Abstract
The recent advent in single-molecule imaging and manipulation methods has made a significant impact on the understanding of molecular mechanisms underlying many essential cellular processes. Single-molecule techniques such as electron microscopy and DNA fiber assays have been employed to study the duplication of genome in eukaryotes. Here, we describe a single-molecule assay that allows replication of DNA attached to the functionalized surface of a microfluidic flow cell in a soluble Xenopus leavis egg extract replication system and subsequent visualization of replication products via fluorescence microscopy. We also explain a method for detection of replication proteins, through fluorescently labeled antibodies, on partially replicated DNA immobilized at both ends to the surface.
1. Introduction
Single-molecule techniques have become popular in the last two decades and have been used to study many biological processes. A major advantage of single-molecule methods is that measurements are made on individual molecules allowing their kinetics to be determined directly. Therefore, one can gain essential information about transient dynamics and heterogeneities, which are not accessible using conventional biochemical approaches. Dynamics of individual molecules are generally studied using fluorescence microscopy [1–3]. Furthermore, mechanical manipulation of individual molecules can be achieved through a number of single-molecule techniques such as optical and magnetic tweezers, atomic force microscopy, and flow stretching, which elucidates the response of biological molecules to an external force [4, 5].
The eukaryotic DNA replication field has benefited from single-molecule methods. Early single-molecule studies of eukaryotic DNA replication involved taking pictures of replication intermediates via electron microscopy [6–8]. DNA fiber autoradiography which relies on pulse-labeling replicating chromosomal DNA with 3H-thymidine provided the first evidence for bidirectional replication in eukaryotic cells [9]. A more convenient DNA fiber assay has been developed which makes use of modified nucleotides such as Bromouridine (BrdU), Chlorouridine (CldU), Iododeoxyuridine (IdU), digoxigenin-dUTP or biotin-dUTP for pulse-labeling DNA and detects their incorporation via fluorescent antibodies [10–14]. These DNA fiber methods have also been combined with immunostaining of chromatin-associated proteins [15] and with fluorescence in situ hybridization (FISH) to visualize DNA replication at specific loci and determine the location of replication origins in the eukaryotic genome [16–18]. In electron microscopy or DNA fiber methods, DNA is first replicated in vivo or in vitro in a test tube and DNA molecules are subsequently spread on a surface and imaged. These methods provide significant information regarding the location and timing of replication initiation, origin density, and fork rates.
Single-molecule methodology has been applied extensively to study DNA replication in prokaryotes. For example, replication of surface-tethered DNA substrates has been studied using fluorescence microscopy, flow stretching, optical tweezers, and magnetic tweezers, providing crucial information regarding molecular mechanisms of replisome components [19–25]. On the other hand, eukaryotic DNA replication has not been investigated using similar single-molecule assays in a microfluidic flow cell, mostly due to the absence of a reconstituted system that recapitulates chromosomal replication in eukaryotes.
Cell-free extracts derived from the eggs of the African clawed frog (Xenopus laevis) have been used as model systems for DNA replication studies in higher eukaryotes. In the classic approach, a low-speed supernatant (LSS) of egg cytoplasm is mixed with sperm chromatin [26]. In this system, formation of a nuclear envelope precedes replication of chromatin. A variation of this system has been developed that eliminates the requirement for nuclear envelope formation [27]. This soluble cell-free extract system consists of two extracts. DNA is first mixed with high-speed supernatant (HSS) of egg cytoplasm that lacks membrane vesicles and supports the ORC-dependent assembly of the MCM2-7 helicase into pre-replication complexes (pre-RC) on DNA (the “licensing” reaction). Licensed DNA is then added into a second, nucleoplasmic extract (NPE), leading to initiation of replication. Thus, HSS mimics the events of replication initiation that occur during the G1 phase of the cell cycle whereas NPE recapitulates the S phase. An important benefit of this approach is that it allows replication of virtually any DNA template such as λ DNA or circular plasmids.
In this review, we describe a technique that allows replication of surface-immobilized DNA in the nucleus-free Xenopus egg extract system. We first give a detailed protocol for construction of a microfluidic flow cell that is used as a platform to immobilize DNA substrates. We then describe end modification of λ DNA and subsequent tethering of DNA to the surface of a flow cell. Next, we define steps required for replication of surface-immobilized DNA using egg extracts, labeling replicated DNA, and subsequent visualization of replication products. Finally, we describe a method that allows immunostaining and imaging of replication proteins on individual, partially replicated and stretched DNA.
2. Description of Methods
2.1. Flow cell preparation
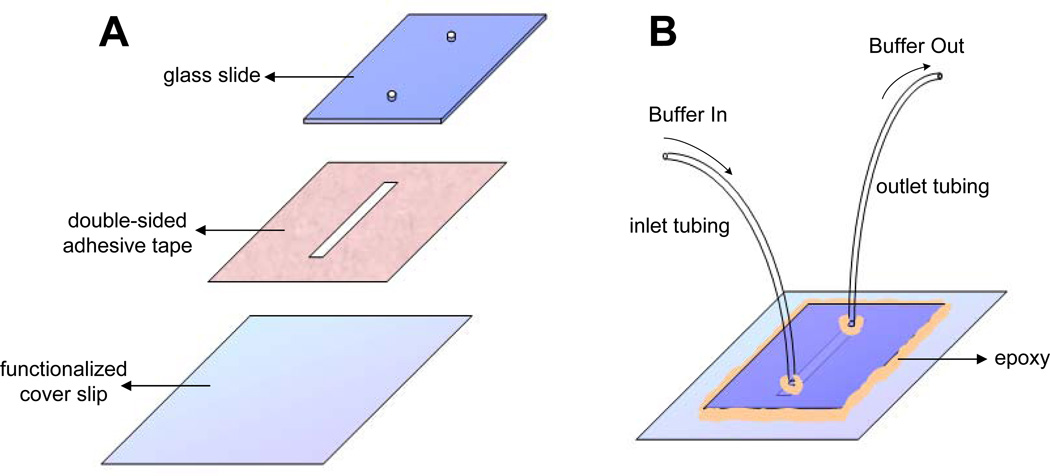
