Abstract
The ability of the GroEL chaperonin to unfold a protein trapped in a misfolded condition was detected and studied by hydrogen exchange. The GroEL-induced unfolding of its substrate protein is only partial, requires the complete chaperonin system, and is accomplished within the 13 seconds required for a single system turnover. The binding of nucleoside triphosphate provides the energy for a single unfolding event; multiple turnovers require adenosine triphosphate hydrolysis. The substrate protein is released on each turnover even if it has not yet refolded to the native state. These results suggest that GroEL helps partly folded but blocked proteins to fold by causing them first to partially unfold. The structure of GroEL seems well suited to generate the nonspecific mechanical stretching force required for forceful protein unfolding.
The GroEL chaperonin (1, 2) captures non-native proteins by means of a ring of hydrophobic residues that line the entrance to the central cavity of its heptameric ring (Fig. 1) (3). When GroEL binds adenosine triphosphate (ATP) and the GroES cochaperonin, a massive structure change doubles the GroEL cavity volume and occludes its hydrophobic binding surface (4, 5). Spectroscopic evidence (6, 7 ), proteinase protection experiments (6, 8), and electron microscopy (4, 9) leave no doubt that the substrate protein is transiently encapsulated in the central cavity under the GroES lid. However, despite much additional structural and biochemical study (1, 2), the manner in which the GroEL structure change promotes protein folding remains to be demonstrated.
Fig. 1.
(Top) The crystal structure of the asymmetric GroEL14-GroES7 complex solved by Xu et al. (5). The two opposed heptameric rings of GroEL are shown in white and yellow. The binding sites for GroES and the substrate protein are in the apical domains between each green and red helix pair (3–5, 33). In the less expanded ring (left), which captures the substrate protein, the binding sites are 25 Å from each other. On addition of ATP and GroES, the apical domain of each GroEL subunit twists upward and outward so that the binding sites move apart to a position 33 Å from one another, as shown in the open conformation at the right with the bound GroES removed for clarity. Neighboring binding sites move apart by 8 Å and non-neighboring sites by larger increments, up to 20 Å. A substrate protein tethered to these sites will be forcibly stretched and partially unfolded. [Figure supplied by Z. Xu and P. B. Sigler; see also figure 1 of (32).] (Bottom) A schematic representation of the mechanism of stretch-induced T-H exchange. In the resting state (left) a segment of the substrate protein is tethered between two of the seven peptide binding sites in the apical domain of GroEL. Within the substrate protein a secondary structural element, for example a β-sheet as shown here by the open arrows, protects the radiolabeled amide hydrogens (T) from exchange. During the encapsulation process the rigid body movement of the apical domains causes the peptide binding sites to move further apart (right), generating a stretch-induced unfolding of the substrate protein and rapid exchange of the amide hydrogens (H).
Two models, not mutually exclusive, are under consideration. The Anfinsen cage model (10) is based on the view that protein folding is limited by intermolecular reactions that produce aggregation. The model proposes that the GroEL cavity provides a sequestered microenvironment where folding to the native state can proceed while the substrate protein is protected from aggregation. However, numerous experiments have shown that the substrate protein is ejected from the cavity with each round of ATP hydrolysis whether it has reached the native state or not (11). The iterative annealing model (12) is based on the view that the rate-limiting step in slow protein folding is the intramolecular reorganization of misfolded and trapped protein segments, dependent on some degree of protein unfolding (13–15). This model proposes that ATP hydrolysis is coupled to a forceful unfolding of the misfolded substrate protein and its release, either into the protected central cavity or to the exterior, so that the misfolding is relieved and forward folding can resume. Incompletely folded proteins undergo further iterations, in the biological equivalent of optimization through annealing (16 ), until they achieve the native state. However, there is no evidence for a GroES- and ATP-dependent unfolding reaction on the 13-s time scale of the GroEL–adenosine triphosphatase cycle.
We explored GroEL function using unfolded ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO, from Rhodospirillum rubrum) labeled by hydrogen-tritium exchange. The role of the individual system components and parameters was studied through their effect on the exchange of the protected RuBisCO hydrogens. Prior studies of GroEL (17–21) used various hydrogen exchange approaches (22). Tritium exchange provides advantages including sensitivity, accuracy, rapidity, and the ability to focus on one protein within a complex, which allowed us to test the entire active chaperonin system and its individual components on the biologically relevant time scale of seconds.
