Abstract
Iron regulation in the brain is both necessary and highly complex. Too little or too much iron can compromise neurological function, yet we still do not know all of the regulatory processes. In our research, we seek to identify genes and gene networks underlying individual differences in brain iron regulation. To this end, we fed mice from 20+ inbred strains a diet low in iron from weaning to 4 months of age. At sacrifice, we measured iron content in the ventral midbrain (VMB). The VMB contains the substantia nigra, a region particularly vulnerable to iron imbalance. The results showed high, inter-strain variability in dietary iron reduction, from almost no loss to more than 40 % vs. control. When we performed quantitative trait loci (QTL) analysis, we observed a significant area on chromosome 2. Within this QTL, we selected glial high-affinity glutamate transporter 1 (Glt1) as the leading candidate. Expression of this gene is both correlated with VMB iron and is also cis-modulated by local sequence variants that segregate in the BXD family. VMB expression differences of Glt1 in six strains covary with differential susceptibility to VMB iron loss.
Keywords: BXD, Ventral midbrain, Recombinant inbred, Quantitative trait loci, Glial glutamate high-affinity transporter
Introduction
Iron plays a number of roles in the brain, from vital contributions to basic cellular metabolism to participation in myelin formation and neurotransmitter synthesis and regulation; however, the same properties that make iron necessary also make it toxic in excess as its redox activity contributes to oxidative stress and neurodegeneration. Despite complex iron regulation in the brain, iron imbalances and consequent disease can develop. One brain region that shows particular vulnerability to iron imbalance and associated neurological disease is the substantia nigra (SN). Dopaminergic neurons of the SN project to the caudate-putamen to form the nigrostriatal dopamine pathway, which regulates voluntary movement. Dietary iron deficiency impairs dopamine (DA) function in this pathway [2]; alternatively, excess iron can lead to neurodegeneration of DA neurons in this nucleus [5]. Diseases resulting from too little iron in the SN include restless legs syndrome (RLS; [11, 12]), and too much iron here is a feature of Parkinson’s disease [32, 33]. While there is considerable evidence to suggest that iron imbalances play a causal role in both of these diseases, there is no clear explanation for why these imbalances develop.
Genetic differences in the ability to regulate iron in the brain are likely to contribute; however, identifying the underlying genes is difficult. Brain iron regulation is region-specific and highly complex, involving many genes and biochemical pathways. These genes act at multiple levels, e.g., transcriptionally and post-transcriptionally, and by different mechanisms, e.g. transport, absorption, storage, and export. Moreover, they may interact with environmental factors such as diet. Iron management in the brain, therefore, is a complex trait.
Complex traits in humans can be modeled in genetic reference populations in animals. One such model consists of recombinant inbred strains such as the BXD strains of mice [36]. The BXD strains, derived from C57BL/6J and DBA/2J progenitors, can be used to quantify variation in a phenotype, such as brain iron concentration; this variation can then be traced back to genomic polymorphisms at specific locations and eventually the underlying genes. This is the basis for quantitative trait loci (QTL) analysis. In the BXD, this is possible because these strains have been genotyped at more than 14,000 markers, including single nucleotide polymorphisms (SNP). Hundreds of genes may lie within a QTL interval, so identifying the underlying genes requires complementary methods. One method is to use BXD gene expression data (a public resource at www.genenetwork.org) to screen for genes within the QTL interval whose expression correlates with the trait of interest [23]. Expression differences in these genes can be hypothesized to underlie the trait variation. This approach has proven successful, an early example being the identification of Kcnj9 expression as a mediator of basal locomotor activity [15]. Here, we have applied this approach in a preliminary search for genes that influence iron concentration and susceptibility to iron loss in the ventral midbrain (VMB), which contains the SN and ventral tegmentum.
We profiled VMB iron concentrations and the response to a long-term, low-iron diet across the BXD strain family. We carried out QTL analysis, expression QTL analysis, and corroborative secondary array analyses to identify candidate genes and genes that modulated by iron loss. This work exposed wide genetic variability in the effects of iron deficiency in the brain and led to the identification of a novel QTL for VMB iron and a candidate gene for this QTL.
Methods
Animals
All experimental protocols were conducted in accordance with the National Institutes of Health Animal Care guidelines and were approved by the Penn State Institutional Animal Care and Use Committee. Mice were from 22 recombinant inbred (RI) strains (BXD) and the parental strains from which they were derived (C57BL/6J and DBA/2J). Mice were reared and housed under a constant light–dark cycle (0600–1800 hours, on–off), ambient temperature (23 ± 2 °C) and relative humidity (40 %). Same-sex littermates were co-housed (maximum of three males or four females per cage).
