Abstract
Detecting genomic structural variants from high-throughput sequencing data is a complex and unresolved challenge. We have developed a statistical learning approach, based on Random Forests, which integrates prior knowledge about the characteristics of structural variants and leads to improved discovery in high throughput sequencing data. The implementation of this technique, forestSV, offers high sensitivity and specificity coupled with the flexibility of a data-driven approach.
Structural variants are a major form of genetic variation1,2. The investigation of structural variants has provided key insights into the genetic basis of common human disease3, most notably in neuropsychiatric disorders, where it has been well established that rare and de novo mutations confer significant risk4.
A variety of tools have been developed to detect structural variation using genomic sequence data. Some approaches rely on information gleaned from changes in read depth5,6, while others also include signals from outlier read pairs7,8. Still other methods rely on split-read signals9 (reads that span a breakpoint). Recently, the 1000 Genomes Project undertook a thorough evaluation10 of these and other structural variant detection methods and found that there was wide variability in terms of sensitivity and specificity among the methods. While some of the tested methods were clear leaders (notably GenomeSTRiP7 for its low false discovery rate), no single method produced results with a sensitivity comparable to a careful merging of calls from all the methods.
The fact that the fusion of structural variant calls from multiple discovery methods yields better results than any single method suggests that existing tools may be individually too narrow in terms of the signals they consider or the methodology they employ. If a single method could effectively capture the collective insight provided by the variety of available tools, it might obviate the need to run all of them and carefully merge the results, saving valuable time and resources.
In this work, we present a first step in this direction by drawing on machine learning ideas to facilitate better structural variant discovery. We used previous studies as a basis for extracting the multidimensional patterns that discriminate calls that validate experimentally from those that do not.
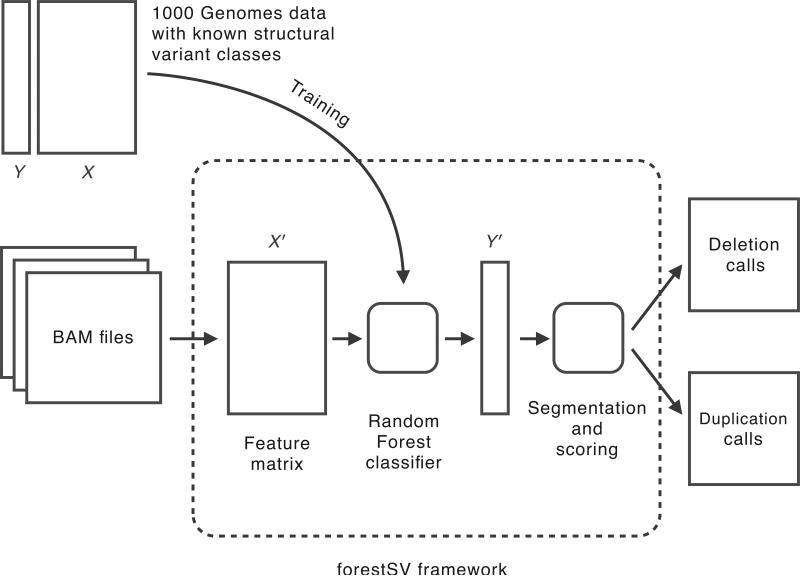
Beyond the typical read depth and read pair signatures used by most structural variant discovery approaches, we constructed additional features from the sequencing data (described in Online Methods), thereby representing the genome in a higher-dimensional space (shown as the feature matrix X in Fig. 1). We trained a Random Forest classifier11 to partition this space in a way that optimizes the classification of deletions, duplications, and false positives whose identities are stored in the class label vector Y (Fig. 1, Supplementary Fig. 1).
Figure 1. The forestSV framework.
The core of the framework is a Random Forest classifier trained to recognize structural variants based on calls and data (Y and X, respectively) from the 1000 Genomes Project. To make calls on new data, BAM files are given as input. The data from the BAM files are used to construct a new feature matrix X'. The classifier provides a mapping from X' to predicted structural variant classes, Y'. The predictions are segmented into structural variant calls and scored according to the confidence in their class assignment. The output of the framework is a pair of lists of predicted deletion and duplication events, with their associated confidence scores.