In nonpermissive conditions RuBisCO folding is blocked. It fails to fold spontaneously (23) and can reach the native state only with the help of the complete GroEL-GroES-ATP system (24). When unfolded RuBisCO is trapped in this way, most of its amide hydrogens exchange rapidly with unlabeled water protons, as expected, but a core of 12 highly protected hydrogens exhibit exchange half-lives of 30 min and longer (detected by tritium label) (Fig. 2). The number of slowly exchanging hydrogens found and their degree of protection ensures that they represent amide groups and not side chains (22). The slowly exchanging hydrogens provide multiple probe sites that are sensitive to structural stability and change and may or may not represent the same sites in different RuBisCO molecules.
Fig. 2.

Hydrogen-tritium exchange of unfolded RuBisCO. Experimental results (36) monitor the exchange behavior of the well-protected amide hydrogens of unfolded RuBisCO (Rb) when Rb is diluted from denaturing urea into native conditions (pH 8, 22° ± 2°C), where Rb cannot fold without the entire GroEL system. (A) to (C) show the effects on the well-protected hydrogens when the blocked Rb is bound to GroEL (EL) with or without GroES (ES), adenosine diphosphate (ADP), and nucleoside triphosphate [ATP or β,α-imidoadenosine 5′-triphosphate (AMP-PNP)].
The conditions used (pH 8, 22° ± 2°C), chosen to promote the rapid exchange of amide hydrogens that might be transiently unmasked by chaperone action [exchange half-life ~10 ms (22)], require that the trapped hydrogens must be highly protected in the non-native protein so that their exchange is slow enough to be measurable. Some other proteins tested provided similar numbers of slow hydrogens but the hydrogens were less protected (maltose-binding protein, malate dehydrogenase, rhodanese) [see also (19)]. It seems likely that the protected RuBisCO hydrogens are sequestered in a partially folded domain. Nevertheless, unfolded RuBisCO retains sufficient non-native structure, perhaps in other domains (25), so that it is efficiently captured by GroEL. The possibility that the slow hydrogens are p rotected by RuBisCO association or complex formation with GroEL was ruled out by cross-linking experiments that failed to detect RuBisCO association under these conditions and by experiments that compared immediate and delayed GroEL addition.
The time course for exchange of the protected hydrogens is the same for RuBisCO free in solution and when bound to GroEL (Fig. 2A). A similar result was found for unfolded, disulfide-reduced α-lactalbumin (18). To focus on the slowly exchanging hydrogens, we incubated labeled RuBisCO in a small excess of GroEL for 10 min to allow replacement of T with H at the rapidly exchanging sites. The binary complex was then mixed with GroES and various nucleotides (Fig. 2B). The addition of a twofold molar excess of GroES alone had no effect on the exchange rate of the highly protected hydrogens, and neither did Mg2+-ADP, Mg2+-ATP, or Mg2+-AMP-PNP in the absence of GroES. Similarly, experiments on β-lactamase (20) and dihydrofolate reductase (21) found no effect when ATP was added to the GroEL complex without GroES.
In contrast, the addition of GroES and Mg2+-ATP together resulted in the rapid exchange of all but 2.5 of the protected hydrogens, signaling some unfolding event (Fig. 2C). Nonhydrolyzable AMP-PNP was as effective as ATP (Fig. 2C), indicating that the energy for substrate protein unfolding is derived from the binding of the nucleoside triphosphate rather than its hydrolysis and also that the unfolding observed does not require repeated system turnovers. The fact that 2.5 slow hydrogens remained suggests that GroEL does not fully unfold the substrate molecule. It is expected that even a partial unfolding of the protecting structure will tend to labilize protected hydrogens to exchange (22).
When a stoichiometric mixture of GroEL, GroES, and labeled RuBisCO was passed through a gel filtration column, all the label emerged at the position of the complex. When ATP was added, the GroEL-bound RuBisCO lost all but about two of its protected hydrogens whereas added ADP had no effect, as in the prior experiments. Thus, the behavior observed here involves the interaction of GroEL with monomeric RuBisCO, consistent with the fact that dimeric RuBisCO will not fit into the GroEL-GroES cavity.