Glass coverlips were cleaned and functionalized with partially biotinylated polyethylene glycol (PEG) as described in [28]. Briefly, 20 coverslips (VWR VistaVision 2.4×6.0cm No. 1.5) were placed in polypropylene staining jars (5 coverslips per jar), rinsed with water and cleaned via sonication in anhydrous ethanol (EtOH) for 30 minutes. Subsequently, coverslips were rinsed with water and sonicated in 1 M potassium hydroxide (KOH) for 30 minutes. Sonication in EtOH and KOH was repeated for one more round. Water has to be completely removed from the jar for subsequent silane treatment. Therefore, coverslips were rinsed with acetone twice in the jar and sonicated in acetone for 10 min on the third rinse. Coverslips were then treated with silane (Sigma A3648) solution (2% volume/volume in acetone) by filling the jar with silane solution and shaking it for 2 minutes. To cure silane, coverslips were dried in an oven at 110 °C for 30 minutes. 150 mg PEG (Lysan, mPEG-SVA-5000) and 3 mg biotin-modified PEG (Lysan, Biotin-PEG-SVA-5000) was dissolved in 1 ml sodium carbonate (1 M, pH 8.2). To functionalize coverslips, the PEG mixture was sandwiched between coverslips (100 µl PEG mixture per two coverlips) and incubated for 3 hours. Finally, coverslips were separated, rinsed thoroughly with water, and air-dried. The coverlips were stored in a desiccator under vacuum for at least one month without significant loss of surface functionality. Before use, a coverslip was cut in half with a diamond-tipped glass scribe (SPI Supplies, 06004-AB) and the unused piece was placed back in the desiccator for later use. 20 µl of streptavidin (1 mg/ml in PBS pH 7.5, Sigma-Aldrich, S4762) was mixed with 100 µl blocking buffer (20 mM Tris pH 7.5, 50 mM NaCl, 2 mM EDTA, 0.2 mg/ml BSA), spread on the biotinylated surface of the coverslip, and incubated for at least 30 minutes. A glass (VWR, 48300-025) or quartz (Technical Glass, 1-mm thick, 2 × 5 cm) slide was cut to dimensions of 1.5 cm × 2 cm (Figure 1A). Using a drill machine (Dremel, Model 395) and a diamond-tipped bit, two holes (1.4 cm apart) were drilled on the slide with appropriate sizes to tightly accommodate inlet (Intramedic, PE20; inner diameter 0.015”, outer diameter 0.043”) and outlet (Intramedic, PE60; inner diameter 0.03”, outer diameter 0.048”) polyethylene tubing. A piece of double-sided tape (Grace Bio-Labs, 0.12 mm thick) that is protected by a transparent plastic on one side and white paper on the other was used to construct the flow cell. A rectangular channel with a width of 1.5–2 mm and a length that is sufficient to cover both holes on the microscope slide was drawn on the paper side and subsequently cut out of the tape with a razor blade. The plastic side was peeled off, the tape was adhered to the slide, and edges of the tape flanking sides of the slide were removed using a razor blade. The streptavidinated coverslip was washed with distilled water, dried using compressed air, and attached to the slide via the double-sided tape after peeling off the paper side of the tape. To form a good seal between the coverslip and the slide, a tip of a plastic tweezer (SPI Supplies, K35A) was pressed against the coverslip surface and air was removed from the interface. The inlet and outlet tubing (both 10 cm in length) were attached to the flow cell (Figure 1B). Outer edges of the slide and tubing were sealed with epoxy (Devcon, 5 minute Epoxy). After the epoxy set, blocking buffer was slowly drawn into the flow cell using a 1 ml syringe and 21 gauge needle through the outlet tubing and incubated for at least 15 minutes before use.
Figure 1.
Assembly of a microfluidic flow cell for single-molecule experiments. (A) A glass slide with two holes, double-sided tape, and streptavidin functionalized coverslip were prepared prior to making a flow cell. (B) After the flow cell is assembled, inlet and outlet tubings were fixed and secured with epoxy as depicted. Edges of the coverslip were sealed to prevent formation of air bubbles within the flow channel.
It is important to avoid air bubbles in the flow cell as they may cause shearing of surface-immobilized DNA. Therefore, blocking buffer (5–10 ml in a 15 ml tube) and other buffers to be drawn into the flow cell at a later stage of the experiment were degassed in a desiccator under vacuum for at least one hour at room temperature. Occasional tapping of the desiccator helps dislodging bubbles from the tube walls and thus removal of air from buffer. This degassed blocking buffer was used in all subsequent washes. After the flow cell was assembled, it was placed on a microscope stage and secured with stage clips. Inlet tubing was then merged into a microcentrifuge tube filled with 1 ml blocking buffer that was supported by a binder clip on the stage. The outlet tubing was attached to a longer (30–40 cm) stretch of tubing via a double-sided needle. This tubing was then connected to a 5 ml syringe that was installed on an automated syringe pump (Harvard Apparatus, Pump 11 Plus Single Syringe). To remove air bubbles within the flow channel and tubes, blocking buffer was drawn in at a rate of 150 µl/min while flicking the outlet tubing. This mechanical perturbing of the tubing was repeated until air bubbles were not visible anymore in the channel. Any small air bubble within the channel can be visualized using bright field illumination and 10x magnification objective through eyepiece.
Microscope slides can be reused after each experiment. To disassemble a flow cell, it was placed in acetone in a glass container and incubated overnight which dissolved the epoxy and double-sided tape. The slide was cleaned thoroughly by wiping with acetone before making a new flow cell. Functionalized coverslips were discarded after use.
2.2. Functionalization and immobilization of λ DNA
λ phage has a 48.5 kb linear genome with 12 base pairs (bp) complementary single-stranded DNA ends (Figure 2A). To biotinylate the left end of λ DNA, 50 µl of λ DNA (0.5 mg/ml, New England Biolabs (NEB), N3011) was 5’ phosphorylated using T4 polynucleotide kinase (T4 PNK, 1 µl of 10 U/µl, NEB, M0201) at 37°C in 1x T4 PNK buffer (70 mM Tris pH 7.6, 10 mM MgCl2, 5 mM DTT) for 3 hours. 10 µl of 10 µM biotinylated oligonucleotide (5’-AGGTCGCCGCC-TEG-Biotin-3’, Integrated DNA Technologies (IDT)) complementary to the left end of λ DNA was also phosphorylated with T4 PNK (1 µl of 10 U/µl) by incubating at 37°C for 3 hours in 1x T4 PNK buffer. For annealing, phosphorylated λ DNA (50 µl) and the biotin-modified oligonucleotide (1 µl of 10 µM) was mixed into 0.45 ml of 1x T4 Ligase buffer (NEB, 50 mM Tris pH 7.5, 10 mM MgCl2, 10 mM DTT). The mixture was then heated to 65°C for 5 minutes, and slowly cooled down to room temperature on a heat block. To ligate the oligonucleotide to λ DNA, ATP (final concentration of 10 mM) and T4 DNA ligase (1 µl of 440 U/µl, NEB, M0202) was added, and the mixture was further incubated at room temperature for 4 hours. To tether λ DNA at only one end to the surface of the flow cell, the right end was subsequently annealed and ligated to a complementary unmodified oligonucleotide (5’-GGGCGGCGACCT- 3’, IDT) using the same method described above. 10-fold excess unmodified oligonucleotide was added to saturate any complementary free biotinylated oligonucleotide remaining from the first ligation.