Dietary treatment
Upon weaning at postnatal day 21, mice were assigned to one of two dietary groups: an iron-deficient (ID) diet or an iron-adequate, control (CN) diet. The ID diet was pelleted and contained 3–5 ppm iron (Teklad, Inc.); the CN diet contained 240 ppm iron but was otherwise identical. Both diets were supplied ad libitum. All mice were weighed weekly. In total, 1,175 mice were included, divided into 88 experimental groups (22 strains, 2 sexes, and 2 diets). The number of mice per experimental group (strain × diet × sex) ranged from 2 to 19, with an average of 11 mice per group. The inclusion of one strain with an n of 2 was based on Belknap [3], who showed that within-strain sample size is secondary to the number of strains in terms of power to detect QTL.
Tissue harvest
At postnatal day 120, all mice were sacrificed via CO2 inhalation, and the brain, liver, and spleen were immediately harvested for tissue iron analysis (first cohort) or gene expression analysis (brain only; second cohort). The VMB and dorsal striatum (caudate-putamen, CP) were dissected as described by [6]. In addition, a blood sample was collected at the time of sacrifice to measure various indices of systemic iron status, including hemoglobin, hematocrit, plasma iron, total iron binding capacity, and transferrin saturation. The analysis of these data can be found elsewhere [38].
Iron assay
Tissue samples for iron assays were stored on ice for 1–2 h until they were weighed and stored at −20 °C. Upon thawing, the samples were wet-digested by adding 400 µL trace metal grade 70 % nitric acid (J.T. Baker Ultrex II, Mallinckrodt Baker, Phillipsburg, NJ). Samples were then heated to 80 °C on a heat block for at least 24 h or until homogenization was complete. After cooling, 400 µL nanopure water was added to each tube (Baker Instra-Analyzed Reagent). Samples were stored at room temperature during digestion until ready for analysis. Samples were stored at all times in 1.5-mL polypropylene microcentrifuge tubes to avoid iron contamination from glassware. Iron was assayed by graphite furnace atomic absorption (AA) spectrophotometry (Perkin Elmer Analyst 600, Perkin Elmer, Norwalk, CT). Standards were prepared by diluting a Perkin Elmer iron standard (PE#N930126) in 0.2 % ultrapure nitric acid, and blanks were prepared with digesting and diluting reagents to control for possible contamination. AA values for iron concentration were normalized to tissue weight (micrograms per gram).
Data analysis
VMB and CP iron concentrations following dietary treatment are reported as strain means (±SEM) for each group (sex and diet). These data were then submitted to GeneNetwork (www.genenetwork.org; [35]) for QTL and systems genetic analysis. Iron data were subjected to analysis of variance (ANOVA) for a three between-subject variables experiment (strain, sex, and diet) and, for each diet, two-way ANOVA (strain and sex). Narrow-sense heritability estimates were calculated from the latter. Change in iron for each strain was also calculated (percent control) and submitted to GeneNetwork for QTL mapping.
QTL mapping
QTL mapping was performed with GeneNetwork, an online bioinformatics resource featuring tools for systems genetic and complex trait analysis [9, 35]. QTL mapping involves entering VMB and CP iron data (strain means and SEM) as quantitative traits; the software generates whole-genome interval maps for each trait. The interval maps graphically illustrate phenotype–genotype associations as peaks (QTL) indicating the strength of association between genomic polymorphisms and the quantitative trait throughout the genome. The magnitude of a QTL is evaluated by a logarithm of the odds ratio (LOD score), which reflects the likelihood that allelic variation at a locus underlies variation in the trait. Significance (p<0.05) and suggestive (p<0.63) thresholds for the LOD scores in GeneNetwork are generated from 1,000 random permutations of the trait data. Six iron-related traits were entered: VMB iron concentration following the CN and ID diet, CP iron concentration following the CN and ID diet, and change in VMB and CP iron (percent control). These traits were submitted separately for males and females and with sexes pooled. In addition, principal components were calculated for VMB iron from both sexes and diets, and QTL for the top principal component were also mapped as general iron regulation QTL.