When protein unfolding is induced by the addition of ATP, exchange of the protected hydrogens occurs within the ~45 s necessary for separation of the protein from the freed tritium label. To obtain greater time resolution, we added ATP to an otherwise complete reaction mixture and EDTA was added 5 to 12 s later to quench the reaction. This allows a GroEL cycle in progress to continue but precludes further cycling (11). Figure 3 shows that the system is committed to the unfolding event that causes the rapid tritium loss within 5 s after ATP addition, well within the 13-s turnover time (26 ) for a single round of ATP hydrolysis. The same conclusion is implied by the effectiveness of AMP-PNP (Fig. 2C).
Fig. 3.
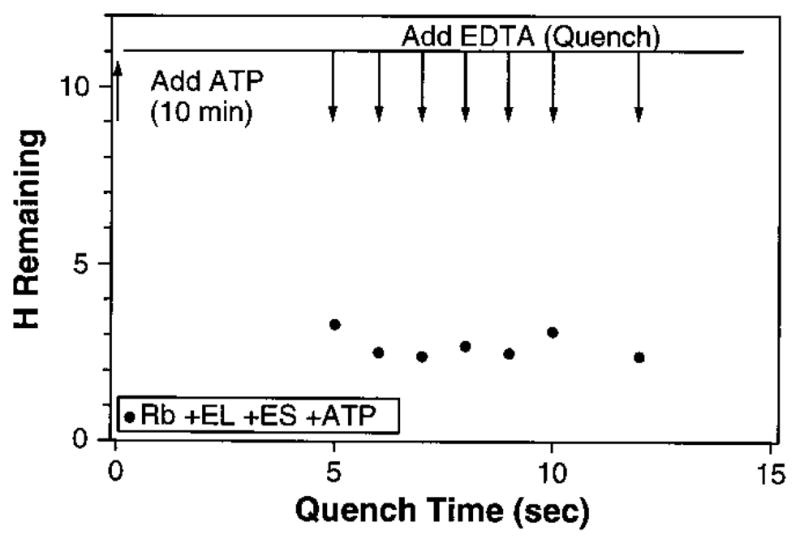
Single-turnover experiment. ATP was added after 10 min of RuBisCO hydrogen exchange. The upper line shows the number of retained, unexchanged tritiums at that point (from Fig. 2). EDTA was then added at the times shown, before one turnover was completed, leaving the system committed to complete one round of ATP hydrolysis but prohibiting further rounds.
To study the release of RuBisCO from the complex, we did experiments with substoichiometric GroEL, with a GroEL:RuBisCO ratio of 1: 20 (Fig. 4A). When ATP is added, tritium loss occurs over a 10-min time period because multiple turnovers of each GroEL complex are required in order to process the excess RuBisCO (13 s per turnover). The upper predicted curve in Fig. 4A assumes that each RuBisCO molecule remains bound to GroEL until it reaches the native state (average of 24 turnovers), losing its tritium label in the process. The lower predicted curve, which matches the data, assumes that each GroEL turnover induces the exchange of the sensitive hydrogens and then releases the not-yet-native protein to compete equally with all of the remaining unfolded RuBisCO molecules for rebinding (whether labeled or unlabeled). Analogous experiments were done with substoichiometric GroES (Fig. 4B; RuBisCO:GroEL:GroES ratio of 1:1.2:0.04). The predicted curve, which matches the data, assumes that GroES cycles through the RuBisCO-GroEL complexes, releasing the sensitive tritium label on each visit (13 s).
Fig. 4.
