Summary
Since the discovery of a single white eyed male in a population of red eyed flies over 100 years ago (Morgan, 1910), the compound eye of the fruit fly, Drosophila melanogaster, has been a favorite experimental system for identifying genes that regulate various aspects of development. For example, a fair amount of what we know today about enzymatic pathways and vesicular transport is due to the discovery and subsequent characterization of eye color mutants such as white. Likewise, our present day understanding of organogenesis has been aided considerably by studies of mutations, such as eyeless, that either reduce or eliminate the compound eyes. But by far the phenotype that has provided levers into the greatest number of experimental fields has been the humble “rough” eye. The fly eye is comprised of several hundred unit-eyes that are also called ommatidia. These unit eyes are packed into a hexagonal array of remarkable precision. The structure of the eye is so precise that it has been compared to that of a crystal (Ready et al., 1976). Even the slightest perturbations to the structure of the ommatidium can be visually detected by light or electron microscopy. The cause for this is two-fold: (1) any defect that affects the hexagonal geometry of a single ommatidium can and will disrupt the positioning of surrounding unit eyes thereby propagating structural flaws and (2) disruptions in genes that govern the development of even a single cell within an ommatidium will affect all unit eyes. In both cases the effect is the visual magnification of even the smallest imperfection. Studies of rough eye mutants have provided key insights into the areas of cell fate specification, lateral inhibition, signal transduction, transcription factor networks, planar cell polarity, cell proliferation and programmed cell death just to name a few. This review will attempt to summarize the key steps that are required to assemble each ommatidium.
Keywords: Drosophila, retina, ommatidium, photoreceptor, cone cell, pigment cell
Structure of the Adult Eye and the Ommatidium
The Drosophila melanogaster compound eye is a simple nervous system consisting of approximately 800 unit eyes or ommatidia that are assembled into a hexagonal array of striking precision (Fig. 1A, B). A typical eye will contain roughly 32–34 interlocking vertical columns of unit eyes. The number of ommatidia per column is graded across the eye and this gives the eye an overall oval or egg shape. The compound eye also contains a set of mechano-sensory bristles, which are located at the anterior vertex of each ommatidium. As is the case in other insects, each bristle complex within the fly eye consists of four cells: the tormogen (socket), trichogen (shaft), thecogen (sheath) and sensory neuron (Wigglesworth, 1953; Waddington and Perry, 1968). The compound eye as well as the surrounding head capsule, antenna, maxillary palps and ocelli are all derived from the eye-antennal disc (Vogt, 1946; Ferris, 1950; Gehring, 1966; Ouweneel, 1970; Haynie and Bryant, 1986).
Figure 1. Structure of the adult compound eye.
(A) Scanning electron micrograph of an adult wild type Drosophila compound eye. (B) Light microscope section of an adult wild type compound eye. Note the change in chirality of the ommatidia at the equator. (C) A high magnification view of a single ommatidium. The photoreceptors are numbered according to the convention established by Dietrich, 1909. Note that the photoreceptors are arranged in an orientation that resembles an assymetrical trapezoid. Anterior is to the right on all images.
The core of the adult ommatidium contains eight photoreceptor neurons, four lens secreting cone cells and two primary pigment cells. Each ommatidium then shares six secondary pigment cells, three tertiary pigment cells and three mechano-sensory bristle complexes with its surrounding neighbors (Fig. 2A; Waddington and Perry, 1960). The eight photoreceptors (R1–R8) extend a highly ordered array of microvilli, called the rhabdomere, into the center of the ommatidium. The R1–R6 photoreceptors occupy the outer regions of the ommatidium while the R7 and R8 neurons are more centrally positioned. In any one given plane only seven rhabdomeres are seen because the R7 cell sits atop the R8. When correctly arranged the positioning of the rhabdomeres within the distal retina (R1–R7) resembles an asymmetric trapezoid (Fig. 1C; Dietrich, 1909). The orientation of the trapezoid (which reflects the positioning of the photoreceptors) abruptly switches at the equator such that the trapezoids in the dorsal and ventral halves of the eye are mirror images of each other. There is four-fold symmetry across the eye fields of both compound eyes (Fig. 2B; Dietrich, 1909). These chiral differences in the adult reflect earlier polarizations of the eye field as well as opposing directions of ommatidial rotation (reviewed by Jenny, 2010).
Figure 2. Cellular composition and chirality of the Drosophila ommatidium.
(A) An illustration describing the cellular composition of a single ommatidium. (B) An illustration describing the four-fold symmetry that exists within the two compound eyes of a single adult fly. Note that the photoreceptors are arranged in an orientation that resembles an asymmetrical trapezoid. Anterior is to the right in all illustrations.
Cellular Development of the Ommatidium
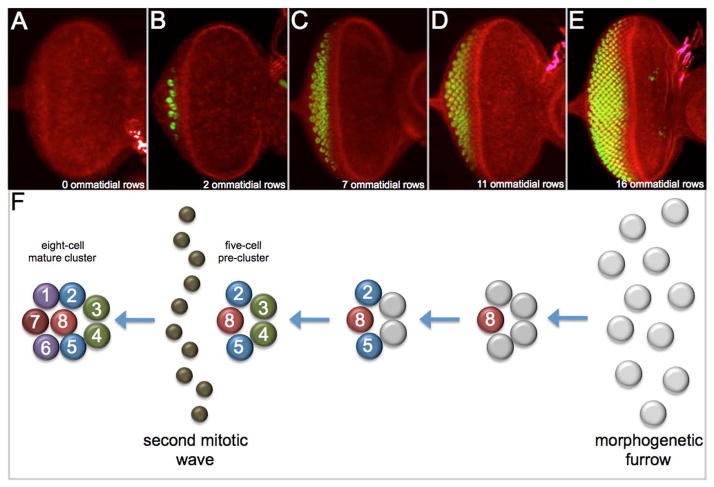
The adult compound eye is derived from a monolayer epithelium called the eye-antennal disc (Vogt, 1946; Ferris, 1950; Poulson, 1950; Gehring, 1966; Ouweneel, 1970; Haynie and Bryant, 1986). During embryogenesis the nascent eye-antennal discs consists of only 8–9 cells (Madhavan and Schneidermann, 1977). During the first two larval instars the eye disc undergoes a tremendous increase in size (reviewed by Kumar, 2011), is subdivided into dorsal and ventral compartments (reviewed by Singh et al., 2005) and is channeled towards an eye fate by the retinal determination network (reviewed by Kumar, 2010). Overt patterning of the eye disc begins at the late second/early third larval transition when a wave of differentiation, called the morphogenetic furrow, initiates at the posterior margin of the disc and sweeps across the eye field transforming a sea of undifferentiated cells into ordered rows of periodically spaced ommatidia (Fig. 3A–E; Ready et al., 1976).
Figure 3. The morphogenetic furrow and the assembly of the ommatidium.
(A–E) Confocal images of developing wild type eye eye discs. The adjacent antennal disc has been removed in each images. red = F-actin, green = ELAV. Note that the morphogenetic furrow initiates at the posterior margin and traverses the disc towards the anterior edge of the epithelium. (F) An illustration depicting the developmental history of an ommatidium. Note that the eight-cell mature cluster has two-fold symmetry. This symmetry will be broken as the ommatidium rotates and adopts the asymmetric trapezoidal orientation of the adult retina. Anterior is to the right in all images and illustrations.
The ommatidium is derived from cell clusters that arise deep within the morphogenetic furrow. As undifferentiated cells are incorporated into the rolling furrow a subset of them are organized into periodic clusterings of approximately twenty cells called rosettes. Several cells from this structure are expelled leaving an arc-like structure of 7–9 cells. These will eventually give rise to the five-cell precluster, the structural beginning of the ommatidium (Wolff and Ready, 1991a). The five cells of the precluster will give rise and correspond to five photoreceptor neurons. The R8 cell is the first cell to be specified followed by the R2/5 and R3/4 pairs (Fig. 3F; Tomlinson and Ready, 1987a). All cells that are not incorporated into the preclusters undergo a single additional round of mitosis. This second mitotic wave is required to generate the remaining cells needed to produce mature ommatidia (Fig. 3F; Ready et al., 1976; Wolff and Ready, 1991a; deNooji et al., 1995). As cells exit the second mitotic wave several cells immediately join the pre-cluster and adopt the fates of the final three photoreceptors (R1/6/7). This addition creates the mature eight-cell cluster (Fig. 3F; Tomlinson and Ready, 1987a).