Figure 2.
Single-molecule manipulation and visualization of λ DNA. (A) The sequence of 12 bp single stranded ends of λ DNA. (B) A cartoon demonstration of the experimental setup used for stretching and imaging DNA. (C) SYTOX images of a singly-tethered λ DNA molecule in the absence (i) and presence (ii) of buffer flow. (iii) SYTOX image of a doubly-tethered λ DNA molecule stretched to 78% of the contour-length in the absence of flow (iv) A λ DNA molecule was stretched to 88% of the contour length by including 100 µM chloroquine in buffer (bar 2 µm). Adapted with permission from [30].
To couple the modified DNA to the surface of the flow cell, biotin-modified λ DNA was diluted 20-fold in blocking buffer, 0.1 ml of diluted DNA was drawn into the flow cell (50 µl/min), and incubated for 5–10 minutes (Figure 2B). Further incubation leads to higher surface coverage with DNA. To remove unbound DNA, the flow cell was washed with 0.5 ml blocking buffer. At this stage, λ DNA can be labeled and visualized using SYTOX Orange (Invitrogen, S11368), a fluorescent dye that intercalates into double-stranded DNA (dsDNA). To label λ DNA, SYTOX was diluted to a concentration of 15 nM in blocking buffer, drawn in to the flow cell at the desired rate, and tethered λ DNA molecules were imaged as described in Section 2.6 (Figure 2 B and C). To remove SYTOX, the flow cell was washed with blocking buffer for 5 minutes at a rate of 100 µl/min. We note that we did not stain DNA with SYTOX prior to replication reaction unless necessary. Other fluorescent DNA intercalating agents such as YOYO-1 can be used to label dsDNA. We prefer to use SYTOX because unlike YOYO-1, it can easily be removed from DNA by washing the flow cell with blocking buffer. In addition, YOYO-1 induces more photo-cleavage of dsDNA than SYTOX Orange upon excitation.
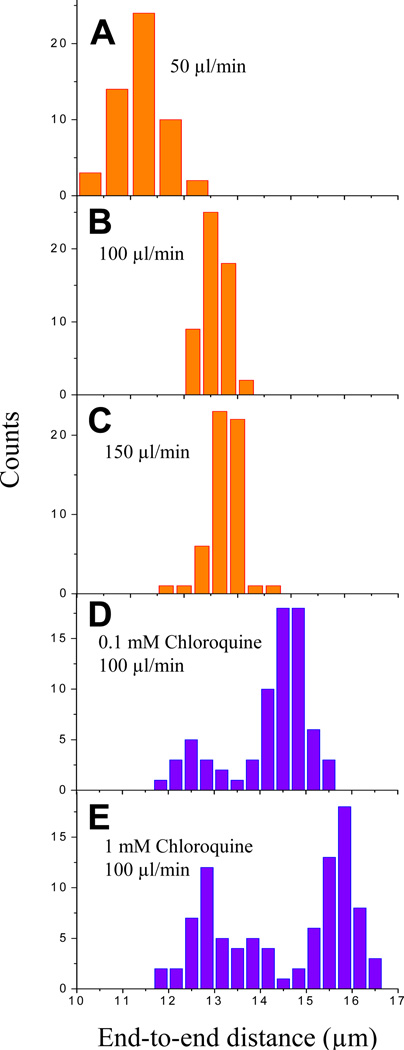
To attach both ends of the DNA to the surface of the flow cell, the left end of λ DNA was biotin modified as described above, while the 12 bp overhang on the right end was initially left unmodified. This substrate was then tethered to the surface as described above. To stretch λ DNA and tether its right end to the surface, biotin modified oligonucleotide complementary to the right end of λ DNA (5’-GGGCGGCGACCT-TEG-Biotin-3’, IDT) was diluted to a concentration of 100 nM in blocking buffer and drawn into the flow cell at a rate of 100 µl/min for 10 minutes. At this flow rate, λ DNA molecules that were attached at the left end to the surface were extended to 70–80% of the contour length of B-form DNA (16.5 µm) by buffer flow and annealed to the biotinylated oligonucleotide on the right end. The right end also attached to the surface via biotin-streptavidin making λ DNA molecules doubly-tethered (Figure 2C-iii and 3B). Increasing the flow rate above 100 µl/min had little further effect on stretching (Figure 3A–C). In order to stretch DNA further, we used chloroquine, which intercalates into and extends the contour length of double-stranded DNA [29]. A 40 mM chloroquine (Sigma, C6628) solution was prepared in distilled water and mixed with biotinylated oligonucleotide in blocking buffer at a final concentration of 100 µM chloroquine and 100 nM oligonucleotide immediately before use. When this mixture was drawn at a rate of 100 µl/min, λ DNA molecules were stretched to 85–95 % of the contour length of B-form DNA (Figure 2C-iv and 3D). Higher chloroquine concentrations can be used to achieve even further stretching of λ DNA (Figure 3E). Care needs to be exercised not to expose the flow cell to light in the presence of chloroquine which may lead to non-specific sticking of DNA to the surface. Finally, chloroquine was removed by washing the flow cell with 0.5 ml blocking buffer (100 µl/min). A significant fraction of λ DNA molecules were less stretched even in the presence of chloroquine probably because they became doubly-tethered after chloroquine was removed (Figure 3 D and E). Increasing the duration for which the chloroquine mixture was drawn (in the presence of biotinylated oligonucleotide) may reduce the number of such less stretched molecules.
Figure 3.
Histograms of extent of DNA stretching. λ DNA was flowed in at a constant rate into microfluidic flow cell at various rates in the absence (A–C) and presence (D–E) of chloroquine. End-to-end distance of individual molecules was determined from SYTOX images. Due to the presence of singly-tethered molecules that can be stretched with buffer flow, images were taken in the absence of flow to confirm that all stretched molecules were doubly-tethered. After washing off chloroquine, a fraction of DNA molecules became doubly-tethered giving rise to bimodal distribution in D and E. Adapted with permission from [30].