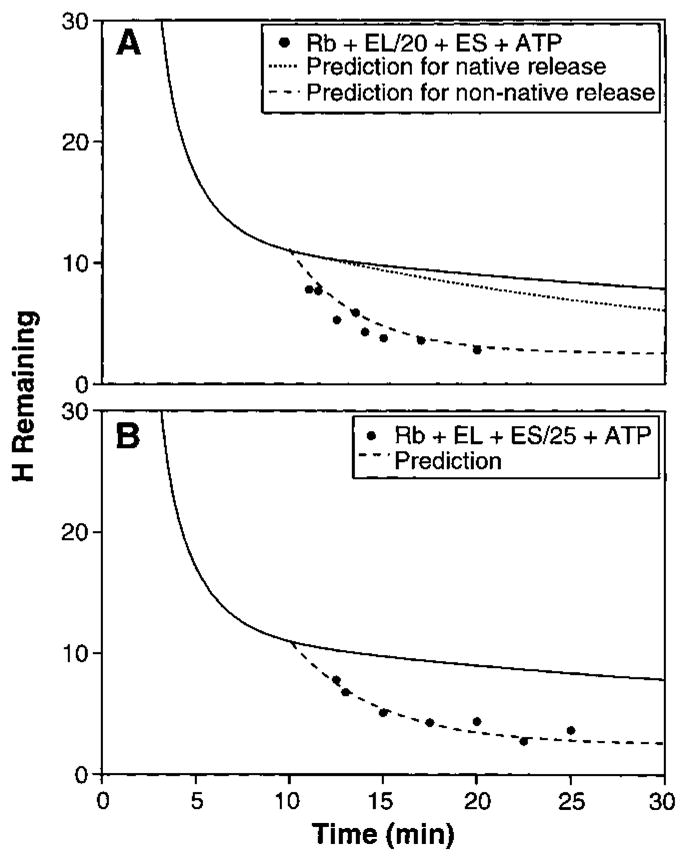
Kinetics of substrate protein release. (A) Time-dependent hydrogen exchange in the presence of limiting GroEL (RuBisCO:GroEL: GroES ratio of 1:0.05:1.2). The solid line is from Fig. 2. The upper predicted curve (dotted line) assumes that RuBisCO is released from the complex only when it reaches the native form after an average of 24 turnovers (10), that it loses its carried tritium (except for 2.5 still protected sites) on the first turnover cycle (13 s), and then does not compete for rebinding. The lower predicted curve (dashed line) assumes that each RuBisCO molecule experiences complete tritium loss on one turnover (except for 2.5 protected sites) and is then ejected from the GroEL complex while still unfolded so that it competes with all the other unfolded molecules for rebinding. (B) Time-dependent hydrogen exchange with limiting GroES (RuBisCO: GroEL:GroES = 1:1.2:0.04). The predicted curve assumes that GroES cycles through the RuBisCO-GroEL complexes and induces exchange of the sensitive tritium label on the first RuBisCO turnover. The fitting equation for hydrogen exchange (HX) with non-native protein release on each turnover is H = Aexp[−kb(t − to)]exp[−kcat(t − to)] + C. The total number of exchangeable hydrogens at the initial 10-min time point (to) is 11, given by 8.5 sensitive hydrogens (A) and 2.5 insensitive ones (C). The background uncatalyzed HX rate during the pertinent time period (10 to 30 min) is kb (0.033 min−1). The chaperone-catalyzed HX rate (kcat) in (A) is 0.23 min−1, given by the 1/20 stoichiometry and 13-s processing time. In (B), kcat is 0.18 min−1, given by the 1/25 stoichiometry and 13-s processing time. The equation for the native protein release curve in (A) was approximated as H = [A − N(t − to)]exp[−kb(t − to)] + C. A, C, and kb are as before. N depends on the linear recovery rate for native protein, expressed in terms of the rate of loss of exchangeable hydrogen label (0.01 min−1), given by the 1/20 stoichiometry, 13-s turnover time, and 24 turnovers per successful RuBisCO folding.
These results show that RuBisCO is unfolded and released on each system turnover, inconsistent with the view that the substrate protein must remain in the cavity until it reaches the native state. In these substoichiometric experiments, the addition of AMP-PNP instead of ATP produced no detectable acceleration of exchange. Thus, AMP-PNP supports protein unfolding but not release and continued processing, whereas ADP supports neither function. Cryo electron microscopy indicates that AMP-PNP induces a smaller structure change in the GroEL complex than does ATP, and ADP causes no detectable change (4).