For training, the features in X were constructed from data available in the BAM files for the high-coverage trios (a total of six individuals) in the 1000 Genomes Project12 (1KG). The structural variant class labels in Y were assigned based on an integration of the calls from refs. 10, 13, 14. The seven class labels “deletion”, “duplication”, “deletion-flanking”, “ duplication-flanking”,” false positive deletion”, “false positive duplication”, and “invariant” are defined in the supplementary information (Supplementary Table 1).
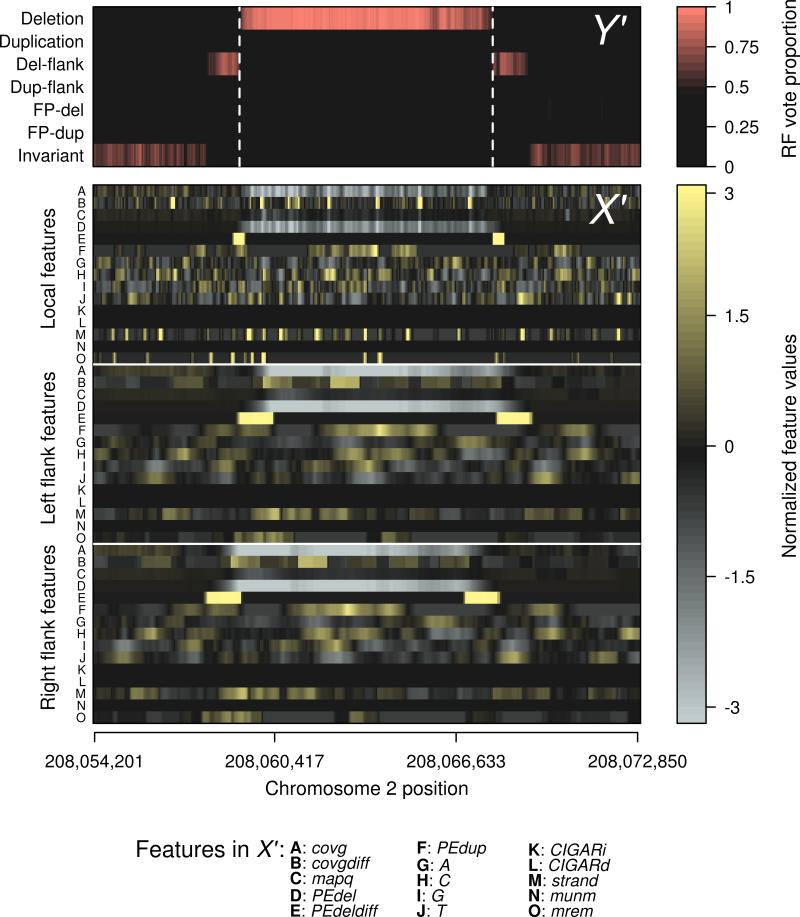
The rows of X (and consequently the elements of Y) correspond to overlapping 100 bp windows of the genome that we call “subjects” here (Supplementary Fig. 1). When presented with new data (X') the trained classifier assigns a predicted class (Y') to each subject (Fig. 2). The final variant calls are then generated by a simple segmentation routine that merges consecutive subjects of the same predicted class into a single event. These events are ranked by a prediction confidence score produced by the classifier (the Random Forest vote proportion, Fig. 2).
Figure 2. Mapping of features to structural variant calls by Random Forests.
Sequence features (A-O in matrix X') from a representative deletion locus on chromosome 2 of the autistic individual 03C14438, and their corresponding classification (Y') by the classifier as a deletion event. The estimated boundaries of the deletion event are shown by the dashed lines. Because the classifier has been trained to recognize regions that flank known structural variants, the presence of these landmarks, independent of the deletion call itself, provides additional support for the presence of an event. Features are labeled according to their designation given in the forestSV package, and are also described in methods. Detailed descriptions of the features are provided in Supplementary Table 2. Features have been standardized to mean 0 and variance 1.