2.3. Replication of immobilized DNA in Xenopus egg extracts
In the following subsections, we give step-by-step descriptions of the protocols used for replication of surface-tethered λ DNA in Xenopus egg extracts from multiple or single initiations on each DNA molecule. Using the protocols described here, we previously addressed whether the two MCM2-7 complexes that assemble at every origin of replication must remain coupled during replication [30] as proposed in some models [31, 32]. According to these models, DNA is pumped towards the interface between the two sister replisomes and newly replicated DNA is extruded laterally. If this model is correct, a DNA constrained at both ends should not serve as an efficient replication template due to the tension that accumulates on the unreplicated portions of the molecule. We showed that doubly-tethered DNA is as efficiently replicated in egg extracts as singly-tethered DNA from multiple initiations (Figure 4), which contrasts to obligatory physical coupling of sister replisomes [30]. In addition, we demonstrated that a single pair of diverging forks can replicate doubly-tethered molecules to an extent greater than the slack originally present in the molecule (Figure 5A), suggesting that sister replisomes can uncouple after initiation and that uncoupling does not affect fork rates (Figure 5B) [30].
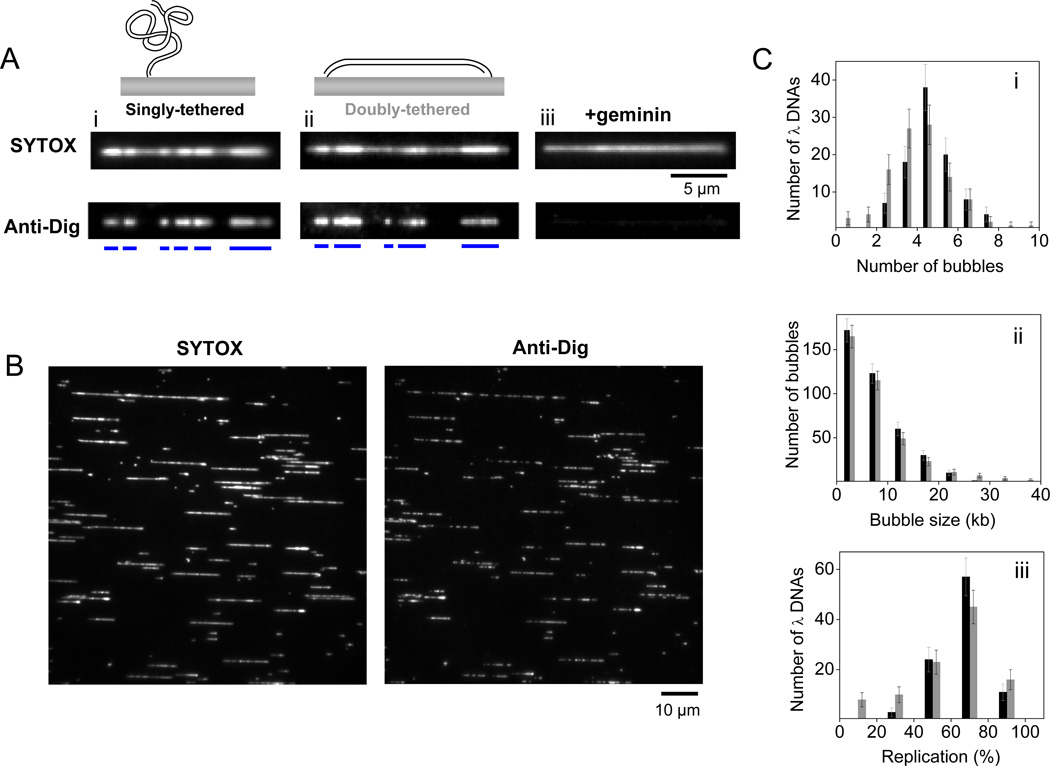
Figure 4.
Replication of surface-immobilized λ DNA in Xenopus egg extracts from multiple initiations. (A) SYTOX (top) and anti-dig (bottom) images of partially replicated singly-tethered (i) and doubly-tethered (ii) λ DNA molecules after 15 minute incubation in NPE. Anti-dig did not always stain replication bubbles evenly. Therefore, an anti-dig tract that was not interrupted by a region with intensity below a certain threshold was assigned to a single replication bubble. Blue bars represent location of individual replication bubbles. (iii) The presence of geminin in HSS inhibits replication of surface-immobilized DNA. (B) SYTOX (left) and corresponding anti-dig (right) images of a representative field of view containing many replicated singly-tethered λ DNA molecules after 15 minute incubation in NPE. Images were recorded in the presence of buffer flow (125 µl/min). Many broken DNA molecules as well as concatemers of λ DNA were present on the surface. We only took full length monomeric λ DNA molecules into account in our analysis. (C) (i) Histogram of the number of replication bubbles per immobilized λ DNA detected through anti-dig tracts. (ii) Length distributions of replication bubbles. (iii) Histogram of replicated fraction of individual λ DNA molecules. Adapted with permission from [30].
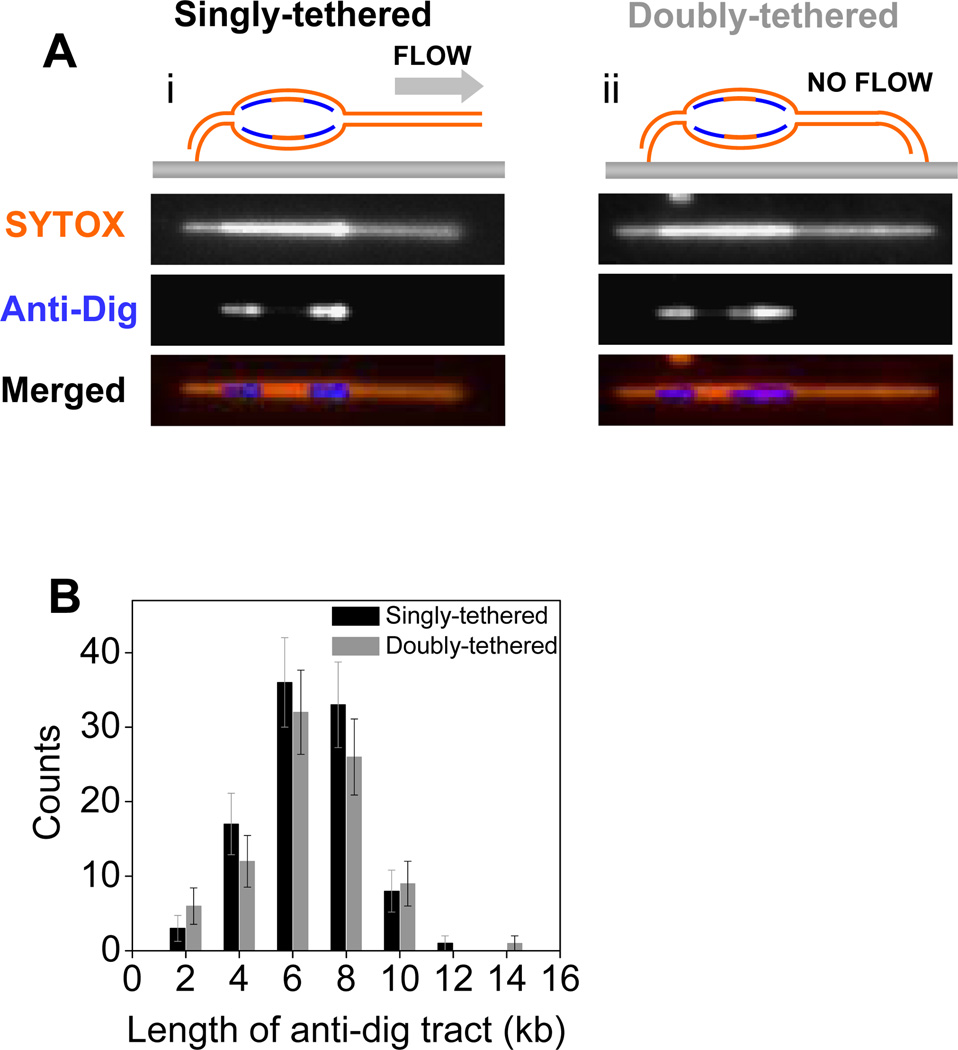
Figure 5.