eQTL mapping
Expression QTL (eQTL) mapping was also performed with GeneNetwork and used for candidate gene selection. The BXD strains exhibit wide genetic-based differences in the basal expression levels of many genes [10]. Expression differences in a gene can be linked back to QTL, termed eQTL, just as any other phenotype. eQTL are driven by polymorphisms that act by altering the expression of a gene, as opposed to protein sequence. Cis-acting eQTL are driven by polymorphisms within or very near to the gene itself. By screening for cis-eQTL within a known QTL region for a trait of interest, one can identify genes whose expression correlates with their trait and is co-regulated by the same QTL as their trait. In this way, eQTL mapping combined with classical QTL mapping can aid in candidate gene selection [23]. GeneNetwork is among other things a public repository for microarray data profiling the BXD panel and offers access to basal expression data on a variety of brain regions and tissues, including the ventral tegmental area (VTA) of the VMB. GeneNetwork also supplies the tools with which to map eQTL for genes of interest within these databases. Our initial QTL mapping experiment yielded a significant QTL for VMB iron concentration. We used the gene expression databases and eQTL mapping tools to aid in candidate gene selection for this QTL. We screened for genes within the QTL interval whose expression correlated with VMB iron levels and exhibited cis regulation, thus regulation by an overlapping eQTL. Any such gene could be hypothesized to contribute to variation in VMB iron levels via differential expression. In other words, the QTL and eQTL could be driven by the same polymorphism. For all eQTL mapping in our study, we used the VTA dataset. The VTA expression data were obtained by Miles and colleagues, using Affymetrix M430 2.0 arrays in mice post-saline injection as part of an ethanol study.
Genetic correlations between phenotypes
GeneNetwork also features a phenotype database, a public repository of data from over 700 traits previously measured across several laboratories in BXD RI (and other) strains. These include behavioral, biochemical, and anatomical traits. The data consist of strain means, not raw data from individual mice, and so we use the term genetic correlation. Using this database, we performed correlation and network analyses to identify relationships with plausible biological significance between VMB and CP iron levels and other phenotypes, including iron- and dopamine-related traits.
Microarray analysis
eQTL mapping led to the nomination of a candidate gene whose expression correlates with VMB iron levels; this correlation was detected using existing gene expression data from other independent studies. For direct observation of this correlation, we performed corroborative secondary array analysis. Six strains were selected, with VMB iron losses encompassing the range observed across the panel. Mice from these strains were subjected to the same dietary treatment as above, with one low iron group and one control group per strain. At 120 days of age, gene expression (transcript abundance) was measured in the VMB. Males and females were both represented in each experimental group and were balanced as much as possible; the selected strains showed no evidence of sex-based differences in iron concentrations in CN or ID conditions. The number of mice per experimental group ranged from three to eight with an average of five mice. Samples were not pooled. Microarray analysis was performed at the Molecular Resource Center at the University of Tennessee. Analysis was performed using the Illumina Mouse WG-6 version 2.0 Expression BeadChip. Balanced randomization was used to position the samples on the arrays. Gene expression was assayed according to manufacturer’s instructions. Normalization of the data set was done using the rank-invariant method [27]. The data were transformed to obtain the log2 of each probe set and standardized using Z scores. The Z scores were doubled, and 8 was added to each to produce a set of Z scores with a mean of 8, a variance of 4, and a standard deviation of 2. The advantage of this modified Z score is that a twofold difference in expression level corresponds approximately to a 1-unit difference in expression. Expression levels below 7 were considered essentially to be background. Normalized data were initially filtered by omitting genes for which expression was lower than 8.0 in >70 % of the samples. Data were then analyzed for gene expression differences among our experimental groups in Nexus Expression™ software (Biodiscovery, El Segundo, CA). To correct for multiple comparisons, we applied the False Discovery Rate [4].