The lens secreting cone cells are the last cells to be added to the ommatidium during the larval instar. Based on their position within the ommatidium they are referred to as the anterior, posterior, equatorial and polar cone cells. The anterior and posterior cone cells join the cluster first followed by the equatorial and polar pair (Fig 4A–C; Ready et al., 1976; Cagan and Ready, 1989a). Any cell that is not specified as either a photoreceptor neuron or a lens secreting cone cell adopts one of three pigment cell fates during the pupal stage of development (Cagan and Ready, 1989b). The primary pigment cells are added first and this completes the formation of the core ommatidium (Fig. 4D; Cagan and Ready, 1989a). The secondary and tertiary pigment cell types are added last and are important for creating the hexagonal lattice that surrounds the core ommatidium (Fig. 4E; Cagan and Ready, 1989a, b). The mechanosensory bristles are the final elements that complete the ommatidium (Fig 4F; Ready et al., 1976). The eye field generates more cells than is required. Since the initial number of R8 neurons determines the number of all other photoreceptors, cones and primary pigment cells, all excess cells wind up adopting either a secondary or tertiary fate. It is therefore thought that these cell types represent the default cell fate choice in the eye. These excess pigment cells must be eliminated or else the precise structure of the eye will be compromised and a rough eye will result. An epoch of apoptosis occurs during the mid-pupal stage and eliminates enough cells such that only a single secondary cell exists along the long faces of each ommatidium and that only a single tertiary pigment cells occupies the vertices (Cagan and Ready, 1989b: Wolff and Ready, 1991b; Miller and Cagan, 1998; Larson et al., 2010).
Figure 4. Order of cone, pigment and bristle cell recruitment in the pupal retina.
(A–F) An illustration depicting the order in which all non-neuronal cells are recruited into the ommatidium. The photoreceptor clusters are marked in pink and sit below the quartet of cone cells in each panel. See figure 2 for a description of each cell type. Anterior is to the right in each illustration.
An early study had suggested that all cells within an ommatidium were descended from a single “mother” cell and were thus related by lineage (Bernard, 1937). This conclusion was called into question when subsequent analyses of rdgB and white mosaic clones revealed that a single clone could encompass any combination of photoreceptor, cone and pigment cells. Two-celled clones were the most informative: these clones could include, for example, either two photoreceptors or a photoreceptor and a pigment cell. Similarly, a two-cell clone could encompass cells from two different ommatidia and even include cells from ommatidia that reside on either side of the dorso-ventral compartment boundary. These results led to the conclusion that cell lineage actually plays no role in ommatidial assembly (Benzer, 1971, 1973; Hanson et al., 1972; Hofbauer and Campos-Ortega, 1976; Ready et al., 1976; Campos-Ortega and Hofbauer, 1977; Lawrence and Green, 1979). The observation that clones could encompass cells from ommatidia on both sides of the equator might lead one to conclude that there is no lineage boundary in the eye. However, it has been shown that the expression of the Iroquois genes in the dorsal half of the eye field does in fact create a lineage restriction boundary and that mosaic clones usually do not cross the midline (Dominguez and de Celis, 1998; Cavodeassi et al., 1999). It should be noted that, unlike the wing in which lineage restriction is absolute, clones can cross the dorsal-ventral boundary (Dominguez and de Celis, 1998).
Laying the first brick: R8 Selection
The studies that established the nomenclature governing the eye of the Diptera preceded by nearly 70 years those that described the sequences of cell fate choices that construct the ommatidium (Dietrich, 1909. Ready et al., 1976). As a result, the photoreceptor that was numbered last (R8) is actually the first photoreceptor within the ommatidium to be born (Fig. 3F). A key issue centered around the question of how are cells within the furrow selected to become the founding members (R8 cell) of each ommatidium. The answer lies in the expression pattern and activity of atonal (ato), which encodes a basic helix-loop-helix transcription factor. The loss of ato leads to a complete elimination of all photoreceptor development (Fig. 10A, B; Jarman et al., 1994). However, it is qualitatively different than mutations that eliminate the activity of retinal determination genes such as eyeless (ey), eyes absent (eya) and sine oculis (so). In these instances, the lack of photoreceptor specification stems from the earlier failure of the morphogenetic furrow to initiate (Bonini et al., 1993; Cheyette et al., 1994; Quiring et al., 1994; Serikaku and O’Tousa, 1994). However, in ato mutants the furrow does in fact initiate but photoreceptor specification does not occur (Jarman et al., 1995).
Figure 10. Schematic of phenotypes associated with loss-of-function mutations in genes that govern ommatidial assembly.
(A–O) Depictions of the mutant phenotypes associated with each gene that has been discussed within this review. Note that although the R8 cell is not shown in most panels it is present in all mutants save ato in panel B. Anterior is to the right in each illustration.
Ato protein is distributed in a classical pro-neural expression pattern. Within the furrow, its initially broad expression pattern is refined first to periodically spaced clusters of 10–15 cells (intermediate groups) then to a cluster of just 2–3 cells (R8 equivalence group) and eventually to a single cell (the presumptive R8) within each pre-cluster. Ato protein is found within the single R8 cell for approximately three ommatidial rows before its expression is extinguished (Fig. 5A; Jarman, 1994; 1995; Dokucu et al., 1996). It should be noted here that the intermediate group corresponds to the rosette while the equivalence group corresponds to three of the five-cells that comprise the precluster. The ato expression pattern can, in and of itself, completely explain how R8 cells are selected. Subsequent studies were focused on explaining how the ato pattern was refined to a single cell per pre-cluster and how the R8s (and by extension the pre-clusters) are spaced apart from each other with such regularity. At least two different mechanisms were initially thought to account for the refinement of the ato expression pattern in the eye disc. This was based in part on the identification of two separate enhancers, one controlling the initial expression stripe ahead of the furrow and the other having command over expression within the intermediate group, the equivalence group and the single R8 cell (Sun et al., 1998).
Figure 5. Proneural selection of the R8 cell requires refinement of the atonal expression pattern.
(A) An illustration documenting the steps in the refinement of the ato expression pattern. Note that Notch is required for the transition from the intermediate group to the R8 equivalence group and that ro refines ato expression with the equivalence group. (B) Description of the genotype of the intermediate group, equivalence group, R8 and R2/5 pair photoreceptor pair. (C) Regulatory circuit between the ro, sens and ato genes within the equivalence group. Anterior is to the right in each illustration.
A study that was published several years prior to the discovery of ato had demonstrated that if Notch signaling in the developing retina is blocked then all cells within the furrow adopt a neuronal fate (Cagan and Ready, 1989b). A re-examination of these experiments (this time using an antibody directed against Ato protein) concluded that the refinement of ato expression from its initial broad stripe down to the 10–15 cell intermediate group (rosette) occurs independently of Notch signaling. However, Notch is important for repressing ato transcription during the transition from the rosette through the arc to the pre-cluster as inhibiting Notch activity results in the failure of the intermediate group to be resolved into the equivalence group (Fig. 5A). Conversely, ubiquitous activation of the Notch pathway eliminates ato expression just within the intermediate group. As ato expression within the equivalence group and the single R8 cells is unaffected by either loss or hyper-activation of the Notch pathway it appears that the latter refinement step also occurs independently of Notch signaling (Baker et al., 1996).
How is a single cell chosen from the equivalence group to maintain ato expression and eventually become the R8 photoreceptor? The answer was thought to initial lie with the activity of the rough (ro) gene. It encodes a homeobox containing DNA binding protein that is first distributed broadly within the morphogenetic furrow but then restricted to four cells (R2/5 and R3/4) of the pre-cluster (Fig. 6B; Kimmel et al., 1990). A comparison of the transcriptional patterns of these two genes indicates that they are never co-expressed within the furrow or pre-cluster (Fig. 5A, B; Dokucu et al., 1996). Ro was suspected of playing a role in repressing ato within the equivalence group after 1–2 additional R8 photoreceptors were seen in ro loss-of-function mutants (Tomlinson et al., 1988; Heberlein et al., 1991). The extra R8s were thought to result from the continued presence of Ato protein in all cells of the equivalence group well after ato transcription would normally have been refined down to a single cell (Dokucu et al., 1996). Furthermore, elevating levels of Ro protein results in a severe down-regulation of ato levels and a subsequent block in ommatidial assembly (Dokucu et al., 1996; Chanut et al., 2000). These results have led to a model in which ro expression is activated in two of the three cells that comprise the equivalence group and as a consequence, the surviving ato positive cell adopts an R8 fate while the other two cells are specified as the R2/5 pair (Dokucu et al., 1996).
Figure 6. The EGF Receptor and a combinatorial code of transcription factors govern cell fate decisions.
(A) A model of how the Egfr pathway regulates cell fate specification within the ommatidium. The original source of the ligand is the R8. It is secreted and received by neighboring cells. As each cell is recruited into the ommatidium it in turn will generate a new Spi signal and the process repeats itself. At the same time an inhibitory ligand, Argos, is also generated. In situations where Spi and Argos are present at high levels the pathway promotes ommatidial assembly. In cells where Argos found at higher levels than Spi then the pathway blocks differentiation. The downstream effectors of the Egfr pathway (Pnt and Yan) are also differentially expressed and these aid in the promotion and inhibition of differentiation respectively. (B) Schematics depicting the expression patterns of several genes that govern the fate of the outer photoreceptors. Only the photoreceptors and cone cells are depicted. Anterior is to the right in each illustration.