The performance of forestSV was first assessed on the 1KG data, and compared with calls of other widely used methods10. We trained the classifier on five of the six 1KG genomes, and then made structural variant calls on the left-out genome, doing this for each of the six individuals in turn. We then combined the scored calls and constructed sensitivity-specificity curves (Supplementary Fig. 2) that reflect forestSV's ability to correctly prioritize the calls in a gold standard set. For comparison, we also marked the sensitivity and specificity of the “donor” call sets (method/group designations are carried over from ref. 10). For both deletion and duplication events, forestSV provided a combination of sensitivity and specificity that, in the gold standard set we examined, was superior to all of the donor call sets and matched the sensitivity and specificity of the merged and genotyped call set provided in ref. 10 (Supplementary Fig. 2). This suggests that integrative methods, such as forestSV, outperform approaches that rely on a single methodology or sequence feature for structural variant discovery. Lastly, we examined whether the inclusion of related individuals in the training set of this leave-one-out scheme might lead to a disproportionate boost in the performance of forestSV, and we did not find evidence for such an effect (Supplementary Fig. 3).
The characteristics of the calls generated by forestSV are in line with the consensus genotyped calls in ref. 10 (Supplementary Results, Supplementary Figs. 4-6). There is a relationship between event size and forestSV event score for duplications (Supplementary Fig. 7), while for deletions scores are unbiased across a range of event sizes. We found that while forestSV was trained on high-coverage data, it also performs reasonably well on low-coverage (5×) data (Supplementary Fig. 8). The accumulation of calls with a relaxing event score is characteristically different for deletions and duplications (Supplementary Fig. 9).
As an independent evaluation of our method, we applied forestSV to high coverage genome sequence data from a five-person family currently under investigation in our laboratory, consisting of a father, mother, monozygotic (MZ) twins concordant for autism, and an unaffected sibling. Performance of forestSV was evaluated based on patterns of Mendelian inheritance in the family and identity of genotypes between MZ twins (Supplementary Fig. 10). “Mendelian inconsistencies” were defined as calls present in any of the children but not in either of the parents (with > 50% reciprocal overlap). “MZ discordances” were defined similarly as events detected in one twin and not found with > 50% reciprocal overlap in the other. We then plotted how rates of Mendelian inconsistency and MZ discordance varied with the event scores in the children (Supplementary Fig. 10). By both measures, error rates are very low among events scoring > 0.65, suggesting that this set is enriched for true structural variants. These results suggest that an event score threshold in the range of 0.65-0.70 will keep error rates below 5% for deletions and ~10% for duplications. At a threshold of 0.65, forestSV discovered 2,300-2,593 unique deletions per individual, and 53-72 unique duplications per individual. Clearly, sensitivity for detecting duplications is lower than for deletions and hence a more relaxed threshold may be needed (see Supplementary Fig. 3d and Supplementary Fig. 9). Because this data set has not yet been as thoroughly characterized as the 1KG high-coverage trio samples, we are unable to reliably estimate sensitivity directly.
forestSV is particularly well suited to the detection of rare variants because it is not reliant on finding variant support in multiple individuals. It can call structural variants effectively in a single genome. This contrasts with earlier methods developed by our group 5 and others7 that require multiple genomes to be analyzed simultaneously and which favor variants with support in multiple individuals. This is a key advantage in light of the knowledge that rare structural variants, with population frequencies of ≤10-4, play an important role in common disease and particularly in diseases of the brain15.
We implemented forestSV as an R package. It includes an executable that, with a single command, takes BAM files as input and produces deletion and duplication call sets. The package source code and documentation, together with a thorough technical tutorial describing its use, are available for download from our website at http://sebatlab.ucsd.edu/software. forestSV will continually improve its discovery abilities as additional training data becomes available (Supplementary Results, Supplementary Fig. 11) and we will periodically provide updates to the trained classifier on our website.