(A) SYTOX (top), anti-dig (middle), and merged (bottom) images of singly-tethered (i) and doubly-tethered (ii) λ phage DNA molecules, which contain a single replication bubble. Experiments were performed under the conditions described in Section 2.3.3. (B) Distribution of the length of anti-dig tracts after 25 minutes dig-dUTP pulse in NPE on singly-tethered (black) and doubly-tethered (grey) λ DNA molecules. Adapted with permission from [30].
2.3.1. Assembly of pre-replication complexes on surface-immobilized λ DNA
Extracts were prepared as described [33] and were stored at −80°C. An ATP regeneration system (ATP mix) was prepared just before use by mixing 5 µl ATP (1 M, pH 7), 10 µl phosphocreatine (1 M), and 0.5 µl creatine phosphokinase (5 mg/ml). 33 µl of HSS was thawed, supplemented with 1 µl ATP mix and 0.5 µl nocadozole (0.5 mg/ml) to avoid microtubule polymerization, and mixed thoroughly with a pipette. Extract was then spun at room temperature at 14,000 rpm for 5 minutes to spin down any membranes and to remove air bubbles. Supernatant was transferred to a new microcentrifuge tube and was further incubated for 5 minutes at room temperature. 0.75 µl of “carrier” plasmid (pBluescript KII-, 200 ng/µl) was added into HSS to achieve a critical DNA concentration, which is required for formation of pre-RCs, including on the immobilized substrates [34]. Before extracts were introduced into the flow cell, inlet tubing was shortened by cutting it with a clean razor blade to a length of 5 cm to reduce the dead volume within the tubing. HSS supplemented with carrier DNA was then drawn into the flow cell at a rate of 10 µl/min for 2 minutes and incubated for a further 10 minutes. The remaining HSS (supplemented with DNA) was stored at room temperature and used as described below. Flow of extract in the flow cell was confirmed by visualizing the movement of residual aggregates within the channel using bright field illumination.
To confirm that subsequent replication of λ DNA was MCM2-7 dependent, we added a final concentration of 4 µM geminin into HSS (supplemented with ATP mix and nocadozole) before introducing the HSS into the flow cell as described above.
2.3.2. Replication from multiple origins
During the HSS incubation, a 15 µl NPE aliquot was thawed, supplemented with 1 µl ATP mix, and centrifuged for 5 minutes. After a further 5 minutes incubation at room temperature, 10 µl of NPE was mixed with 10 µl HSS, 9 µl ELB (10 mM Hepes-KCl pH 7.7, 2.5 mM MgCl2, 50mM KCl) and 1 µl of digoxigenin labeled dUTP (dig-dUTP, 200 mM, Roche Applied Science, 11093088910). This replication mixture was immediately drawn into the flow cell (2 min at 10 µl/min), which led to replication initiation, and allowed to incubate for 15 minutes. The dig-dUTP was included to label replicated regions (Figure 4). DNA was stained with SYTOX, dig-dUTP was labeled with fluorescent antibody as described in Section 2.4, and replicated DNA was visualized (see Section 2.6 for details). SYTOX intensity was doubled in the replicated regions due to the presence of two daughter dsDNA molecules (Figure 4A and B). Dig-dUTP incorporated tracts exactly matched the high intensity SYTOX regions (Figure 4A and B). We note that replication bubbles labeled with dig-dUTP smaller than a diffraction-limited region (<1 kb) will not contain an obvious double SYTOX intensity due to the presence of unreplicated and replicated DNA within the same diffraction-limited region. The presence of geminin in HSS completely inhibited the double intensity SYTOX tracts as well as dig-dUTP incorporation, indicating that DNA synthesis occurred in an MCM2-7 dependent manner in our assay (Figure 4A-iii). The average number of replication bubbles per λ DNA molecule (Figure 4C-i) and the size of replication bubbles (Figure 4C-ii) were consistent with replication kinetics observed in Xenopus egg extracts using DNA fiber assays ([11, 13], see [30] for more details).
2.3.3. Replication from single origins
To limit the number of initiations to a single event per DNA template, we used p27Kip, which inhibits initiation by blocking Cdk2 actitivity [35]. Replication was initiated as described above with the exception that a smaller (15 µl) volume of replication mixture was drawn into the flow cell for 1 minute at a flow rate of 10 µl/min. A second replication mixture was prepared by mixing 5 µl NPE, 5 µl HSS, 4 µl ELB and 1 µl p27Kip (2 mg/ml). This second mixture was drawn into the flow cell 2 minutes after the first replication mixture to prevent further initiations while allowing elongation and was incubated for 10 minutes. Under these conditions, the majority of molecules contained no replication bubble (~85–90 %), ~10% had a single replication bubble, and a small fraction (<5%) contained multiple replicons. To confirm bidirectional replication, a third extract that contains p27Kip and dig-dUTP (5 µl NPE, 5 µl HSS, 3 µl ELB, 1 µl p27Kip (2 mg/ml), 1 µl dig-dUTP (100 mM)) was introduced (1 minute at 10 µl/min) and incubated for 25 minutes. Two dig-dUTP tracts were visible at the ends of each replication bubble consistent with bidirectional replication with the origin residing in the middle (Figure 5).