Our results parallel a fluorescence study of RuBisCO encapsulation and folding. Rye et al. (7 ) found that ATP or AMP-PNP but not ADP causes a rapid decrease in RuBisCO fluorescence intensity and anisotropy (~1 s) within the GroEL-GroES complex. With ATP this is followed by a slower rise in both fluorescence parameters at about the rate expected for native RuBisCO formation (~5 min). It seems likely that the fast change detected by fluorescence corresponds to the same unfolding event detected by tritium exchange. Rye et al. also found that the addition of ATP allows RuBisCO to fold within the GroEL cavity when the ejection mechanism is disabled. One assumes however that proteins that are able to may also fold outside the cavity after normal ejection.
GroEL can unfold proteins in a passive mass action sense, without ATP and GroES (27 28). This function depends on selective binding of the more unfolded protein form out of a reversible equilibrium mixture in which protein unfolding occurs spontaneously. This can occur even when the starting material is the native protein (17, 18, 28, 29) because protein molecules spontaneously unfold and refold even under native conditions (30). The binding of partially unfolded proteins might serve to sequester them while they fold toward the native state on the chaperonin surface. Chaperone molecules that passively assist protein folding may function in this way. This seems less likely for GroEL in active Escherichia coli where the ratio ATP:GroES:GroEL is about 5000:2:1.
In our experiments, the passive binding of blocked RuBisCO to GroEL alone had no unfolding effect, even over the time scale of 1 hour (Fig. 2). The induced structure change seen here and by Rye et al. (7 ) requires the complete energy-dependent chaperonin system and occurs on the biologically relevant time scale of seconds. The observation of ATP-dependent unfolding seems clearly relevant for GroEL function, and especially so in view of the rate-limiting nature for blocked protein folding of an unfolding-dependent reorganization process (13).
The demonstration that RuBisCO molecules bound in the GroEL-GroES complex experience an energy-dependent unfolding reaction fits well with available structural information. The sequence of molecular events includes a dramatic structure change, illustrated in Fig. 1 (2, 4, 5, 31). When ATP and GroES are bound, the equatorial domains of each GroEL subunit in the heptameric ring remain in tight contact but the apical binding domains twist upward and outward, causing the GroEL binding sites to move away from each other (32). The multisite ring structure of GroEL is well suited to bind an unfolded substrate protein at several points. Its stable equatorial platform together with the expansion of the distance between its binding sites (Fig. 1) appears well suited to generate a nonspecific stretching force, using the free energy of ATP binding (33). Furthermore, the movement of the apical domains together with the binding of GroES occludes the hydrophobic protein-binding surfaces and can be expected to displace the substrate protein into the cavity. Folding may then proceed within the cavity or subsequently when the GroES cap is removed and the protein escapes.
The ability of a stretching force to unfold protein structure has been demonstrated (34). The requirement for a partial unfolding has previously been implicated as the rate-limiting step in the folding of blocked proteins (13). A mechanical unfolding device of the sort we envisage here could operate equally well on a variety of different substrate proteins, thus accounting for GroEL’s lack of specificity. Other molecular chaperones and protein systems known to be similarly constructed (35) could exploit the same operating principle to unfold their substrate proteins.
Acknowledgments
We thank Z. Xu and P. B. Sigler for Fig. 1 and A. Horwich and C. Frieden for helpful discussion and information. Supported by NIH grant GM31847 to S.W.E.