Results
Iron in the VMB
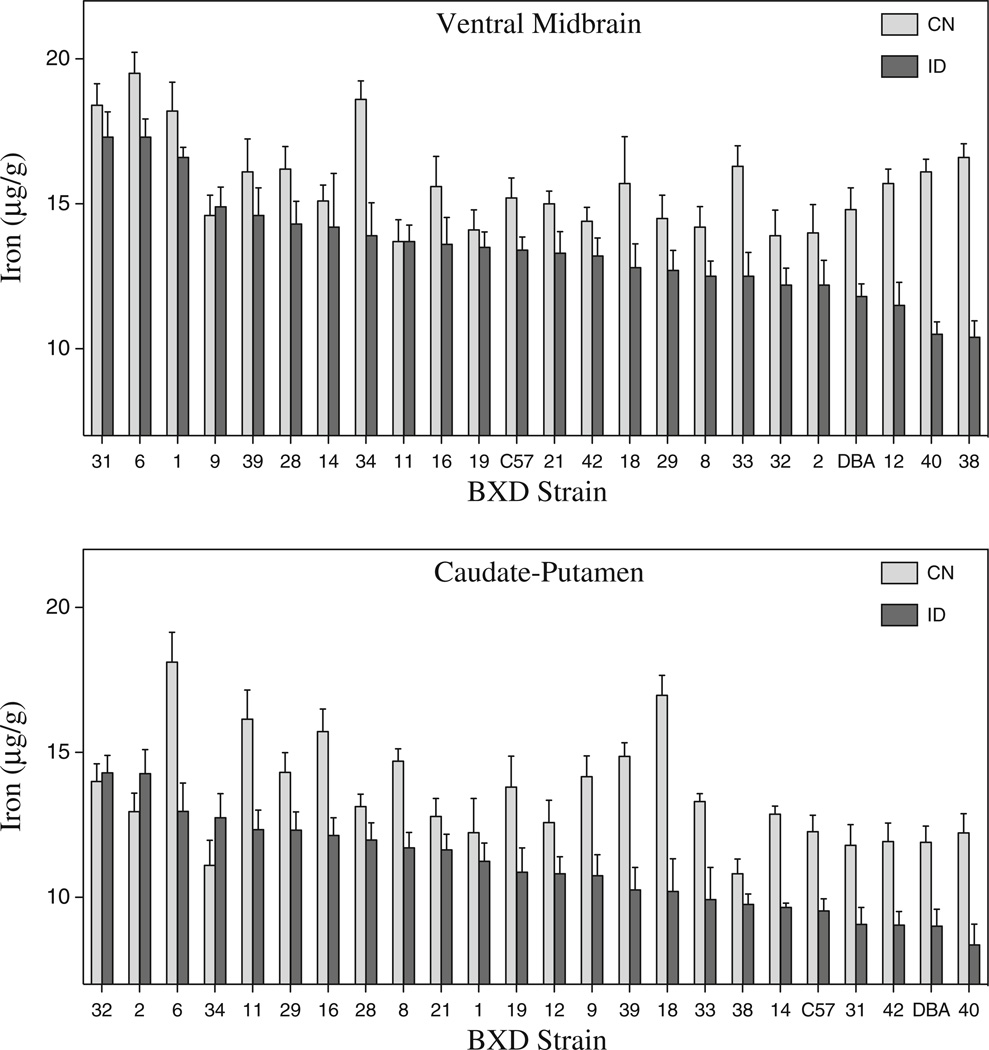
The BXD strains differed widely in both baseline VMB iron concentrations and iron loss following the low iron dietary treatment (Fig. 1a). Mean iron losses ranged from no change to 37 %. There was a main effect of diet (F1,1007=95.802, p< 0.001) and strain (F21,1007=7.898, p<0.001) There was also a significant strain × diet interaction (F21,1007=2.603, p<0.001). There was no main effect of sex, but there was a significant strain by sex interaction (F21,1007=2.038, p<0.004). Male and female VMB iron concentrations were correlated in both the CN (r=0.557, p<0.01) and ID (r=0.572, p<0.01) groups. Male and female results are not shown separately but can be accessed on GeneNetwork (genenetwork.org). To determine narrow-sense heritability estimates for VMB iron concentration, two-way ANOVAs for each dietary condition (strain and sex) were performed. Heritability, or the percentage of the variance owing to genetic factors, was determined to be 14 % for CN mice and 23 % for ID mice, suggesting stronger dependence of VMB iron concentrations on genetic factors following a challenge to the system. Mean ID iron concentrations were correlated with iron concentrations in the CN mice (males, r=0.50, p<0.02; females, r=0.50, p<0.02), and more so with iron loss (males, r=0.62, p<0.01; females, r=0.74, p<0.001).
Fig. 1.
Iron concentration in the VMB and CP in the BXD strains and their parental strains. Male and female mice were fed either a low iron (ID) or control (CN) diet from weaning to 120 days of age. a Ventral midbrain iron. b Caudate-putamen iron
Iron in the CP
Dopaminergic neurons of the SN project to the CP, thus forming the nigrostriatal DA pathway; for this reason, we measured iron in the CP as well. The effects of the ID diet were similar to those seen in the VMB; iron losses ranged from 0 to 44 %, depending on the strain (Fig. 1b). ANOVA showed significant main effects for strain and diet (strain, F21, 919=9.497, p<0.001; diet, F1, 919=110.554, p<0.001) There was also a significant strain by diet interaction (F21, 919=3.478, p<0.001) and strain by diet by sex interaction (F21, 919=1.657, p<0.05). CP iron content for males and females was also significantly correlated in both the CN group (r=0.728, p<0.001) and to a lesser extent in the ID group (r=0.522, p<0.01). The narrow-sense heritability of CP iron concentrations for the CN group was 22 % and for the ID group was 26 %.