As explained in Section 2.3.1, licensing of surface-immobilized λ DNA requires the addition of a carrier plasmid, which will also undergo replication in the flow cell. To avoid this, a duplex linear DNA as short as 29 bp at a final concentration of 10 ng/µl was used instead of plasmid to increase the effective DNA concentration in HSS. Since pre-RC’s cannot assemble on a 29 bp DNA [36], this short duplex is not licensed in HSS. Replacing plasmid DNA with the short duplex during the licensing reaction had no visible effect on replication. However, we observed slower fork rates when plasmid was omitted from the replication mixture containing NPE. Therefore, after licensing reaction with HSS (supplemented with the short duplex), we used a replication mixture that contained plasmid DNA. To prevent licensing of this plasmid and its subsequent replication, we pre-incubated HSS with NPE before addition of plasmid. The high concentration of geminin in NPE was sufficient to prevent replication of this plasmid [37]. In summary, we used the following protocol to replicate surface immobilized λ DNA from a limited number of initiations in the absence of replicating plasmid:
Mix 33 µl of HSS with 1 µl ATP mix and 0.5 µl nocadozole. Centrifuge 14,000 rpm for 5 minutes.
Mix 0.5 µl 29 bp duplex DNA (300 ng/µl) with 15 µl HSS. Draw into the flow cell at 10 µl/min for 1 minute. Incubate for 10 minutes.
Mix 16 µl NPE with 1 µl ATP mix and spin down for 5 minutes.
Mix 17 µl HSS (no DNA) with 17 µl NPE. Incubate 5 minutes. Add 4 µl ELB and 1 µl plasmid (75 ng/µl diluted in ELB) into 11 µl HSS/NPE mix. Draw into the flow cell at a rate of 10 µl/min for 1 minute. Incubate for 2–3 minutes.
Add 3.5 µl ELB, 1 µl plasmid (75 ng/µl), and 0.5 µl p27Kip (2 mg/ml) into 11 µl HSS/NPE mix. Draw into the flow cell at a rate of 10 µl/min for 1 minute. Incubate for 12 minutes.
Add 2.5 µl ELB, 1 µl plasmid (75 ng/µl), 0.5 µl p27Kip (2 mg/ml), and 1 µl dig-dUTP (0.1 mM diluted in ELB) into 11 µl HSS/NPE mix. Draw into the flow cell at a rate of 10 µl/min for 1 minute. Incubate for 25 minutes.
We note that HSS can be omitted from the replication mixture and that this does not inhibit replication. However, we found that the absence of HSS can affect fork elongation rates (and also initiation timing). A 1:1:1 ratio of HSS:NPE:ELB usually leads to the most initiations occurring 2 minutes after introduction. This ratio also yielded optimal fork rates (0.27 kb/min on average). However, the ratio may slightly change for different extract preparations. For example, another extract sample yielded similar fork rates at a ratio of 1.5:1:0.5 (HSS:NPE:ELB). In general, reducing the fraction of HSS in the mixture leads to lower fork rates and faster initiations. Although we do not know why HSS stimulates elongation kinetics while slowing initiation, it is likely due to the fact that the two extracts contain different concentrations of replication factors that may also differ for each extract preparation.
Finally, we should emphasize that the temperature can affect replication kinetics. We performed our experiments at room temperature (22°C). Lower temperatures can lead to delayed origin firing and possibly slower fork rates [38].
2.4. Labeling replicated DNA
After incubating with replication mixture, the flow cell was flushed with SDS-containing buffer (20 mM Tris pH 7.5, 50 mM NaCl, 12 mM EDTA, 0.1% SDS, 10 minutes at 25 µl/min) to remove any protein bound to DNA. As the presence of SDS may interfere with subsequent labeling of DNA, we removed SDS by washing the flow cell with blocking buffer for 10 minutes at a rate of 25 µl/min. We further passivated the surface by introducing casein (50 µl of 0.4 mg/ml in ELB, Sigma, C-3400) and incubating for 10 minutes to reduce subsequent non-specific sticking of DNA to the surface. To label nascent DNA containing dig-dUTP, 0.25 µl fluorescein labeled anti-digoxigenin Fab fragments from sheep (anti-dig, 200 ng/µl, Roche Applied Sciences, 11207741910) was mixed into 200 µl ELB supplemented with 50 µl of casein (2 mg/ml) and drawn into the flow cell at a rate of 25 µl/min for 8 minutes. The flow cell was then immediately flushed with blocking buffer (5 minutes at 25 µl/min). To label λ DNA, SYTOX (15 nM in blocking buffer) was drawn into the flow cell. We used low concentrations of SYTOX to prevent its detection in the fluorescein channel which is used for visualizing anti-dig. Singly-tethered DNA was imaged in the presence of buffer flow (SYTOX containing buffer at a rate of 100 µl/min) to stretch DNA, while doubly-tethered DNA can be imaged in the absence of buffer flow.
2.5. Immunostaining of replication proteins on stretched DNA
2.5.1 Motivation
Proteins associated with the doubly-tethered λ DNA during replication can be detected after extract removal. For example, proteins that bind at the replication fork are expected to colocalize with replication bubble ends. Others, like PCNA (see Figure 6), may be distributed behind the replication fork (Anna B. Loveland et al., unpublished results). Using the immunostaining technique described here, their spatial distribution along the DNA can be determined.
Figure 6.
Immunostaining proteins bound to replicated DNA. (A) Scheme for immunostaining fork-bound PCNA. PCNA is first detected with a Rabbit anti-PCNA primary antibody. Second, the primary antibody is detected with an anti-Rabbit IgG conjugated to the fluorophore Alexa Fluor® 647. (B) Costaining of DNA, replicated DNA and PCNA. Following licensing in HSS, a 1:1:1 mix of HSS:NPE:ELB supplemented with ATP mix and 7 µM dig-dUTP was drawn into the flow cell for 1 minute at 10 µl per minute. Immediately afterwards, the original mix was replaced with an identical mix also supplemented with 66 µg/ml p27Kip to limit further initiation. Replication elongation was allowed to proceed to 25 minutes after initial NPE addition. Then, the extracts were washed out; incorporated dig-dUTP followed by PCNA was immunostained; and lastly SYTOX Orange was drawn into the flow cell as described in the text. Fluorescence of DNA, replication bubbles and PCNA were imaged with the appropriate excitation. The PCNA signals were observed to flank replication bubbles along DNA.