However, it was later show that the difference in the number of ommatidia containing extra ato positive cells in wild type and ro null mutant discs is not statistically significant. Furthermore, the presence of extra ato positive cells in both genotypes only persists to the first ommatidial column. By the second column, ato expression is resolved to a single R8 cell in all ommatidia. These data suggest that ro actually is not required for the initial refinement of ato expression (Pepple et al., 2008). So what role if any does Ro play in the specification of the R8 neuron? It directly binds to and inhibits the expression of senseless (sens), which encodes a zinc finger transcription factor that is important for the activation of pro-neural genes such as ato (Nolo et al., 2000; Pepple et al., 2008). sens transcription is activated within all cells of the equivalence group, is resolved along with ato to the single presumptive R8 and is never coincident with ro expression (Fig. 5A, B). It is likely that the loss of Ato protein in discs in which ro was over-expressed is an indirect consequence of down-regulating sens expression.
How is ro expression kept in abeyance within the presumptive R8? The answer lies in a inhibitory loop between Ro and Sens (Fig. 5C). In the absence of sens, ro expression is activated within the presumptive R8 and it is forced into adopting an R2/5 fate. Conversely, over-expression of sens within the outer photoreceptors leads to an inhibition of ro expression and the adoption of the R8 fate (Frankfort et al., 2001). These results are suggestive of a role for Sens in repressing ro expression within the R8 cell. Within the presumptive R2/5 photoreceptor pair the role of Ro is to in turn suppress sens expression. This mutual antagonism ensures that the three cells of the equivalence group are specified as the R8/2/5 photoreceptor neurons (Fig. 5C; Pepple et al., 2008).
Life After the R8: EGF Receptor Signaling and the Rough, Seven-Up, Spalt and Bar Genes
Since the cells of the ommatidium are not related by lineage it was proposed that the ommatidium was built by a series of inductive events. Based on the developmental history of the ommatidium and the physical cell-cell contacts that are made between the different cell types, the R8 was predicted to recruit R2/5 which in turn would recruit R3/4 and so on (Tomlinson and Ready, 1987a). An inductive signal would, by necessity, involve cell-cell communication and the use of a ligand-receptor signaling complex. A candidate for just such a signal was the EGF Receptor (Egfr) pathway. Egfr itself is expressed broadly within the developing eye disc and loss-of-function null clones were devoid of any photoreceptor neurons (Zak and Shilo, 1992; Xu and Rubin, 1993). It should be noted; however, that Egfr is required for cell proliferation in the eye disc and that null clones are extremely small thereby complicating any analysis of the role that Egfr may or may not play in ommatidial assembly. But by using genetic manipulations that allow for the generation of large Egfr null clones and two different types of conditional knockouts it was conclusively determined that R8 development and ato transcription are regulated independently of Egfr signaling (Freeman, 1996; Dominguez et al., 1998; Kumar et al., 1998; Lesokhin et al., 1999; Yang and Baker, 2001; Frankfort and Mardon, 2004). However, reducing the levels of Egfr or its main activating ligand Spitz (Spi) adversely affected the specification of all other photoreceptor, cone and pigment cells types (Fig. 10C–D; Freeman, 1994; Tio et al., 1994; Tio and Moses, 1997; Freeman, 1996; Kumar et al., 1998). This led to a model in which secretion of Spi by the R8 would activate Egfr signaling in the neighboring R2/5 pair. This process would repeat itself stepwise until all cells within the ommatidium were specified (Fig. 6A; Freeman, 1997). One of the main players through which Egfr activity is channeled is the Ets-related transcription factor Pointed (Pnt). It is expressed in nearly all cells within and posterior to the furrow and its removal leads to a block in photoreceptor recruitment (O’Neill et al., 1994). Interestingly, Egfr signaling is simultaneously used to limit the number of cells that can join the growing cluster. Undifferentiated cells that are interspersed between ommatidia express a second Ets-related transcription factor called Yan. yan loss-of-function mutants are characterized by the presence of supernumerary photoreceptors within each cluster (Lai and Rubin, 1992; Rebay and Rubin, 1995). In developing photoreceptors, Egfr signaling leads to the phosphorylation of Yan by MAPK and this leads to its export from the nucleus and subsequent degradation. Therefore the simultaneous inhibition of Yan and the activation of Pnt by the EGF Receptor pathway lead to photoreceptor differentiation (Fig. 6A).
It is not clear if the EGF Receptor plays a permissive or instructive role in cell recruitment. This determination has been problematic since a connection between Pnt and several factors that regulate individual recruitment steps has not been established. To date several transcription factors have been identified that regulate the fate of the R2/5, R3/4 and R1/6 photoreceptor pairs. The first of these to be discovered was the Ro homeobox transcription factor. As we have discussed earlier, ro functions within the equivalence group to narrow ato expression from 2–3 cells to the presumptive R8 photoreceptor (Dokucu et al., 1996). In addition, ro is also expressed in the R3/4 photoreceptor pair (Fig. 6B). ro also appears to specify and recruit the R2/5 photoreceptor pair (Fig. 3F, 5A). Initial studies indicated that in ro mutants the presumptive R2/5 pair expressed general photoreceptor markers and were part of the adult ommatidium (Meyerowitz and Kankel, 1978; Tomlinson et al., 1988). Since no other photoreceptors are recruited it was hypothesized that Ro regulated the production of a secreted signal that was necessary to recruit the R3/4 cell types. However, upon closer inspection it was revealed that in ro mutants these cells ectopically express seven-up (svp), a gene that partly defines the fate of the R3/4 and R1/6 cell types (Fig. 6B). This suggests that under these circumstances the presumptive R2/5 neurons are transformed into one of the other outer photoreceptor cell types (Fig. 10E; Heberlein et al., 1991).
The recruitment of the R3/4 pair completes the pre-cluster (Fig. 3F). One of the factors that control this specification step is the product of the svp gene, a homolog of the human COUP transcription factor. Svp is distributed in the R3/4 and R1/6 photoreceptor cells (Fig. 6B). Loss of svp leads to the transformation of these outer class photoreceptor cells into a class of photoreceptors that were assumed to be R7-like neurons (Fig. 10F). This was based on the small morphology of the rhabdomeres and the ectopic expression of the Rh4 rhodopsin, which is expressed within just the R7 cells (Mlodzik et al., 1990). A more recent analysis that considered the projection profile of axons and used antibodies to R8 specific rhodopsins however indicates that the R3/4 and R1/6 neurons can be transformed into both R7 and R8 type photoreceptors (Miller et al., 2008). This result suggests that the role of svp in eye development is to prevent the R3/4 and R1/6 cells from adopting an inner photoreceptor cell fate fate. Consistent with this hypothesis, ubiquitous expression of svp is capable of transforming the presumptive R7 into an outer photoreceptor. The effect that ectopic Svp protein has on the R8 cell has not been reported so it is unclear if the Svp can transform the R8 into an outer photoreceptor. To a lesser degree the four non-neuronal cone cells can also be forced into adopting an outer photoreceptor cell fate (Hiromi et al., 1993). The ability to repress R7 development in the R3/4 and R1/6 photoreceptor pairs appears to depend on EGF Receptor pathway activity as loss-of-function mutations in the Egfr pathway strongly suppress the svp over-expression phenotypes (Begemann et al., 1995; Kramer et al., 1995). Since these results also suggest that the Egfr pathway is genetically downstream of svp it is possible that svp plays a role in generating the Spi ligand, which as we have seen above is required for each recruitment step. Another gene that appears to lie downstream of svp is pipsqueak (psq), which encodes a BTB-domain containing transcription factor (Horowitz and Berg, 1996). It was first identified as a suppressor of the svp over-expression phenotype. It is transcribed within the R3/4 pair and its expression within this pair is dependent upon svp. Removal of psq results in the specific loss of the R3/4 pair (Fig. 10G; Weber et al., 1995).