References and Notes
- 1.Fenton WA, Horwich AL. Protein Sci. 1997;6:743. doi: 10.1002/pro.5560060401. [DOI] [PMC free article] [PubMed] [Google Scholar]; Coyle JE, Jaeger J, Groβ M, Robinson CV, Radford SE. Folding Des. 1997;2:R93. doi: 10.1016/S1359-0278(97)00046-1. [DOI] [PubMed] [Google Scholar]; Braig K. Curr Opin Struct Biol. 1998;8:159. doi: 10.1016/s0959-440x(98)80033-x. [DOI] [PubMed] [Google Scholar]
- 2.Sigler PB, et al. Annu Rev Biochem. 1998;67:581. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- 3.Fenton WA, Kashi Y, Furtak K, Horwich AL. Nature. 1994;371:614. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]; Braig K, et al. :578. ibid. [Google Scholar]
- 4.Roseman AM, Chen S, White H, Braig K, Saibil HR. Cell. 1996;87:241. doi: 10.1016/s0092-8674(00)81342-2. [DOI] [PubMed] [Google Scholar]
- 5.Xu Z, Horwich AL, Sigler PB. Nature. 1997;388:741. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 6.Weissman JS, et al. Cell. 1995;83:577. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]; Weissman JS, Rye HS, Fenton WA, Horwich AL. 1996;84:481. doi: 10.1016/s0092-8674(00)81293-3. ibid. [DOI] [PubMed] [Google Scholar]
- 7.Rye HS, et al. Nature. 1997;388:792. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 8.Mayhew M, et al. 1996;379:420. doi: 10.1038/379420a0. ibid. [DOI] [PubMed] [Google Scholar]
- 9.Sparrer H, Rutkat K, Buchner J. Proc Natl Acad Sci USA. 1997;94:1096. doi: 10.1073/pnas.94.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]; Llorca O, Marco S, Carrascosa JL, Valpuesta JM. FEBS Lett. 1997;405:195. doi: 10.1016/s0014-5793(97)00186-5. [DOI] [PubMed] [Google Scholar]
- 10.Ellis RJ, Hartl FU. FASEB J. 1996;10:20. doi: 10.1096/fasebj.10.1.8566542. [DOI] [PubMed] [Google Scholar]
- 11.Todd MJ, Viitanen PV, Lorimer GH. Science. 1994;265:659. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]; Weissman JS, Kashi Y, Fenton WA, Horwich AL. Cell. 1994;78:693. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]; Smith KR, Fisher MT. J Biol Chem. 1995;270:21517. doi: 10.1074/jbc.270.37.21517. [DOI] [PubMed] [Google Scholar]; Taguchi H, Yoshida M. FEBS Lett. 1995;359:195. doi: 10.1016/0014-5793(95)00041-7. [DOI] [PubMed] [Google Scholar]; Burston SG, Weissman JS, Farr GW, Fenton WA, Horwich AL. Nature. 1996;383:96. doi: 10.1038/383096a0. [DOI] [PubMed] [Google Scholar]; Sparrer H, Lilie H, Buchner J. J Mol Biol. 1996;258:74. doi: 10.1006/jmbi.1996.0235. [DOI] [PubMed] [Google Scholar]; Ranson NA, Burston SG, Clarke AR. 1997;266:656. doi: 10.1006/jmbi.1996.0815. ibid. [DOI] [PubMed] [Google Scholar]
- 12.Todd MJ, Lorimer GH, Thirumalai D. Proc Natl Acad Sci USA. 1996;93:4030. doi: 10.1073/pnas.93.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]; Corrales FJ, Fersht AR. :4509. doi: 10.1073/pnas.93.9.4509. ibid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sosnick TR, Mayne L, Hiller R, Englander SW. Nature Struct Biol. 1994;1:149. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 14.Thirumalai D. In: Statistical Mechanics. Doniach S, editor. Plenum; New York: 1994. pp. 15–134. [Google Scholar]; Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Proteins Struct Funct Genet. 1995;21:167. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]; Dill KA, et al. Protein Sci. 1995;4:561. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guo ZY, Thirumalai D. Biopolymers. 1995;36:83. [Google Scholar]