2.5.2 Basic Protocol
To maintain protein-DNA interactions for immunostaining, NPE was removed from the flow cell differently than for DNA staining alone. For immunostaining of PCNA on the DNA, we stopped the replication reaction with a flow of wash buffer 2 (10 mM Hepes pH 7.7, 300 mM KCl, 2.5 mM MgCl2 supplemented with 0.1 mg/ml BSA (New England Biolabs)) for 5 minutes at a rate of 20 µl/min. Note that while this buffer is appropriate for PCNA, whose binding to chromatin is very salt-resistant [39], it may not be appropriate for proteins whose DNA binding is sensitive to high salt. Next, a 25-minute wash (10 µl/min) with ELB++ (10 mM Hepes-KCl pH 7.7, 100 mM KCl, 2.5 mM MgCl2, 1 mg/ml BSA Fraction V (OmniPur Cat. # 2930, EMD)), a buffer containing a high concentration of BSA, was used to wash and block the surface of the flow cell.
Once the extract was removed and the surface blocked, both replicated DNA and DNA-bound protein were immunostained as follows. First, replication bubbles were stained for incorporated dig-dUTP by flowing in 0.2 ng/µl anti-dig in ELB++ at a flow rate of 10 µl/min for 5 minutes. Afterwards, the Fab fragments were washed out with ELB++ at a flow rate of 10 µl/min for 5 minutes. At this point, the presence of replication bubbles was verified by imaging the fluorescein channel. Next, a primary antibody recognizing the native-folded state of the protein of interest was introduced into the flow cell. Anti-Xenopus laevis PCNA IgG was isolated from immune serum [40] on a Protein A Sepharose Fast Flow resin (GE Healthcare) and acid-eluted following manufacturer’s protocol, dialyzed against 1 × PBS, and flash frozen in liquid nitrogen. For immunostaining, a fresh aliquot of the frozen stock was diluted to a concentration of 1.2 ng/ml in ELB++ and introduced into the flow cell at a rate of 10 µl/min for 15 minutes. After a 5-minute wash with ELB++, the anti-PCNA primary antibody was fluorescently detected with 2 ng/µl of Alexa Fluor® 647 goat anti-rabbit IgG (H+L) (Invitrogen, A21244) in ELB++ for 5 minutes at a flow rate of 10 µl/min. The secondary antibody was removed with flow of ELB++ for 5 minutes at a flow rate of 10 µl/min. Finally, dsDNA was co-stained by flowing in 25 nM SYTOX in ELB++ at a flow rate of 10 µl/min and was continually refreshed at this flow rate for the subsequent imaging period.
Lastly, the fluorescence of replication bubbles, DNA, and the replication protein was imaged and the three fluorescent channels were co-aligned. For each field of view, images were collected in three different fluorescent channels (see Section 2.6 for details). Multiple fields are imaged until more than 50 λ DNA molecules were recorded under a given condition. Due to an imperfect overlap of the three emission paths and sample drift, the three imaged channels were not perfectly aligned [41]. To register or co-align them, two or more diffraction-limited objects visible in all of the three channels were localized relative to each other. The difference vectors were used to translationally and/or rotationally shift the channels relative to one another to yield a co-aligned three channel image utilizing software packages like the MatLAB Image Processing Toolbox (The Mathworks Inc., Natick, MA) or ImageJ [42]. One excellent fiducial marker for registration was replicated, singly-tethered λ DNA which was costained with both antibodies and SYTOX Orange and which appeared as an almost diffraction-limited object. Alternatively, we also routinely flowed in 1:1000 TetraSpeck Beads (100 nm, Invitrogen, T7279) in ELB++ for 20 seconds at a rate of 10 µl/min and immediately washed them out to limit binding to 5–10 per field of view. Finally, the co-aligned images were displayed as an RGB image revealing the relative position and distribution of the protein, replication bubbles, and DNA.
2.5.3 Other considerations
When designing single-molecule immunostaining experiments, care must be taken to include appropriate controls to recognize non-specific antibody binding and to limit replication to λ DNA. Negative controls should be used to ascertain the dependence of secondary antibody binding on the presence of the replication protein studied. The most straightforward control is to limit the extent of DNA replication and verify that the protein colocalizes only with anti-dig stained regions of DNA. Alternatively, the experiment can be repeated in the presence of a replication inhibitor, the staining procedure repeated and relative signals compared. Finally, we have occasionally observed binding of carrier plasmid DNA to λ DNA. The wash conditions described for the protein immunostaining experiments are not stringent enough to remove all bound plasmid DNA from λ DNA. If the carrier plasmid DNA was allowed to replicate, proteins bound to replication forks on plasmid and λ DNA would be immunostained. Thus, to restrict immunostaining to the individual replication forks on λ DNA, we only license λ DNA and not the carrier plasmid as described in Section 2.3.3.
TIPS
Never freeze the anti-digoxigenin-fluorescein Fab fragments (Roche) as this increases nonspecific binding. Instead, we have successfully stored the reagent at 4°C in the presence of 0.02% Sodium Azide for over a year.
Always use SYTOX Orange stain last as it appears to crosslink protein to DNA and may interfere with wash out efficiency if used early on.
Take care not to draw any air bubbles into the flow cell as this shears λ DNA off of the coverslip surface.
TetraSpeck beads do not stick to the PEG functionalized glass but after exposure to extracts and subsequent wash-out, they exhibit increased non-specific binding to the surface.
If DNA is not washed with SDS, as for replication protein immunostaining, the SYTOX intensity along the DNA will be less uniform and intensity doubling of replication bubbles will be less reproducible.
2.6. Microscopy
Fluorescence imaging was performed on an Olympus IX-71 inverted microscope equipped with a 60× oil objective (PlanApo, NA=1.65, Olympus) and a 1.6× magnification unit. The microscope and optics were built on a vibration-isolated optical table (Newport, RS1000). 488-nm, 568-nm, and 647-nm laser beams from a multi-wavelength Ar-Kr ion laser (Innova 70C-Spectrum, Coherent Inc.) were separately aligned to achieve total internal reflection mode through the rear port of the microscope. Intensity of each excitation wavelength was adjusted via continuously variable optical neutral density filters (Thorlabs, NDC-50C-2M). Anti-digoxigenin-fluorescein Fab fragments, marking replicated DNA, were imaged with 488-nm excitation (2.3 W/cm2, 100 ms) through a standard FITC filter set (Excitation: D480/30, Emission: D535/40, Dichroic: 505dclp; Chroma). SYTOX Orange, staining double-stranded DNA, was imaged with 568-nm excitation (3.5 W/cm2, 100 ms) through a custom filter set (Excitation: z568/10, Emission: ET620/60 Dichroic: T585lp; Chroma). Finally, the α-PCNA signal was imaged with 647-nm excitation (0.16 W/cm2, 100 ms) through a standard Cy5 filter set (Excitation: D640/20, Emission: D680/30, Dichroic: 660dclp; Chroma). Exposure duration was controlled by computer-operated shutters (Vincent Associates, VS14S2T0) placed in the optical path of each laser line. Fluorescent images were captured with a back-illuminated electron-multiplying CCD camera (Andor Technology, Andor iXon) at varying rates controlled by Andor iQ 1.8.1 software.