Online Methods
The methodology described in this work has been assembled as an R package called forestSV, which is available for download at http://sebatlab.ucsd.edu/software. This package is distributed with a tutorial that will enable the user to reproduce the type of analysis we have demonstrated.
Features
A coverage vector is a one-dimensional data structure whose elements map to positions in the genome, and whose entries indicate the number of times the corresponding position is covered by a read. Such a vector gives an indication of read depth. The features that the Random Forest classifier uses to predict the class identity of a genomic window are weighted coverage vectors. As the name implies, a weighted coverage vector has weights applied to the individual reads before the coverage is computed. The weights may be logical (binary) or continuous. In the work described here, weights are extracted from the BAM files, and convey information on such attributes as outlier read pairs, base content, mapping quality, CIGAR operations, and mapping flags. Descriptions of the weighting procedures for these features is given below, and their explicit implementation can be found in the source code of the featMat() function in the forestSV package.
To construct features, the weighted coverage vectors are evaluated at three scales: local (100 bp window), left flank (1 kb window to the left of the local window), and right flank (1 kb window to the right of the local window). The mean value of each weighted coverage vector is taken at these three scales, and the position defined by the local window constitutes a row in the feature matrix. Throughout the text, we refer to these row entries in the feature matrix as “subjects”. The columns of the matrix are the features themselves (15 total), evaluated at each of the three scales (for a total of 45 columns). The next row in the matrix is offset by 50 bp, and the features are evaluated again, and so on. The 15 principal features are given in Supplementary Table 2. The process of constructing a feature matrix from a BAM16 file is implemented in the featMat() function provided in the forestSV package.
Not all features are equally informative, and a feature's information content varies by event class, i.e. some features may be useful for discerning duplications but less useful for deletions, etc. We summarized the contribution of each feature by using the permutation importance measures available in Random Forests11 (Supplementary Fig. 12). These importance measures quantify the degradation in predictive accuracy of the classifier when the values of a feature are permuted randomly. The degree to which a classifier's performance is degraded by the random permutation of a feature reflects the importance of that feature. Unsurprisingly, the most important features are those already used by other structural variant discovery algorithms: read depth and paired end signatures. However, several of the additional features we introduced did in fact contribute to improved discovery. For instance, false duplication calls are more accurately identified when the mapq feature is included in the model, and identification of false deletion calls is improved by information contained in the strand, munm, and mrem features. It should be noted that Random Forest classifiers are generally robust against the inclusion of uninformative features, so the less informative features (such as CIGARd and CIGARi) do not adversely impact the accuracy of the classifier. We retain them in the classifier for the possibility that they may prove to be useful when including future data and structural variant types.
It is common practice in structural variant discovery methods to impose a global correction for GC content5,6, since such regions have been known to have biased read depth signals 17. Here we do not impose a global correction, but rather include base content as a predictor in the training process. This has the advantage of avoiding the assumption that extreme GC content will lead universally to false positives, independent of all other variables. If GC content is found to have predictive power in discriminating true variants from false positives in the training set, it is incorporated into the classifier in a way that allows for (though does not require) its effect to be conditional on the other variables used. Such an allowance for the dependence of the predictive features cannot be achieved with a single, genome-wide GC content correction.
To demonstrate that high-confidence calls generated by forestSV take GC content into account, we compared the distribution of GC content among high-confidence calls (event score > 0.65) with that of low-confidence calls (event score < 0.65). The results (Supplementary Fig. 13) show that GC content does indeed vary with the event score, with higher-scoring calls tending more toward intermediate GC content (mean: 0.43), and particular depletion in the tail tending toward high GC content. The two distributions differ significantly (P < 10-10 by the Kolmogorov-Smirnov test).
Training data
Training examples were defined using experimentally validated (or invalidated) structural variant calls from individuals NA12878, NA12891, NA12892, NA19240, NA19238, and NA19239 in and refs. 10, 13, 14. Subjects were given one of seven class labels: deletion, duplication, deletion-flanking, duplication-flanking, false positive deletion, false positive duplication, or invariant. Descriptions of these classes are given in Supplementary Table 1.