- 15.Radford SE, Dobson CM, Evans PA. Nature. 1992;358:302. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]; Jennings PA, Finn PA, Jones BE, Matthews CR. Biochemistry. 1993;32:3783. doi: 10.1021/bi00065a034. [DOI] [PubMed] [Google Scholar]; Kiefhaber T. Proc Natl Acad Sci USA. 1995;92:9029. doi: 10.1073/pnas.92.20.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]; Konermann L, Collings BA, Douglas DG. Biochemistry. 1997;36:5554. doi: 10.1021/bi970046d. [DOI] [PubMed] [Google Scholar]; Matagne A, Radford SE, Dobson CM. J Mol Biol. 1997;267:1068. doi: 10.1006/jmbi.1997.0963. [DOI] [PubMed] [Google Scholar]; Wildegger G, Kiefhaber T. 1997;270:294. doi: 10.1006/jmbi.1997.1030. ibid. [DOI] [PubMed] [Google Scholar]; Hammack B, Godbole S, Bowler BE. 1998;275:719. doi: 10.1006/jmbi.1997.1493. ibid. [DOI] [PubMed] [Google Scholar]; Freund C, Gehring P, Baici A, Holtak TA, Plückthun A. Folding Des. 1998;3:39. doi: 10.1016/S1359-0278(98)00007-8. [DOI] [PubMed] [Google Scholar]; Shastry MCR, Udgaonkar JB. J Mol Biol. 1998;247:1013. doi: 10.1006/jmbi.1994.0196. [DOI] [PubMed] [Google Scholar]; Burton RE, Myers JK, Oas TG. Biochemistry. 1998;37:5337. doi: 10.1021/bi980245c. [DOI] [PubMed] [Google Scholar]
- 16.Kirkpatrick S, Gelatt CD, Jr, Vecchi MP. Science. 1983;220:671. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 17.Zahn R, Spitzfaden C, Ottiger M, Wuthrich K, Pluckthun A. Nature. 1994;368:261. doi: 10.1038/368261a0. [DOI] [PubMed] [Google Scholar]; Zahn R, Perrett S, Fersht AR. J Mol Biol. 1996;261:43. doi: 10.1006/jmbi.1996.0440. [DOI] [PubMed] [Google Scholar]
- 18.Okazaki A, Ikura T, Nikaido K, Kuwajima K. Nature Struct Biol. 1994;1:4396. doi: 10.1038/nsb0794-439. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg MS, et al. Proc Natl Acad Sci USA. 1997;94:1080. doi: 10.1073/pnas.94.4.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gervasoni P, Gehrig P, Pluckthun A. J Mol Biol. 1998;275:663. doi: 10.1006/jmbi.1997.1481. [DOI] [PubMed] [Google Scholar]
- 21.Gross M, Robinson CV, Mayhew M, Hartl FU, Radford SE. Protein Sci. 1996;5:2506. doi: 10.1002/pro.5560051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Englander SW, Kallenbach NR. Q Rev Bio-phys. 1984;16:521. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]; Englander SW, Mayne L. Annu Rev Biophys Biomol Struct. 1992;21:243. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]; Bai Y, Milne JS, Mayne L, Englander SW. Proteins Struct Funct Genet. 1993;17:75. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]; Connelly GP, Bai Y, Jeng M-F, Mayne L, Englander SW. 1993;17:87. doi: 10.1002/prot.340170111. ibid. [DOI] [PubMed] [Google Scholar]; Englander SW, Sosnick TR, Englander JJ, Mayne L. Curr Opin Struct Biol. 1996;6:18. doi: 10.1016/s0959-440x(96)80090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Vies SM, Viitanen PV, Gatenby AA, Lorimer GH, Jaenicke R. Biochemistry. 1992;31:3635. doi: 10.1021/bi00129a012. [DOI] [PubMed] [Google Scholar]; Schmidt M, Buchner J, Todd MJ, Lorimer GH, Viitanen PV. J Biol Chem. 1994;269:10304. [PubMed] [Google Scholar]
- 24.Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Nature. 1989;342:884. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 25.Schneider G, Lindqvist Y, Lundqvist T. J Mol Biol. 1990;211:989. doi: 10.1016/0022-2836(90)90088-4. [DOI] [PubMed] [Google Scholar]
- 26.Viitanen PV, et al. Biochemistry. 1990;29:5665. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- 27.Walter S, Lorimer GH, Schmid FX. Proc Natl Acad Sci USA. 1996;93:9425. doi: 10.1073/pnas.93.18.9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark AC, Frieden C. J Mol Biol. 1999;285:1777. doi: 10.1006/jmbi.1998.2403. [DOI] [PubMed] [Google Scholar]
- 29.Zahn R, Perrett S, Stenberg G, Fersht AR. Science. 1996;271:642. doi: 10.1126/science.271.5249.642. [DOI] [PubMed] [Google Scholar]
- 30.Bai Y, Sosnick TR, Mayne L, Englander SW. 1995;269:192. doi: 10.1126/science.7618079. ibid. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chamberlain AK, Handel TM, Marqusee S. Nature Struct Biol. 1996;3:782. doi: 10.1038/nsb0996-782. [DOI] [PubMed] [Google Scholar]; Hiller R, Zhou ZH, Adams MWW, Englander SW. Proc Natl Acad Sci USA. 1997;94:11329. doi: 10.1073/pnas.94.21.11329. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fuentes EJ, Wand AJ. Biochemistry. 1998;37:3687. doi: 10.1021/bi972579s. [DOI] [PubMed] [Google Scholar]
- 31.Xu Z, Sigler PB. personal communication.