3. Concluding Remarks
Here, we described detailed protocols for replicating λ DNA attached at either one or both ends to the surface of a microfluidic flow cell in Xenopus egg extracts and subsequent detection of replicated DNA and immunodetection of replication factors. We note that singly-tethered DNA molecules are readily chromatinized in egg extracts (data not shown). In contrast, since doubly-tethered molecules are constrained at both ends, chromatin assembly is minimal. Thus, studying replication kinetics of singly and doubly-tethered DNA using this new single-molecule assay will allow investigation of the role of chromatinization in clustering [13] and sequence-preference [43] of replication initiation. The ability to immunodeplete proteins from egg extracts [33] will also make it possible to examine effects of different factors on the timing of replication initiation, origin-to-origin distance, and fork elongation rates. For example, one could test whether specific chromatin remodeling factors are required for fork progression through singly-tethered DNA. In addition, immunostaining of proteins on stretched DNA can be performed to understand how other replication factors such as MCM2-7 or RPA are distributed on replicating DNA. We believe that the ability to use the precision and sensitivity of single-molecule manipulation and imaging techniques in cell-free extracts will allow the characterization of complex enzymatic reactions in environments that closely mimic those found inside living cells, without the need for biochemical reconstitution. The fact that single, stretched DNA molecules are excellent templates for eukaryotic DNA replication suggests that in the future, it will be possible to image the movement of single replisomes in real time. Indeed, we recently developed an imaging approach that allows real-time, single-molecule imaging of specific proteins at individual replication forks (Loveland et al., submitted). Live monitoring of replication will provide in-depth information about the dynamics of replication initiation and elongation. In particular, the static imaging method described here may conceal events such as merging of replication bubbles as well as pausing or stalling of replication forks.
Acknowledgements
The work described was funded by grants from the NIH (GM077248), ACS (RSG0823401GMC), and the Netherlands Organization for Scientific Research (NWO; Vici 680-47-607) to A.M.v.O, and grants from the NIH (GM62267 and GM80676) and ACS (RSG0823401GMC) to J.C.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 2.van Oijen AM. Curr. Opin. Biotechnol. 2011;22:75–80. doi: 10.1016/j.copbio.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Toprak E, Kural C, Selvin PR. In: Methods Enzymol. Walter NG, editor. Academic Press; 2010. pp. 1–26. [DOI] [PubMed] [Google Scholar]
- 4.Neuman KC, Nagy A. Nature Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Oijen AM. Biopolymers. 2007;85:144–153. doi: 10.1002/bip.20624. [DOI] [PubMed] [Google Scholar]
- 6.Funderud S, Andreassen R, Haugli F. Nucleic Acids Res. 1979;6:1417–1431. doi: 10.1093/nar/6.4.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Truett MA, Gall JG. Chromosoma. 1977;64:295–303. doi: 10.1007/BF00294937. [DOI] [PubMed] [Google Scholar]
- 8.Newlon CS, Petes TD, Hereford LM, Fangman WL. Nature. 1974;247:32–35. doi: 10.1038/247032a0. [DOI] [PubMed] [Google Scholar]
- 9.Huberman JA, Riggs AD. J. Mol. Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- 10.Lengronne A, Pasero P, Bensimon A, Schwob E. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrick J, Stanislawski P, Hyrien O, Bensimon A. J. Mol. Biol. 2000;300:1133–1142. doi: 10.1006/jmbi.2000.3930. [DOI] [PubMed] [Google Scholar]
- 12.Marheineke K, Hyrien O. J. Biol. Chem. 2001;276:17092–17100. doi: 10.1074/jbc.M100271200. [DOI] [PubMed] [Google Scholar]
- 13.Blow JJ, Gillespie PJ, Francis D, Jackson DA. J. Cell Biol. 2001;152:15–26. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson DA, Pombo A. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailis JM, Luche DD, Hunter T, Forsburg SL. Mol. Cell. Biol. 2008;28:1724–1738. doi: 10.1128/MCB.01717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. Mol. Biol. Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norio P, Schildkraut CL. Science. 2001;294:2361–2364. doi: 10.1126/science.1064603. [DOI] [PubMed] [Google Scholar]
- 18.Pasero P, Bensimon A, Schwob E. Genes Dev. 2002;16:2479–2484. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Nature. 2009;457:336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, van Oijen AM. Nature Structural and Molecular Biology. 2008;15:170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao NY, Georgescu RE, Finkelstein J, O'Donnell ME. Proc. Natl. Acad. Sci. U.S.A. 2009;106 doi: 10.1073/pnas.0906157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 23.Maier B, Bensimon D, Croquette V. Proc. Natl. Acad. Sci. U.S.A. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christian TD, Romano LJ, Rueda D. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21109–21114. doi: 10.1073/pnas.0908640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. Nature. 2009;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blow JJ, Laskey RA. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 27.Walter J, Sun L, Newport J. Mol. Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 28.Tanner NA, van Oijen AM. In: Single Molecule Tools, Part B: Super-Resolution, Particle Tracking, Multiparameter, and Force Based Methods. Walter NG, editor. Academic Press; 2010. pp. 259–278. [Google Scholar]
- 29.Cohen SN, Yielding KL. J. Biol. Chem. 1965;240:3123–3131. [PubMed] [Google Scholar]
- 30.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Mol. Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sclafani RA, Fletcher RJ, Chen XS. Genes Dev. 2004;18:2039–2045. doi: 10.1101/gad.1240604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi TS, Wigley DB, Walter JC. Trends Biochem. Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Lebofsky R, Takahashi TS, Walter JC. Methods Mol. Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- 34.Lebofsky R, van Oijen AM, Walter JC. Nucleic Acids Res. 2011;39:545–555. doi: 10.1093/nar/gkq739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter J, Newport J. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 36.Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. J. Biol. Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 37.Arias EE, Walter JC. Genes Dev. 2005;19:114–126. doi: 10.1101/gad.1255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pacek M, Walter JC. EMBO. J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bravo R, Macdonald-Bravo H. J. Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kochaniak AB, Habuchi S, Loparo JJ, Chang DJ, Cimprich KA, Walter JC, van Oijen AM. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy DB. Fundamentals of Light Microscopy and Electronic Imaging. New York: Wiley-Liss; 2001. [Google Scholar]
- 42.Abramoff M, Magelhaes P, Ram S. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 43.Stanojcic S, Lemaitre J-M, Brodolin K, Danis E, Mechali M. Mol. Cell. Biol. 2008;28:5265. doi: 10.1128/MCB.00181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]