In addition to being important for the construction of an ommatidium, the correct specification of the R3/4 pair is also critical for establishing planar polarity across the retina. Mutations within several genes result in ommatidia that often contain either two R3 or two R4 cells (Zheng et al., 1995; Fanto et al., 1998; Fanto and Mlodzik, 1999). These ommatidia do not adopt a normal asymmetric trapezoid arrangement, which means that they have incorrect chirality. These ommatidia also fail to rotate in the proper direction or to the correct degree. It has been determined that the fate of the R3 is determined by frizzled (fz), a component of the Wingless (Wg) pathway, while the fate of the R4 photoreceptor falls under the control of the Notch pathway. Communication between these cells is critical for establishing the two different cell fates. Fz signaling in the R3 activates the expression of Delta (Dl), which in turn activates the Notch receptor in the adjacent R4 (Fanto and Mlodzik, 1999). The activation of Dl within the R3 also requires the spalt (sal) locus, which encodes a pair of zinc finger transcription factors that are distributed in both the R3 and R4 and the cone cells. Loss of sal produces ommatidia that contain either two R3s or two R4s, a phenotype that is nearly identical to that seen in fz mutants (Fig. 6B, 10H, I; Barrio et al., 1999; Mollereau et al., 2000; Domingos et al., 2004)
The last three photoreceptors (R1/6/7) are born within the second mitotic wave and are distinct from those that comprise the precluster (Ready et al., 1976; Wolff and Ready, 1991). The fate of the R1/6 pair of photoreceptors is dependent upon the product of the Bar locus, which encodes a pair of homeobox transcription factors (Kojima et al., 1991; Higashijima et al., 1992). Originally Bar was of interest because it was located on the X-chromosome and mutations resulted in a particularly severe rough eye (Tice, 1914). The rough eye phenotype is the result of hyper-expression of both Bar proteins (Kojima et al., 1991). The BarH1/2 proteins are found exclusively within the R1/6 photoreceptor pair and the primary pigment cells (Fig. 6B; Higashijima et al., 1992). When ectopically expressed within developing cone cells, the anterior/posterior pair is converted into R1/6-like outer photoreceptors while the polar/equatorial pair is transformed into primary pigment cells (Fig. 10J; Hayashi et al., 1998). These results suggest that Bar is required for the specification of both the R1/6 photoreceptor and the primary pigment cells.
And the last shall be first: The R7 and the Sevenless Pathway
The origins of today’s interest in retinal cell fate specification can be traced back to the late 1960s when a mutant called sevenless (sev) was discovered in Seymour Benzer’s laboratory during the search for phototaxis mutants (Benzer, 1967). sev mutants are completely normal save the loss of a single cell, the R7, which is transformed into an equatorial cone cell (Fig. 7A, B, 10K; Harris et al., 1976; Tomlinson and Ready, 1986; 1987). During normal development the R7 cell is the last photoreceptor to be specified (Fig. 3F; Ready et al., 1976; Tomlinson, 1985; Tomlinson and Ready, 1987). The extreme specificity of the sev mutant phenotype was the main attraction for those interested in using the fly eye as a model for studying cell fate decisions and the sev gene provided a window into understanding how cells adopt different fates. A tremendous amount of excitement surrounded the cloning of the sev gene as it turned out to encode a receptor tyrosine kinase (Fig. 8A; Banerjee et al., 1987a; Hafen et al., 1987; Bowtell et al., 1988; Basler and Hafen, 1988; Simon et al., 1989). This immediately suggested that other cells of the ommatidium were signaling towards the presumptive R7 and that the Sev receptor played a major role in receiving and transmitting that inductive cue. Immediate questions about the expression pattern and timing of activation were raised. Surprisingly, the Sev receptor is found within multiple cell types and not just the R7. It is also expressed within the R3/R4 pair, the four cone cells and the two mystery cells (Fig. 8B; Banerjee et al., 1987b; Tomlinson et al., 1987b; Basler et al., 1989; Bowtell et al., 1989a). Despite its broad transcriptional profile, sev is only required 9–10 rows behind the morphogenetic furrow (Basler and Hafen, 1989a, b; Bowtell et al., 1989b). At this point in the developmental history of the ommatidium all other photoreceptor cell fates have already been specified. How were these cells prevented from acquiring an R7 fate? A partial answer lies in fact that one of the functions of the Ro and Svp proteins is to repress the Sevenless pathway in the R3/4 cells (Basler et al., 1990; Kimmel et al., 1990; Mlodzik et al., 1990). As these proteins are also distributed within the R2/5 and R1/6 pairs (Fig. 6B) respectively they prevent these cells from being transformed into R7s under experimental circumstances in which either an activated Sev receptor or its ligand is ubiquitously expressed (van Vactor et al., 1991; Dickson et al., 1992a). As you will see below, the ligand for the Sev receptor is expressed solely within the R8 and is membrane bound. As the cone cells never contact the R8 directly they are never provided with access to the ligand. Therefore even though sev is expressed in multiple cell types it can only be activated within the presumptive R7.
Figure 7. The R7 fails to form in sevenless mutant ommatidia.
(A) Light microscope section of an adult wild type retina. Note the presence of seven photoreceptors in each ommatidium. The R8 cell sits below the R7. (B) Light microscope section of an adult sevenless retina. Note that in nearly all ommatidia only six photoreceptors are present. The R7 has been converted into an equatorial cone cell. The R8 is not seen within this plane of field. Note that a single ommatidium has an intact R7 cell (upper left hand corner). Anterior is to the right in both images
Figure 8. Specification of the R7 neuron by the Sevenless signal transduction pathway.
(A) An illustration depicting the core of the Sevenless pathway. The small dark blue spheres represent phosphorylation events. (B) Schematics depicting the expression patterns of several genes that control the fate of the R7 photoreceptor. Only the photoreceptors and cone cells are depicted. Anterior is to the right panel B.
Mutations in the gene encoding the ligand were predicted to mimic the sev loss-of-function phenotype and therefore mutants could be isolated based on the missing R7 cell. A screen looking for new “sevenless” flies successfully identified a gene called bride of sevenless (boss). It is expressed specifically within the R8 cell and encodes a large multi-pass trans-membrane bound protein (Fig. 8B; Reinke and Zipursky, 1988; Hart et al., 1990). Loss of boss also eliminates the R7 cell (Fig. 10L). The status of Boss as the Sev receptor was confirmed when Boss and Sev were first shown to physically interact at the cell surface and were then internalized into the presumptive R7 as it is being specified (Kramer et al., 1991; Cagan et al., 1992). Thus a local tethered signal from the already specified R8 is received and interpreted by the presumptive R7 as an instruction to adopt the R7 fate.
At the time of these discoveries many laboratories were also racing to identify genes that lay genetically and biochemically downstream of the Sev receptor. In order to find downstream factors several novel approaches were employed including searches for (1) gain-of-function mutations that could restore the R7 cell to the sev loss-of-function mutant; (2) loss-of-function mutations that could enhance the frequency of R7 loss caused by a temperature sensitive Sev receptor; and (3) loss-of-function mutations that could block the transformation of cone cells into R7s that results from ubiquitous expression of an activated Sev receptor (Basler et al., 1991; Mullins and Rubin, 1991; Rogge et al., 1991). Within just a few years these screens identified nearly every component of what is today known as the Ras/ERK pathway (Fig. 8A). These include the downstream of receptor kinase (drk) which links receptor tyrosine kinases to downstream effectors, Ras1 itself, two factors (Gap1 and GNEF) that balance the GDP/GTP bound states of Ras and the three cytoplasmic kinases (Raf1, MEK and MAPK) that constitute the MAPK/ERK pathway (Rogge et al., 1991; Simon et al., 1991; Bonfini et al., 1992; Dickson et al., 1992b; Fortini et al., 1992; Gaul et al., 1992; Oliver et al., 1993; Biggs et al., 1994; Brunner et al., 1994; Hsu and Perrimon, 1994). Further biochemical and genetic studies have identified many additional cytoplasmic components of the Sevenless pathway. Their identification has provided us with a more complete understanding of this signaling system (reviewed in Raabe, 2000). As more and more effort was put into identifying new links in the chain connecting the Sev receptor to transcription factors in the nucleus, questions were raised as to the potential universality of the Ras/ERK cassette. Satisfyingly, this cassette was found to be common to all receptor kinase pathways in flies and homologous components were present in vertebrate systems (Diaz-Benjumea and Hafen, 1994; reviewed in Hafen et al., 1993).
But how is a specific outcome, the specification of the R7, achieved by using a common signaling cassette? Although the answer to that question still eludes us today, a picture of what happens in the presumptive R7 nucleus has started to emerge. One of the earliest transcriptional targets of the Sevenless pathway is the phyllopod (phyl) gene. It encodes a novel protein that is expressed and required in R1/6 and R7 (Fig. 8B). Loss of phyl results in the transformation of these three photoreceptors into cone cells (Fig. 10M; Chang et al., 1995; Dickson et al., 1995). The ability of Phyl to promote neuronal specification depends upon its ability to form a complex with Seven in absentia (Sina), an E3 ligase that is also important for promoting R7 development (Fig. 10N; Carthew and Rubin, 1990). Together the Phyl-Sina complex targets substrate proteins for ubiquitinylation and degradation. One of its targets is an isoform of the transcriptional repressor Tramtrack (Fig. 8A; Ttk88: Tang et al., 1997; Li et al., 1997; Li et al., 2002). Ttk88 protein is normally absent from differentiating neurons of the ommatidium and loss of function mutants result in the conversion of cone cells into R7s (Fig. 10O; Xiong et al., 1993; Lai et al., 1996). Removal of either phyl or sina results in ectopic expression of ttk88 in the R1/6 and R7 precursors. Ultimately, these cells are forced to adopt a cone cell fate (Xiong et al., 1993; Lai et al., 1996; Li et al., 1997). So it appears that the Boss signal from the R8 binds and activates the Sev receptor in the presumptive R7. The effects of triggering the Sevenless pathway are seen in the nucleus when the Phyl-Sina complex forms in response and targets the Ttk88 transcriptional repressor for degradation (Fig. 8A). Thus the default cell fate choice, the non-neuronal cone cell, is averted and the R7 fate is selected.