Only Illumina data acquired from the 1000 Genomes Project website were used for construction of the training set features. MAQ-aligned 1KG data were used in the performance comparison with the calls from ref. 10 and BWA-aligned 1KG data were used to train the classifier that was used for structural variant discovery in the BWA-aligned autism data. Size distributions and event counts for each of the classes in the gold standard set are shown in Supplementary Figure 14. The regions used as the basis for the training set are available in Supplementary Data 1.
forestSV offers an open-ended framework that could be re-trained to identify additional classes of structural variation that are not currently well ascertained with any available method. The detection of a new structural variant class could be facilitated by adding new features to the matrix X that are indicative of the class. Training examples of the new class could then be included in the training data and a new classifier constructed. Such extension of forestSV is discussed in greater detail in the tutorial that accompanies the forestSV package.
Donor call sets
The call sets comprising the work in ref. 10 are referred to throughout this work by the two-letter abbreviation originally given there. These are: Applied Biosystems/Life Technologies (AB), Boston College (BC), BGI-Shenzhen (BG), Broad Institute (BI), Leiden University Medical Center (LN), University of Oxford (OX), University of California, San Diego (SD), Wellcome Trust Sanger Institute (SI), University of Washington (UW), Washington University, St. Louis (WU), and Yale University (YL).
Random Forest classifier
The R reference implementation18 of Random Forests11 was used to build the classifier. Forests with 50 trees were grown, and the bootstrap resampling was stratified according to the classes, with a maximum within-class sample size of 500,000 subjects. All other arguments took on the default value.
Segmentation and scoring of structural variant calls
When presented with new data, the trained classifier outputs seven numeric values for every subject, one for each class, that together sum to 1. These are the class vote proportions, and are an indication of the identity of the subject, as predicted by the classifier. A higher value indicates higher confidence that the subject belongs to the corresponding class. Since these seven scores are distributed in space, that is, along the length of a chromosome, the way in which they co-vary along the chromosome can be used to identify regions of structural variation. For instance, in regions where vote proportion for class deletion is consistently higher than the vote proportions of all the other classes, this is indicative of a deletion event. The extent of the event is defined by the extent over which the vote proportion for deletion exceeds that of all the other classes (i.e. simple majority vote).
More concretely, events (deletions and duplications) are defined as chromosomal regions where contiguous subjects have deletion or duplication as their maximally-scoring class. Events of the same class are merged if separated by 1 kb or less. The final score of the merged events is the mean score of the majority class over the subjects within the event boundaries. This process is implemented in the svScore() and svCall() functions provided by the forestSV package.
The breakpoint resolution of forestSV calls is limited by the fact that subjects are offset by 50 bp, meaning that all event predictions have boundaries in increments of 50 bp. This is the result of design decisions that favored efficient discovery over high resolution of breakpoints, but can be addressed by supplementing forestSV calls with breakpoint assembly methods, such as TIGRA_SV (Chen, L. et al., unpublished, http://genome.wustl.edu/software/tigra_sv).
Performance assessment
For each of the six 1KG individuals (NA12878, NA12891, NA12892, NA19240, NA19238, and NA19239), a classifier was trained with the training data from that individual omitted. Structural variants were then called and scored, as described above, for the left-out individual. Calls for all six of the individuals are provided in Supplementary Data 2.
Sensitivity-specificity curves for deletions and duplications were then calculated on the gold standard set (Supplementary Fig. 2), which consisted of the previously described gold standard deletions and duplications (positives), as well as all false positive deletion calls and false positive duplication calls (true negatives). Assessed calls were required to have > 50% reciprocal overlap with gold standard calls (both positives and negatives) to be used in the performance calculation. For example, if none of the calls from a method had > 50% reciprocal overlap with any of the gold standard negatives, this would lead to 100% specificity; likewise, if none of a method's calls had > 50% reciprocal overlap with the gold standard positives, this would lead to 0% sensitivity.