Although results for the VMB and CP were similar in terms of strain and sex effects, iron concentrations in these brain regions were not correlated; in some strains, the VMB was more heavily impacted, but in others, the CP showed greater losses. Regional differences have been previously demonstrated in iron loss in the brain [8, 14, 28]; this is the first evidence that the relative impact among the regions is genetically influenced. The CP also differed from the VMB in that baseline iron concentrations were not correlated with ID iron concentrations, but change in iron was related to both the baseline and ID iron concentrations (males CN, r=−0.57, n=20, p<0.01; males ID, 0.75, n=20, p<0.001).
Systemic iron
To determine whether iron loss in the brain was related to changes in systemic iron status and/or anemia, we also assessed a number of indices of systemic iron status, including hematocrit, hemoglobin, plasma iron, and total iron binding capacity. These data are published elsewhere [38]; however, all measures showed wide variability among the BXD strains. No systemic iron parameters were correlated with VMB or CP iron concentrations in either dietary condition; however, in males, variables that were correlated with the ID-related change in VMB iron concentration are as follows: Hematocrit ([21]; r=0.55, p<0.01), CN plasma Fe (r=−0.67, p<0.001), CN spleen weight (males r=0.52, p< 0.05), and CN total iron binding capacity (micrograms per deciliter; r=−0.62, p<0.01). Thus, for males, strains with lower baseline hematocrit and spleen weight but higher plasma Fe concentration and total iron binding capacity tended to be the most susceptible to VMB iron loss following iron deficiency. For female mice, the change in VMB iron was unrelated to any systemic variables, including difference scores.
QTL analysis
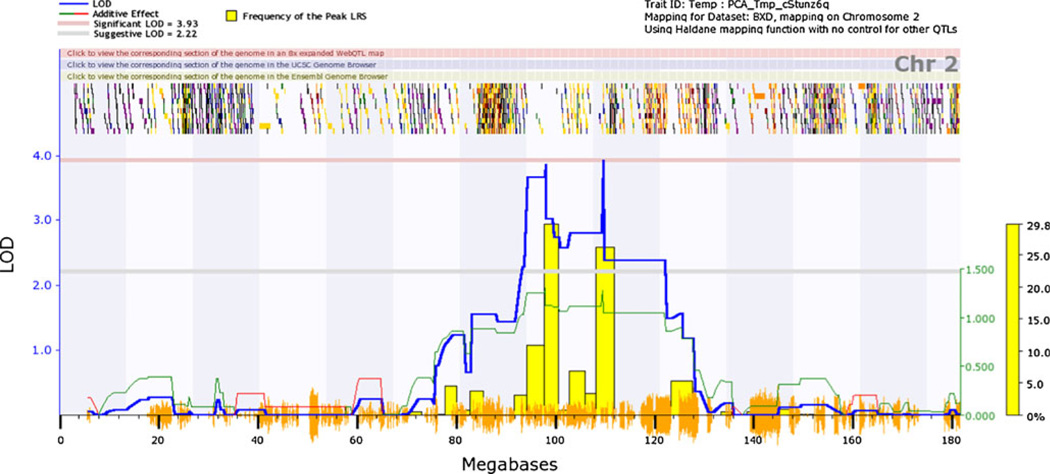
For VMB iron, we detected significant and suggestive QTL on chromosome 2 under basal and iron-deficient conditions, respectively. In control mice, the C57BL/6 allele at the peak markers was associated with 14 % higher iron concentration compared to the DBA/2 allele. Also of interest was a suggestive QTL on chromosome 17 for VMB iron in control males (~12.5–23.4 Mb) that lies in the vicinity of a QTL previously reported for iron, copper, and zinc in various brain regions [18] and near Btbd9, whose human homologue is associated with periodic limb movements, restless legs syndrome and serum ferritin [34, 37]. Low iron in the substantia nigra is a key pathophysiological sign in restless legs syndrome. No significant QTL were seen for iron concentration in the CP in either treatment condition or treatment by sex combination or for change in iron concentration in either region. Because we were interested in the QTL on chromosome 2, we performed a principal components analysis on iron concentration, combining both dietary treatments and sexes. The QTL map is presented in Fig. 2. The first and only principal component, accounting for 62 % of total variance, revealed a QTL peak at 109.62 Mb to yield a significant LOD score of 3.94. The associated SNP marker is rs6208879. Because this principal component is a latent variable, we consider it to be relevant to iron homeostasis overall in the ventral midbrain.
