat anti-mouse (Invitrogen, A11017, A11072) and Alexa Fluor 488 or 594 goat anti-rabbit (Invitrogen, A11070, A11020). Rapamycin (Sigma-Aldrich, 53123-88-9) was used at a final concentration of 10 µm.
Cell culture
The crypt-like human intestinal epithelial cells, HIEC, were generated and grown as described previously.24,49 These cells exhibit all the morphological and functional characteristics of normal human proliferative/stem cell crypt cells and are considered to be undifferentiated crypt-like progenitor cells.24,49 The HIEC(IndHNF1α/Cdx2) + GATA-4 cells were generated as previously described.24 HIEC(IndHNF1α/Cdx2) + GATA-4 cells exhibit characteristics of differentiated intestinal epithelial cells.24 Colon cancer cell lines were obtained and cultured according to ATCC guidelines (www.ATCC.org).
RNA extraction and RT-PCR
RNA extraction and reverse transcription were performed as previously described.50 The primer pairs used were: for SQSTM1, p62-F 5′-CCCGTCTACAGGTGAACTCC-3′ and p62-R 5′-CTGGGAGAGGACTCAATCA-3′, for BECN1, becn1-F 5′-AGGTTGAGAAAGGCGAGACA-3′ and becn1-R 5′-AATTGTGAGGACACCCAAGC-3′. Amplification was for 35 cycles with an annealing temperature of 55°C. Other primers used were for RPLP0 (ribosomal protein, large, P0), CDH1/E-cadherin and TNC/tenascin C as previously described.51,52
Quantitative RT-PCR analyses
Total RNA from tissue of colon cancer specimens and corresponding resection margins (RM) were extracted using the RNeasy Plus Mini Kit (Qiagen, 74136). RT-PCR analyses were performed using AMV-RT (Roche Diagnostics, 10109118001) according to the manufacturer’s instructions. Quantitative PCR was performing using an MXP3000P Real-Time System (Stratagene) as previously described.53 The reference gene used for these experiments was B2M (β-2-microgobulin) based on a previous study regarding appropriate housekeeping genes in normal intestine and colon cancer.53 The primer pairs used for SQSTM1 were the same as above and as previously described for B2M.53 The annealing temperature of the reactions was 60°C and the amplification efficiencies of the primers were 97% for SQSTM1 and 102% for B2M as determined by standard curve analysis. Relative mRNA levels were determined by normalizing levels from colon cancer samples to those from matching RM and calculated according to the Pfaffl mathematical model.54 All samples were run in triplicate and the no template control (NTC) did not show an amplification product.
Western blot
Western blot analyses were performed on SDS-PAGE gels under denaturing conditions as previously described.51 Total protein (50 µg/ml) preparations were separated on 12% or 15% gels and electrotransferred onto a nitrocellulose membrane (BioRad, 162-0115). Nonspecific protein binding was blocked using 10% Blotto-0.1% Tween followed by incubation with primary antibodies diluted in the blocking solution, overnight at 4°C. Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (anti-mouse, anti-rabbit, GE Healthcare Amersham Bioscience, NA934V, NA931V) and developed using the Immobilon Western® kit (Millipore, WBKLSO100).
Indirect immunofluorescence
For standard indirect immunofluorescence, human adult colon and colorectal samples (well to moderately differentiated) were cut in 3-µm thin sections with a Leica CM3050S cryostat on silane-coated glass slides (Fisher Scientific, 125523).51 For immunofluorescence of BECN1 and SQSTM1, sections were fixed in MeOH for 10 min at −20°C and nonspecific sites were blocked for 1 h at room temperature with 10% Blotto-PBS (pH 7.4). Primary antibodies were diluted 1:1000 in blocking solution containing 0.05% azide and incubated overnight at 4°C. The secondary antibody used was diluted in the blocking solution 1:400 at room temperature for 1 h. Tissue was stained with 0.01% Evan’s blue in PBS (pH 7.4), the slides were mounted as previously described26,51 and viewed with a DMRXA microscope (Leica) equipped for epifluorescence and digital imaging (RTE/CCD Y/Hz-I300 cooled camera). Images were acquired using MetaMorph software (Universal Imaging Corporation) with 20× and 40× objectives and images were modified using Photoshop software (Adobe). BECN1 expression was estimated by counting positive granules in the lower third, middle third and upper third/surface epithelium in a minimum of 3 crypts per section from samples obtained from eight patients.
Supplementary Material
Acknowledgments
The work was supported by the Canadian Institutes of Health Research Grants MOP-57727 and MOP-97836 (to J.F.B.). J.F.B. is the recipient of a Canadian Research Chair in Intestinal Physiopathology and is a member of the FRSQ-funded Centre de Recherche Clinique Étienne Lebel of the CHUS.