A Second Signal: The Notch Pathway Promotes the R7 Fate
Due to the unique contacts that the R7 makes with both the R8 and the R1/6 pair, it was postulated that the R7 fate requires two distinct signals: one from the R8 and from the R1/R6 cells. As we have seen above, signaling from the R8 is mediated by Boss-Sev interactions. Hints to the nature of the second signal came from experiments in which the ommatidia lacking the Notch pathway lacked an R7 and gained an extra outer photoreceptor (Cagan and Ready, 1989; Cooper and Bray, 2000). Consistent with this result, activation of the Notch pathway resulted in ommatidia with fewer outer photoreceptors and extra R7-like cells (Fortini et al., 1993; Cooper and Bray, 2000). One the of the Enhancer of split genes was shown to be expressed within the R7 providing evidence for active Notch signaling in this cell (Cooper and Bray, 2000). Subsequent experiments analyzing Delta (Dl) mutant and Noch over-expression mosaic ommatidia determined that the R1/6 pair produce the Dl signal, which is then received by Notch within the presumptive R7 (Tomlinson and Struhl, 2001). An obvious question is why does the R7 need two signals to adopt its fate? A recent set of findings indicates that Notch signaling is used for at least three different steps within the R7. The first is to block photoreceptor development from occurring. In the second step, the Notch pathway will activate the expression of sev thus allowing for the receptor of the Boss signal. And finally, Notch functions to prevent the R7 from adopting an R1/6 fate (Tomlinson et al., 2011). Thus it appears that the two pathways cooperate to ensure that the R7 equivalence group is properly specified.
Finishing the Ommatidium: Cone Cell Recruitment
Each ommatidium contains a quartet of non-neuronal lens secreting cone cells (Waddington and Perry, 1960). The cone cells, along with the R1/6 and R7 photoreceptors, are born within the second mitotic wave and collectively these seven cells are referred to as the “R7 equivalence” group (Ready et al., 1976; Tomlinson, 1985; Tomlinson and Ready, 1987; Wolff and Ready, 1991). This term is appropriate as mutations affecting R1/6 and R7 such as sev, sina and phyl transform these photoreceptors into cone cells (Tomlinson and Ready, 1986, 1987; Carthew and Rubin, 1990; Chang et al., 1995; Dickson et al., 1995). Conversely, activation of the Sevenless pathway within the cone cells results in their transformation into R7-like photoreceptors (Basler et al., 1991; Fortini et al., 1992). Once all photoreceptors have been specified the remaining undifferentiated cells read the identities of adjacent differentiated cells and adopt the next most appropriate fate. As a consequence ommatidia that contain extra photoreceptors, like those in Notch and argos mutants, also have supernumerary cone and pigment cells (Cagan and Ready, 1989a; Freeman et al., 1992).
Although many mutants are characterized by the transformation of photoreceptors into cone cells, none of the genes that have been discussed so far are required per se for cone cell formation. Insight into the development of this cell type was finally provided by the characterization of DPax2, which prior to its cloning had been referred to by some as sparkling (based on its glossy eye) and as shaven by others (based on the lack of thoracic bristles). The disparate names and phenotypes are due to mutations in two enhancers of DPax2 (Fu et al., 1998). DPax2 is expressed within and is the main regulator of cone cell development (Fu and Noll, 1997; Shi and Noll, 2009). The sparkling enhancer, which appears to control all cone cell expression of DPax2, is bound by transcription factors that are terminal members of the EGF Receptor and Notch signaling pathways as well as by the product of the lozenge (lz) locus (Fig 8A). All three inputs are required for DPax2 expression within cone cell precursors (Flores et al., 2000). The loss of a single input will result in the failure of cone cell formation (Batterham et al., 1996; Daga et al., 1996; Flores et al., 1998; 2000). A recent detailed analysis of the sparkling enhancer suggests that in addition to the three known inputs several additional transcription factors (yet to be identified) are required for proper DPax2 expression within the cone cells (Fig. 9A; Swanson et al., 2010).
Figure 9. Molecular mechanisms of cone and primary pigment cell development.
(A) A depiction of the regulatory inputs into the sparkling enhancer of the DPax2 gene. Several transcription factors such as Lz, Su(H), Pnt and Yan are known to bind and are required for proper expression of DPax2 in the cone cells. A recent molecular analysis of the sparkling enhancer has identified additional sequences that are important for the proper functioning of the enhancer (Swanson et al., 2010). The identity of the predicted DNA binding proteins (X, Y, Z) that interact with these sites are still unknown. (B) The source of the ligands for both EGF and Notch receptors are the developing photoreceptors. (C) The Notch pathway controls the fate of the primary pigment cells. The source of the activating ligand is the lens secreting cone cells. Anterior is to the right in panels B and C.
How the cone cells are specifically recruited from the R7 equivalence group is not a completely well understood system although significant progress has been made. For instance, the ligands for the EGF Receptor and Notch receptors are generated from within the eight-cell photoreceptor cluster (Fig. 9B; Freeman, 1996; Pickup et al., 2009). Likewise, several transcription factors such as Sine oculis (So), Glass (Gl) and Hairless (H) have been shown to bind to enhancers within lz and direct its expression in the eye (Fig. 8A; Yan et al., 2003; Potzer et al., 2008). There is likely to be competition amongst the seven cells of the equivalence group for the various fates. Whether you consider the winning fate to be that of the photoreceptor or cone cell is likely to be a matter of personal choice.
Turning the Ommatidium Into a Hexagon: The Pigment Cells
As we have discussed above each ommatidium contains three types of pigment cells. Two primary pigment cells surround the four cone cells and contribute to the formation of the lens and to the red eye color of the adult eye. Any cell that fails to adopt a photoreceptor, cone or primary pigment cell fate will go on to become either a secondary or tertiary pigment cell. Six secondary pigment cells surround each ommatidium while the tertiary pigment cells and bristle complexes will occupy positions at each vertex (Fig. 2A: Waddington and Perry, 1960). Although pigment cells are born within the second mitotic wave their fates are not specified until the mid-pupal stage of development (Ready et al., 1976; Cagan and Ready, 1989). It appears that the secondary/tertiary pigment cell fate is the default fate in the eye. This conclusion is based on the observation if cell death is blocked in the pupal retina all of the excess cells that are produced during larval development become secondary or tertiary pigment cells (Cagan and Ready, 1989a; Wolff and Ready, 1991b; Larson et al., 1998).
The molecular determinants of pigment cell fates, particularly that of the secondary and tertiary pigment cells, have not been as well defined as those for the other cell types. However, some progress has been made at least towards understanding how primary pigment cells are specified. These cells appear to be directly recruited by the previously specified cone cells. The EGF Receptor pathway stimulates production of the Dl ligand within the four cone cells, which in turn is used to non-autonomously activate the Notch pathway in the two cells that will ultimately adopt a primary pigment cell fate (Fig. 9C; Nagaraj and Banerjee, 2007). Loss of Notch activity reduces the number of primary pigment cells (Cagan and Ready, 1989b). The transcriptional targets of the Notch pathway within the primary pigment cells are yet to be identified. Several genes such as DPax2 and Bar, which control the fates of other cells types, are also expressed and required within the primary pigment cells and could be potential targets (Higashijima et al., 1992; Fu and Noll, 1997). The identity of the signaling pathways and genes that determine the fates of the secondary and tertiary pigment cells are even more elusive. Several laboratories have demonstrated that the EGF Receptor and Notch pathways as well as many chromatin remodeling proteins play roles in discriminating between cells that live and join the ommatidium and those that are eliminated from the pupal retina by apoptosis (Larson et al., 1998; Tenanbaum et al., 2000; Brachmann and Cagan, 2003; Dos-Santos et al., 2008). Further experimental investigations are needed before we will have a complete picture of how the fates of these cells are specified.