The curves (Supplementary Fig. 2) represent the pooled performance of forestSV over all six individuals. Because the calls released in ref. 10 do not have scores associated with them, we were only able to calculate single points of sensitivity and specificity for each method, rather than entire curves. In Supplementary Figure 3, we showed that inclusion of related individuals in the training set does not lead to inflated estimates of predictive performance. For each of the six 1KG individuals, we trained a classifier on 1, 2, 3, 4, and 5 of the remaining genomes, then assessed performance (area under the ROC curve, AUC) using the gold standard deletions as described previously. In every case, related individuals (parents or offspring) were added to the training set last. We performed a within-individual Z-transformation to normalize the AUC values, and then regressed these values on the number of genomes used in the training set. Only data from training sets consisting entirely of unrelated individuals were used in the regression. Prediction intervals for this linear model are shown at levels of 90%, 95%, and 99%. The observed performance values for training sets that include related individuals all fell well within the 90% prediction interval, suggesting that the inclusion of related individuals while training does not lead to a disproportionate increase in performance compared to including unrelated individuals. This means that our cross-validated performance estimates are not unduly influenced by having related individuals in the training set.
Annotation information used to calculate the metrics depicted in Supplementary Figure 4 were derived from hg18 tracks downloaded from http://genome.ucsc.edu. Proportions of total called sequence occupied by genomic features were calculated by first summing the total unique genomic sequence (in bp) called by each method, then assessing the percentage of that sequence that was covered by the genome feature annotations (segmental duplications, Alu, SINE, LINE, LTR). For centromeres and telomeres, we extended their boundaries by 500 kb and calculated the percentage of calls in each set that intersected these regions.
For the application to the autism data, we used a classifier trained on BWA-aligned Illumina sequence data from all six 1KG individuals. We used the same events (with their class labels) as in the previous 1KG validation, but had to lift over coordinates from hg18 to hg19, since the training BWA BAM files were aligned to this reference assembly. Using the forestSV framework, we produced genome-wide calls in the family 74-0352, which consists of a mother, father, two MZ twins, and a sibling, sequenced to ~40× depth (Illumina, paired-end, BWA aligned to hg18). Mendelian inconsistencies were considered to be calls in a child for which no call in the parents had greater than 50% reciprocal overlap. Similarly, discordant events between the twins were those that failed to achieve > 50% reciprocal overlap.
Computational considerations
We found that processing a single genome (~40× coverage) in a single thread (resulting in both deletion and duplication calls) took about 8 GB of RAM and 8 hours on an Intel Core i7 980X processor. Much of this time represents disk I/O while accessing BAM files. The run time can be significantly reduced if the BAM files are on a local (rather than a network-attached) disk.
Supplementary Material
Acknowledgments
This work was performed under NIH grants HG005725 and MH076431 and with support from the Beyster Family Foundation. We also thank the 1000 Genomes Project for access to data, and J. Wang, H. Zheng, Y. Li, X. Jin, and Y. Shi from BGI-Shenzhen for their roles in producing the unpublished autism sequencing data.
Footnotes
Author Contributions
J.J.M. conceived of and implemented forestSV. J.J.M. and J.S. wrote the manuscript.
References
- 1.Sebat J, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 2.Iafrate AJ, et al. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 3.Stankiewicz P, Lupski JR. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 4.Sebat J, Levy DL, McCarthy SE. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abyzov A, Urban AE, Snyder M, Gerstein M. Genome Res. 2011 doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Nat. Genet. 2011;43:269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K, et al. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills RE, et al. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breiman L. Machine Learning. 2001;45:5–32. [Google Scholar]
- 12.1000 Genomes Project Consortium Nature. 2010;467:1061–1073. [Google Scholar]
- 13.Conrad DF, et al. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarroll SA, et al. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra D, Sebat J. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, et al. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentley DR, et al. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liaw A, Wiener M. R News. 2002;2:18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.