Glossary
Abbreviations:
- RM
resection margin
- ATG
autophagy related
- LC3
microtubule-associated protein1 light chain 3
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/19738
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/19738
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 3.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–49. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 5.Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–62. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 6.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichimura Y, Kominami E, Tanaka K, Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 2008;4:1063–6. doi: 10.4161/auto.6826. [DOI] [PubMed] [Google Scholar]
- 8.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–63. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klionsky DJ. Crohn’s disease, autophagy, and the Paneth cell. N Engl J Med. 2009;360:1785–6. doi: 10.1056/NEJMcibr0810347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization ofbeclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 17.Gao X, Zacharek A, Salkowski A, Grignon DJ, Sakr W, Porter AT, et al. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995;55:1002–5. [PubMed] [Google Scholar]
- 18.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 19.Li BX, Li CY, Peng RQ, Wu XJ, Wang HY, Wan DS, et al. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy. 2009;5:303–6. doi: 10.4161/auto.5.3.7491. [DOI] [PubMed] [Google Scholar]
- 20.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, et al. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–84. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 21.Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, et al. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol. 2008;33:461–8. [PubMed] [Google Scholar]
- 22.Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, et al. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344–9. doi: 10.1111/j.1600-0463.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- 23.Houri JJ, Ogier-Denis E, Trugnan G, Codogno P. Autophagic degradation of N-linked glycoproteins is downregulated in differentiated human colon adenocarcinoma cells. Biochem Biophys Res Commun. 1993;197:805–11. doi: 10.1006/bbrc.1993.2550. [DOI] [PubMed] [Google Scholar]
- 24.Benoit YD, Paré F, Francoeur C, Jean D, Tremblay E, Boudreau F, et al. Cooperation between HNF-1alpha, Cdx2, and GATA-4 in initiating an enterocytic differentiation program in a normal human intestinal epithelial progenitor cell line. Am J Physiol Gastrointest Liver Physiol. 2010;298:G504–17. doi: 10.1152/ajpgi.00265.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang C, Feng P, Ku B, Oh BH, Jung JU. UVRAG: a new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- 26.Dydensborg AB, Teller IC, Groulx JF, Basora N, Paré F, Herring E, et al. Integrin alpha6Bbeta4 inhibits colon cancer cell proliferation and c-Myc activity. BMC Cancer. 2009;9:223. doi: 10.1186/1471-2407-9-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanida I, Ueno T, Kominami E. LC3 and Autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 28.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–7. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Liu Y, Zhu J, Wright E, Ding I, Rodgers GP. Reduced hGC-1 protein expression is associated with malignant progression of colon carcinoma. Clin Cancer Res. 2008;14:1041–9. doi: 10.1158/1078-0432.CCR-07-4125. [DOI] [PubMed] [Google Scholar]
- 30.Besson D, Pavageau AH, Valo I, Bourreau A, Bélanger A, Eymerit-Morin C, et al. A quantitative proteomic approach of the different stages of colorectal cancer establishes OLFM4 as a new nonmetastatic tumor marker. Mol Cell Proteomics. 2011;10:M111.009712. doi: 10.1074/mcp.M111.009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivridis E, Koukourakis MI, Zois CE, Ledaki I, Ferguson DJ, Harris AL, et al. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Am J Pathol. 2010;176:2477–89. doi: 10.2353/ajpath.2010.090049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos RX, Cardoso S, Correia S, Carvalho C, Santos MS, Moreira PI. Targeting autophagy in the brain: a promising approach? Cent Nerv Syst Agents Med Chem. 2010;10:158–68. doi: 10.2174/187152410791196350. [DOI] [PubMed] [Google Scholar]
- 35.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 36.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 37.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–27. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 40.Edelblum KL, Yan F, Yamaoka T, Polk DB. Regulation of apoptosis during homeostasis and disease in the intestinal epithelium. Inflamm Bowel Dis. 2006;12:413–24. doi: 10.1097/01.MIB.0000217334.30689.3e. [DOI] [PubMed] [Google Scholar]
- 41.Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5:713–6. doi: 10.4161/auto.5.5.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–6. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knævelsrud H, Ahlquist T, Merok MA, Nesbakken A, Stenmark H, Lothe RA, et al. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6:863–70. doi: 10.4161/auto.6.7.13033. [DOI] [PubMed] [Google Scholar]
- 44.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 46.Koukourakis MI, Giatromanolaki A, Sivridis E, Pitiakoudis M, Gatter KC, Harris AL. Beclin 1 over- and underexpression in colorectal cancer: distinct patterns relate to prognosis and tumour hypoxia. Br J Cancer. 2010;103:1209–14. doi: 10.1038/sj.bjc.6605904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pardo R, Lo Ré A, Archange C, Ropolo A, Papademetrio DL, Gonzalez CD, et al. Gemcitabine induces the VMP1-mediated autophagy pathway to promote apoptotic death in human pancreatic cancer cells. Pancreatology. 2010;10:19–26. doi: 10.1159/000264680. [DOI] [PubMed] [Google Scholar]
- 49.Perreault N, Beaulieu JF. Use of the dissociating enzyme thermolysin to generate viable human normal intestinal epithelial cell cultures. Exp Cell Res. 1996;224:354–64. doi: 10.1006/excr.1996.0145. [DOI] [PubMed] [Google Scholar]
- 50.Teller IC, Auclair J, Herring E, Gauthier R, Ménard D, Beaulieu JF. Laminins in the developing and adult human small intestine: relation with the functional absorptive unit. Dev Dyn. 2007;236:1980–90. doi: 10.1002/dvdy.21186. [DOI] [PubMed] [Google Scholar]
- 51.Gagné D, Groulx JF, Benoit YD, Basora N, Herring E, Vachon PH, et al. Integrin-linked kinase regulates migration and proliferation of human intestinal cells under a fibronectin-dependent mechanism. J Cell Physiol. 2010;222:387–400. doi: 10.1002/jcp.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francoeur C, Bouatrouss Y, Seltana A, Pinchuk IV, Vachon PH, Powell DW, et al. Degeneration of the pericryptal myofibroblast sheath by proinflammatory cytokines in inflammatory bowel diseases. Gastroenterology. 2009;136:268–77, e3. doi: 10.1053/j.gastro.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 53.Dydensborg AB, Herring E, Auclair J, Tremblay E, Beaulieu JF. Normalizing genes for quantitative RT-PCR in differentiating human intestinal epithelial cells and adenocarcinomas of the colon. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1067–74. doi: 10.1152/ajpgi.00234.2005. [DOI] [PubMed] [Google Scholar]
- 54.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.