Summary and Perspectives
It has been 35 years since Don Ready, Thomas Hanson and Seymour Benzer published their seminal paper on the development of the Drosophila eye (Ready et al., 1976). In this paper they described the simple structure and stereotyped developmental history of the ommatidium. Ten years later Andrew Tomlinson and Don Ready reported that in sevenless mutants the R7 cell is transformed into a non-neuronal equatorial cone cell (Tomlinson and Ready, 1986). This marked the beginning of what has been a remarkable quest to understand the process by which cell fate decisions are made. Studies on ommatidial assembly have provided us with significant insight into the role that signaling pathways play in the communication between cells. Similarly, through the ommatidium we now have a better understanding of how complex tissues containing multiple unique cell types use a combinatorial code of transcription factors to execute the instructions that are sent back and forth between neighboring cells. The impact of the genes discussed in this review is not limited to the fly eye, as recent studies have shown that vertebrate genomes including those of mice and men contain homologs to nearly all of the genes that are known to function within the Drosophila ommatidium. Furthermore, similar to the situation that exists in flies, these genes are also expressed and function outside of the retina. Indeed, mutations within many homologs are associated with numerous diseases and developmental syndromes. It is a near certainty that the humble ommatidium will continue to have an impact on our understanding of how cell fate choices are made in both the fly retina and other developmental systems.
Acknowledgments
I would like to thank Abby Anderson, Richard Hardy, Soni Lacefield, Scott Michaels, Carrie Spratford, Bonnie Weasner and Brandon Weasner for comments on this manuscript. I also would like to thank all those who have contributed to our understanding of the mechanisms by which the ommatidium is constructed. Without their efforts there would not have been a need for this summary. And I apologize to those whose work has not been cited or discussed here. Justin P. Kumar is supported by a grant from the National Eye Institute (2R01 EY014863).
References
- Baker NE, Yu S, Han D. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr Biol. 1996;6:1290–1301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- Banerjee U, Renfranz PJ, Hinton DR, Rabin BA, Benzer S. The sevenless+ protein is expressed apically in cell membranes of developing Drosophila retina; it is not restricted to cell R7. Cell. 1987a;51:151–158. doi: 10.1016/0092-8674(87)90020-1. [DOI] [PubMed] [Google Scholar]
- Banerjee U, Renfranz PJ, Pollock JA, Benzer S. Molecular characterization and expression of sevenless, a gene involved in neuronal pattern formation in the Drosophila eye. Cell. 1987b;49:281–291. doi: 10.1016/0092-8674(87)90569-1. [DOI] [PubMed] [Google Scholar]
- Barrio R, de Celis JF, Bolshakov S, Kafatos FC. Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev Biol. 1999;215:33–47. doi: 10.1006/dbio.1999.9434. [DOI] [PubMed] [Google Scholar]
- Basler K, Christen B, Hafen E. Ligand-independent activation of the sevenless receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell. 1991;64:1069–1081. doi: 10.1016/0092-8674(91)90262-w. [DOI] [PubMed] [Google Scholar]
- Basler K, Hafen E. Control of photoreceptor cell fate by the sevenless protein requires a functional tyrosine kinase domain. Cell. 1988;54:299–311. doi: 10.1016/0092-8674(88)90193-6. [DOI] [PubMed] [Google Scholar]
- Basler K, Hafen E. Dynamics of Drosophila eye development and temporal requirements of sevenless expression. Development. 1989a;107:723–731. doi: 10.1242/dev.107.4.723. [DOI] [PubMed] [Google Scholar]
- Basler K, Hafen E. Ubiquitous expression of sevenless: position-dependent specification of cell fate. Science. 1989b;243:931–934. doi: 10.1126/science.2493159. [DOI] [PubMed] [Google Scholar]
- Basler K, Siegrist P, Hafen E. The spatial and temporal expression pattern of sevenless is exclusively controlled by gene-internal elements. EMBO J. 1989;8:2381–2386. doi: 10.1002/j.1460-2075.1989.tb08367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler K, Yen D, Tomlinson A, Hafen E. Reprogramming cell fate in the developing Drosophila retina: transformation of R7 cells by ectopic expression of rough. Genes Dev. 1990;4:728–739. doi: 10.1101/gad.4.5.728. [DOI] [PubMed] [Google Scholar]
- Batterham P, Crew JR, Sokac AM, Andrews JR, Pasquini GM, Davies AG, Stocker RF, Pollock JA. Genetic analysis of the lozenge gene complex in Drosophila melanogaster: adult visual system phenotypes. J Neurogenet. 1996;10:193–220. doi: 10.3109/01677069609083463. [DOI] [PubMed] [Google Scholar]
- Begemann G, Michon AM, vd Voorn L, Wepf R, Mlodzik M. The Drosophila orphan nuclear receptor seven-up requires the Ras pathway for its function in photoreceptor determination. Development. 1995;121:225–235. doi: 10.1242/dev.121.1.225. [DOI] [PubMed] [Google Scholar]
- Benzer S. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc Natl Acad Sci U S A. 1967;58:1112–1119. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S. From the gene to behavior. JAMA. 1971;218:1015–1022. [PubMed] [Google Scholar]
- Benzer S. Genetic dissection of behavior. Sci Am. 1973;229:24–37. doi: 10.1038/scientificamerican1273-24. [DOI] [PubMed] [Google Scholar]
- Biggs WH, 3rd, Zavitz KH, Dickson B, van der Straten A, Brunner D, Hafen E, Zipursky SL. The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J. 1994;13:1628–1635. doi: 10.1002/j.1460-2075.1994.tb06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard F. Rescherches sur la morphogenese des yeux composes d’arthropodes. Bull Biol Fr Belg. 1937;23(Suppl) [Google Scholar]
- Bonfini L, Karlovich CA, Dasgupta C, Banerjee U. The Son of sevenless gene product: a putative activator of Ras. Science. 1992;255:603–606. doi: 10.1126/science.1736363. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Bowtell DD, Kimmel BE, Simon MA, Rubin GM. Regulation of the complex pattern of sevenless expression in the developing Drosophila eye. Proc Natl Acad Sci U S A. 1989a;86:6245–6249. doi: 10.1073/pnas.86.16.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowtell DD, Simon MA, Rubin GM. Nucleotide sequence and structure of the sevenless gene of Drosophila melanogaster. Genes Dev. 1988;2:620–634. doi: 10.1101/gad.2.6.620. [DOI] [PubMed] [Google Scholar]
- Bowtell DD, Simon MA, Rubin GM. Ommatidia in the developing Drosophila eye require and can respond to sevenless for only a restricted period. Cell. 1989b;56:931–936. doi: 10.1016/0092-8674(89)90626-0. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Cagan RL. Patterning the fly eye: the role of apoptosis. Trends Genet. 2003;19:91–96. doi: 10.1016/S0168-9525(02)00041-0. [DOI] [PubMed] [Google Scholar]
- Brunner D, Oellers N, Szabad J, Biggs WH, 3rd, Zipursky SL, Hafen E. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Kramer H, Hart AC, Zipursky SL. The bride of sevenless and sevenless interaction: internalization of a transmembrane ligand. Cell. 1992;69:393–399. doi: 10.1016/0092-8674(92)90442-f. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989a;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes & Dev. 1989b;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Rubin GM. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, del Corral RD, Campuzano S, Dominguez M. Compartments and organizing boundaries in the Drosophila eye: the role of the homeodomain Iroguois proteins. Development. 1999;128:4933–4942. doi: 10.1242/dev.126.22.4933. [DOI] [PubMed] [Google Scholar]
- Chang HC, Solomon NM, Wassarman DA, Karim FD, Therrien M, Rubin GM, Wolff T. phyllopod functions in the fate determination of a subset of photoreceptors in Drosophila. Cell. 1995;80:463–472. doi: 10.1016/0092-8674(95)90497-2. [DOI] [PubMed] [Google Scholar]
- Chanut F, Luk A, Heberlein U. A screen for dominant modifiers of ro(Dom), a mutation that disrupts morphogenetic furrow progression in Drosophila, identifies groucho and hairless as regulators of atonal expression. Genetics. 2000;156:1203–1217. doi: 10.1093/genetics/156.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Cooper MTD, Bray SJ. R7 photoreceptor specification requries Notch activity. Current Biology. 2000;10:1507–1510. doi: 10.1016/s0960-9822(00)00826-5. [DOI] [PubMed] [Google Scholar]
- Daga A, Karlovich CA, Dumstrei K, Banerjee U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996;10:1194–1205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Hariharan IK. Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science. 1995;270:983–985. doi: 10.1126/science.270.5238.983. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Hafen E. The sevenless signalling cassette mediates Drosophila EGF receptor function during epidermal development. Development. 1994;120:569–578. doi: 10.1242/dev.120.3.569. [DOI] [PubMed] [Google Scholar]