- 32.Lorimer GH. Nature. 1997;388:720. doi: 10.1038/41892. [DOI] [PubMed] [Google Scholar]
- 33.Buckle AM, Zahn R, Fersht AR. Proc Natl Acad Sci USA. 1997;94:3571. doi: 10.1073/pnas.94.8.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Science. 1997;276:1109. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]; Kellermayer MSZ, Smith SB, Ganzier HL, Bustamante C. :1112. ibid. [Google Scholar]
- 35.Petosa C, et al. Nature. 1997;385:833. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]; Groll M, et al. 1997;386:463. doi: 10.1038/386463a0. ibid. [DOI] [PubMed] [Google Scholar]; Ditsel L, et al. Cell. 1998;93:125. [Google Scholar]
- 36.Unfolded RuBisCO [5 M urea, 10 mM HCl, 1 mM dithiothreitol (DTT, pH 2] was initially labeled to hydrogen exchange equilibrium in tritiated water (~10 mCi/ml). To begin the exchange of hydrogen, we diluted RuBisCO (1:20) into conditions that do not permit folding (20 mM tris buffer, pH 8.0, 2 mM magnesium acetate, 2 mM potassium acetate, 1 mM DTT, 0.01% Tween-20, 22° ± 2°C, RuBisCO at 2 μM), with or without a small excess of GroEL14. Free solvent tritium was immediately removed by centrifuging the RuBisCO solution through a Sephadex G-25 spin column [<1 min, 0.5 ml through a 1 cm × 5 cm column (37)] equilibrated with the nonpermissive refolding buffer. After hydrogen exchange for the desired time, free tritium was removed by a second spin column. The tritium label remaining bound was counted by liquid scintillation and computed in terms of the number of hydrogens per RuBisCO molecule not yet exchanged. For this calculation, 100% recovery of the known initial RuBisCO was assumed. Control experiments showed that GroEL does not account for any of the bound label; therefore, the analysis does not require the separation of GroEL from the labeled substrate protein. To avoid tritium contamination of samples, it is necessary to remove the initial free tritium (~1010 cpm/ml) by a large factor (~108), and it is advisable to spatially separate experimental operations to avoid minuscule splash and volatility problems, which accounts for the data spread seen in our early data. In the absence of tritium contamination, accuracy is at the level of a few percent. RuBisCO was prepared as described before (38). GroEL and GroES were overexpressed in E. coli and purified as described before (39). Protein concentration was measured spectrophotometrically at 280 nm using extinction coefficients of 9600 M−1 cm−1 per GroEL monomer, 1200 M−1 cm−1 per GroES monomer (calculated from sequence), and 67,100 M−1 cm−1 for RuBisCO (38). GroEL and GroES concentrations were confirmed by quantitative amino acid analysis.
- 37.Jeng MF, Englander SW. J Mol Biol. 1991;221:1045. doi: 10.1016/0022-2836(91)80191-v. [DOI] [PubMed] [Google Scholar]
- 38.Schloss JV, et al. Methods Enzymol. 1984;90:522. doi: 10.1016/s0076-6879(82)90179-3. [DOI] [PubMed] [Google Scholar]; Pierce J, Gutteridge S. Appl Environ Microbiol. 1986;49:1094. doi: 10.1128/aem.49.5.1094-1100.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark AC, Ramanathan R, Frieden C. Methods Enzymol. 1998;290:100. doi: 10.1016/s0076-6879(98)90010-6. [DOI] [PubMed] [Google Scholar]