- Dickson B, Sprenger F, Hafen E. Prepattern in the developing Drosophila eye revealed by an activated torso--sevenless chimeric receptor. Genes Dev. 1992a;6:2327–2339. doi: 10.1101/gad.6.12a.2327. [DOI] [PubMed] [Google Scholar]
- Dickson B, Sprenger F, Morrison D, Hafen E. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature. 1992b;360:600–603. doi: 10.1038/360600a0. [DOI] [PubMed] [Google Scholar]
- Dickson BJ, Dominguez M, van der Straten A, Hafen E. Control of Drosophila photoreceptor cell fates by phyllopod, a novel nuclear protein acting downstream of the Raf kinase. Cell. 1995;80:453–462. doi: 10.1016/0092-8674(95)90496-4. [DOI] [PubMed] [Google Scholar]
- Dietrich W. Die Fazettenaugen der Dipteren. Z Wiss Zool. 1909;92:465–539. [Google Scholar]
- Dokucu ME, Zipursky SL, Cagan RL. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–4147. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Domingos PM, Mlodzik M, Mendes CS, Brown S, Steller H, Mollereau B. Spalt transcription factors are required for R3/R4 specification and establishment of planar cell polarity in the Drosophila eye. Development. 2004;131:5695–5702. doi: 10.1242/dev.01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M, Wasserman JD, Freeman M. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol. 1998;8:1039–1048. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- Dominguez M, De Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–279. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- Dos-Santos N, Rubin T, Chalvet F, Gandille P, Cremazy F, Leroy J, Boissonneau E, Theodore L. Drosophila retinal pigment cell death is regulated in a position-dependent manner by a cell memory gene. Int J Dev Biol. 2008;52:21–31. doi: 10.1387/ijdb.072406nd. [DOI] [PubMed] [Google Scholar]
- Fanto M, Mayes CA, Mlodzik M. Linking cell-fate specification to planar polarity: determination of the R3/R4 photoreceptors is a prerequisite for the interpretation of the Frizzled mediated polarity signal. Mech Dev. 1998;74:51–58. doi: 10.1016/s0925-4773(98)00063-x. [DOI] [PubMed] [Google Scholar]
- Fanto M, Mlodzik M. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature. 1999;397:523–526. doi: 10.1038/17389. [DOI] [PubMed] [Google Scholar]
- Ferris GF. External morphoogy of the adult. In: Demerec M, editor. Biology of Drosophila. J Wiley; New York: 1950. pp. 368–419. [Google Scholar]
- Flores GV, Daga A, Kalhor HR, Banerjee U. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development. 1998;125:3681–3687. doi: 10.1242/dev.125.18.3681. [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Simon MA, Rubin GM. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Mardon G. Senseless represses nuclear transduction of Egfr pathway activation. Development. 2004;131:563–570. doi: 10.1242/dev.00941. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Nolo R, Zhang Z, Bellen H, Mardon G. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron. 2001;32:403–414. doi: 10.1016/s0896-6273(01)00480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech Dev. 1994;48:25–33. doi: 10.1016/0925-4773(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- Freeman M, Klambt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–975. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Fu W, Duan H, Frei E, Noll M. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development. 1998;125:2943–2950. doi: 10.1242/dev.125.15.2943. [DOI] [PubMed] [Google Scholar]
- Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul U, Mardon G, Rubin GM. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- Gehring W. Cell heredity and changes of determination in cultures of imaginal discs in Drosophila melanogaster. J Embryol Exp Morphol. 1966;15:77–111. [PubMed] [Google Scholar]
- Hafen E, Basler K, Edstroem JE, Rubin GM. Sevenless, a cell-specific homeotic gene of Drosophila, encodes a putative transmembrane receptor with a tyrosine kinase domain. Science. 1987;236:55–63. doi: 10.1126/science.2882603. [DOI] [PubMed] [Google Scholar]
- Hafen E, Dickson B, Raabe T, Brunner D, Oellers N, van der Straten A. Genetic analysis of the sevenless signal transduction pathway of Drosophila. Dev Suppl. 1993:41–46. [PubMed] [Google Scholar]
- Hanson TE, Ready DF, Benzer S. Use of mosaics in the analysis of pattern formation in the retina of Drosophila. Caltech Biol Annu Rept. 1972;40 [Google Scholar]
- Harris WA, Stark WS, Walker JA. Genetic dissection of the photoreceptor system in the compound eye of Drosophila melanogaster. J Physiol. 1976;256:415–439. doi: 10.1113/jphysiol.1976.sp011331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Kramer H, Van Vactor DL, Jr, Paidhungat M, Zipursky SL. Induction of cell fate in the Drosophila retina: the bride of sevenless protein is predicted to contain a large extracellular domain and seven transmembrane segments. Genes Dev. 1990;4:1835–1847. doi: 10.1101/gad.4.11.1835. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Kojima T, Saigo K. Specification of primary pigment cell and outer photoreceptor fates by BarH1 homeobox gene in the developing Drosophila eye. Dev Biol. 1998;200:131–145. doi: 10.1006/dbio.1998.8959. [DOI] [PubMed] [Google Scholar]
- Haynie JL, Bryant PJ. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. The Journal of experimental zoology. 1986;237:293–308. doi: 10.1002/jez.1402370302. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Mlodzik M, Rubin GM. Cell-fate determination in the developing Drosophila eye: role of the rough gene. Development. 1991;112:703–712. doi: 10.1242/dev.112.3.703. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Kojima T, Michiue T, Ishimaru S, Emori Y, Saigo K. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 1992;6:50–60. doi: 10.1101/gad.6.1.50. [DOI] [PubMed] [Google Scholar]
- Hiromi Y, Mlodzik M, West SR, Rubin GM, Goodman CS. Ectopic expression of seven-up causes cell fate changes during ommatidial assembly. Development. 1993;118:1123–1135. doi: 10.1242/dev.118.4.1123. [DOI] [PubMed] [Google Scholar]
- Hofbauer A, Campos-Ortega JA. Cell clones and pattern formation: Genetic eye mosaics in Drosophila melanogaster. Wilhelm Roux Arch. 1976;179:275–289. doi: 10.1007/BF00848237. [DOI] [PubMed] [Google Scholar]
- Horowitz H, Berg CA. The Drosophila pipsqueak gene encodes a nuclear BTB-domain-containing protein required early in oogenesis. Development. 1996;122:1859–1871. doi: 10.1242/dev.122.6.1859. [DOI] [PubMed] [Google Scholar]
- Hsu JC, Perrimon N. A temperature-sensitive MEK mutation demonstrates the conservation of the signaling pathways activated by receptor tyrosine kinases. Genes Dev. 1994;8:2176–2187. doi: 10.1101/gad.8.18.2176. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Jenny A. Planar cell polarity signaling in the Drosophila eye. Curr Topics Dev Biol. 2010;93:190–219. doi: 10.1016/B978-0-12-385044-7.00007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel BE, Heberlein U, Rubin GM. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev. 1990;4:712–727. doi: 10.1101/gad.4.5.712. [DOI] [PubMed] [Google Scholar]
- Kojima T, Ishimaru S, Higashijima S, Takayama E, Akimaru H, Sone M, Emori Y, Saigo K. Identification of a different-type homeobox gene, BarH1, possibly causing Bar (B) and Om(1D) mutations in Drosophila. Proc Natl Acad Sci U S A. 1991;88:4343–4347. doi: 10.1073/pnas.88.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer H, Cagan RL, Zipursky SL. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature. 1991;352:207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- Kramer S, West SR, Hiromi Y. Cell fate control in the Drosophila retina by the orphan receptor seven-up: its role in the decisions mediated by the ras signaling pathway. Development. 1995;121:1361–1372. doi: 10.1242/dev.121.5.1361. [DOI] [PubMed] [Google Scholar]
- Kumar JP. Retinal determination: the beginning of eye development. Curr Top Dev Biol. 2010;93:1–28. doi: 10.1016/B978-0-12-385044-7.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. My what big eyes you have: How the Drosophila retina grows. Dev Neurobiol. 2011 doi: 10.1002/dneu.20921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo BZ, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Harrison SD, Karim F, Li Y, Rubin GM. Loss of tramtrack gene activity results in ectopic R7 cell formation, even in a sina mutant background. Proc Natl Acad Sci U S A. 1996;93:5025–5030. doi: 10.1073/pnas.93.10.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Rubin GM. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell. 1992;70:609–620. doi: 10.1016/0092-8674(92)90430-k. [DOI] [PubMed] [Google Scholar]
- Larson DE, Johnson RI, Swat M, Cordero JB, Glazier JA, Cagan RL. Computer simulation of cellular patterning within the Drosophila pupal eye. PLoS Comput Biol. 2010;6:e1000841. doi: 10.1371/journal.pcbi.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Green SM. Cell lineage in the developing retina of Drosophila. Dev Biol. 1979;71:142–152. doi: 10.1016/0012-1606(79)90088-5. [DOI] [PubMed] [Google Scholar]
- Lesokhin AM, Yu SY, Katz J, Baker NE. Several levels of EGF receptor signaling during photoreceptor specification in wild-type, Ellipse, and null mutant Drosophila. Dev Biol. 1999;205:129–144. doi: 10.1006/dbio.1998.9121. [DOI] [PubMed] [Google Scholar]
- Li S, Li Y, Carthew RW, Lai ZC. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- Li S, Xu C, Carthew RW. Phyllopod acts as an adaptor protein to link the sina ubiquitin ligase to the substrate protein tramtrack. Mol Cell Biol. 2002;22:6854–6865. doi: 10.1128/MCB.22.19.6854-6865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhaven MM, Schneiderman HA. Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development of Drosophila melanogaster. Wilhelm Roux Archive. 1977;183:269–305. doi: 10.1007/BF00848459. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM, Kankel DR. A genetic analysis of visual system development in Drosophilia melanogaster. Dev Biol. 1978;62:112–142. doi: 10.1016/0012-1606(78)90096-9. [DOI] [PubMed] [Google Scholar]
- Miller DT, Cagan RL. Local induction of patterning and programmed cell death in the developing drosophila retina. Development. 1998;125:2327–2335. doi: 10.1242/dev.125.12.2327. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y, Weber U, Goodman CS, Rubin GM. The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell. 1990;60:211–224. doi: 10.1016/0092-8674(90)90737-y. [DOI] [PubMed] [Google Scholar]
- Mollereau B, Wernet MF, Beaufils P, Killian D, Pichaud F, Kuhnlein R, Desplan C. A green fluorescent protein enhancer trap screen in Drosophila photoreceptor cells. Mech Dev. 2000;93:151–160. doi: 10.1016/s0925-4773(00)00287-2. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Sex limited inheritance in Drosophila. Science. 1910;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- Mullins MC, Rubin GM. Isolation of temperature-sensitive mutations of the tyrosine kinase receptor sevenless (sev) in Drosophila and their use in determining its time of action. Proc Natl Acad Sci U S A. 1991;88:9387–9391. doi: 10.1073/pnas.88.21.9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 2007;134:825–831. doi: 10.1242/dev.02788. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Olivier JP, Raabe T, Henkemeyer M, Dickson B, Mbamalu G, Margolis B, Schlessinger J, Hafen E, Pawson T. A Drosophila SH2–SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell. 1993;73:179–191. doi: 10.1016/0092-8674(93)90170-u. [DOI] [PubMed] [Google Scholar]
- Ouweneel WJ. Normal and abnormal determination in the imaginal discs of Drosophila, with special reference to the eye discs. Acta Embryol Exp (Palermo) 1970;1:95–119. [PubMed] [Google Scholar]
- Pepple KL, Atkins M, Venken K, Wellnitz K, Harding M, Frankfort B, Mardon G. Two-step selection of a single R8 photoreceptor: a bistable loop between senseless and rough locks in R8 fate. Development. 2008;135:4071–4079. doi: 10.1242/dev.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup AT, Ming L, Lipshitz HD. Hindsight modulates Delta expression during Drosophila cone cell induction. Development. 2009;136:975–982. doi: 10.1242/dev.027318. [DOI] [PubMed] [Google Scholar]
- Poulson DF. Histogensis, organogenesis and differentiation in the embryo of Drosophila melanogaster. In: Demerec M, editor. Biology of Drosophila. J Wiley; New York: 1950. pp. 368–419. [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans [see comments] Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Raabe T. The sevenless signaling pathway: variations of a common theme. Biochim Biophys Acta. 2000;1496:151–163. doi: 10.1016/s0167-4889(00)00020-3. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Reinke R, Zipursky SL. Cell-cell interaction in the Drosophila retina: the bride of sevenless gene is required in photoreceptor cell R8 for R7 cell development. Cell. 1988;55:321–330. doi: 10.1016/0092-8674(88)90055-4. [DOI] [PubMed] [Google Scholar]
- Rogge RD, Karlovich CA, Banerjee U. Genetic dissection of a neurodevelopmental pathway: Son of sevenless functions downstream of the sevenless and EGF receptor tyrosine kinases. Cell. 1991;64:39–48. doi: 10.1016/0092-8674(91)90207-f. [DOI] [PubMed] [Google Scholar]
- Serikaku MA, O’Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Noll M. Determination of cell fates in the R7 equivalence group of the Drosophila eye by the concerted regulation of D-Pax2 and TTK88. Dev Biol. 2009;331:68–77. doi: 10.1016/j.ydbio.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- Simon MA, Bowtell DD, Rubin GM. Structure and activity of the sevenless protein: a protein tyrosine kinase receptor required for photoreceptor development in Drosophila. Proc Natl Acad Sci U S A. 1989;86:8333–8337. doi: 10.1073/pnas.86.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Lim J, KWC Dorsoventral boundary for organizing growth and planar polarity in the Drosophila eye. Adv in Dev Biol. 2005;14:59–90. [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Swanson CI, Evans NC, Barolo S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev Cell. 2010;18:359–370. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum SB, Gorski SM, Rusconi JC, Cagan RL. A screen for dominant modifiers of the irreC-rst cell death phenotype in the developing Drosophila retina. Genetics. 2000;156:205–217. doi: 10.1093/genetics/156.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- Tice SC. A new sex-linked character in Drosophila. Biol Bull, Wood’s Hole. 1914;26:221–230. [Google Scholar]
- Tio M, Ma C, Moses K. spitz, a Drosophila homolog of transforming growth factor-alpha, is required in the founding photoreceptor cells of the compound eye facets. Mech Dev. 1994;48:13–23. doi: 10.1016/0925-4773(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Tio M, Moses K. The Drosophila TGF alpha homolog Spitz acts in photoreceptor recruitment in the developing retina. Development. 1997;124:343–351. doi: 10.1242/dev.124.2.343. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J Embryol Exp Morp. 1985;89:313–331. [PubMed] [Google Scholar]
- Tomlinson A, Bowtell DD, Hafen E, Rubin GM. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell. 1987;51:143–150. doi: 10.1016/0092-8674(87)90019-5. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Kimmel BE, Rubin GM. rough, a Drosophila homeobox gene required in photoreceptors R2 and R5 for inductive interactions in the developing eye. Cell. 1988;55:771–784. doi: 10.1016/0092-8674(88)90133-x. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. sevenless: a cell specific homeotic mutation of the Drosophila eye. Science. 1986;231:400–402. doi: 10.1126/science.231.4736.400. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Cell fate in the Drosophila ommatidium. Dev Biol. 1987a;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in the Drosophila ommatidium. Dev Biol. 1987b;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Struhl G. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Molecular Cell. 2001;7:487–495. doi: 10.1016/s1097-2765(01)00196-4. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Mavromatakis YE, Struhl G. Three distinct roles for Notch in Drosophila R7 photoreceptor specification. PloS Biology. 2011;9:e100132. doi: 10.1371/journal.pbio.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vactor DL, Jr, Cagan RL, Kramer H, Zipursky SL. Induction in the developing compound eye of Drosophila: multiple mechanisms restrict R7 induction to a single retinal precursor cell. Cell. 1991;67:1145–1155. doi: 10.1016/0092-8674(91)90291-6. [DOI] [PubMed] [Google Scholar]
- Vogt M. Zur labilen Determination der Imagin-alscheiben von Drosophila. I. Verhalten verschieden-altriger Imaginalanlagen bei operativer Defektsetzung. Biol Zbl. 1946;65:223–238. [Google Scholar]
- Waddington CH, Perry MM. The ultrastructure of the developing eye of Drosophila. Proc Roy Soc Biol Sci. 1960;153:155–178. [Google Scholar]
- Weber U, Siegel V, Mlodzik M. pipsqueak encodes a novel nuclear protein required downstream of seven-up for the development of photoreceptors R3 and R4. EMBO J. 1995;14:6247–6257. doi: 10.1002/j.1460-2075.1995.tb00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigglesworth VB. The origin of sensory neurones in an insect, Rhodnius prolixus (Hemiptera) Quart J Microscop Sci. 1953;94:93–112. [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991a;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991b;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Montell C. tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev. 1993;7:1085–1096. doi: 10.1101/gad.7.6.1085. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development. 2001;128:1183–1191. doi: 10.1242/dev.128.7.1183. [DOI] [PubMed] [Google Scholar]
- Zak NB, Shilo BZ. Localization of DER and the pattern of cell divisions in wild-type and Ellipse eye imaginal discs. Dev Biol. 1992;149:448–456. doi: 10.1016/0012-1606(92)90299-v. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhang J, Carthew RW. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development. 1995;121:3045–3055. doi: 10.1242/dev.121.9.3045. [DOI] [PubMed] [Google Scholar]