Fig. 2.
QTL for VMB iron concentration on chromosome 2. Mice were assigned either a low iron or control diet upon weaning; iron concentrations were assayed at 120 days of age. Principal components analysis was performed on VMB iron under both dietary conditions and for both sexes. Genome-wide interval mapping plots above show a significant QTL on chromosome 2 (right peak at approximately 109 Mb)
eQTL analysis
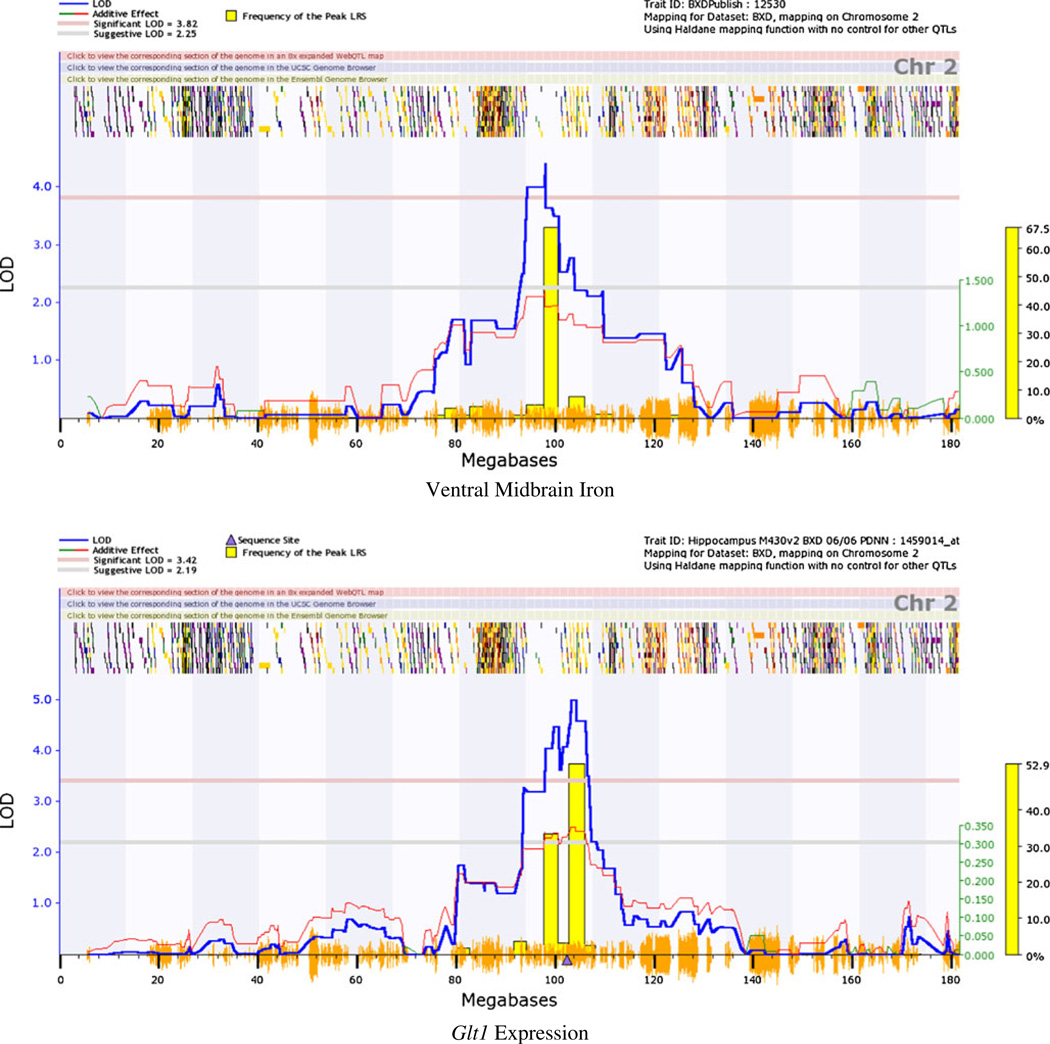
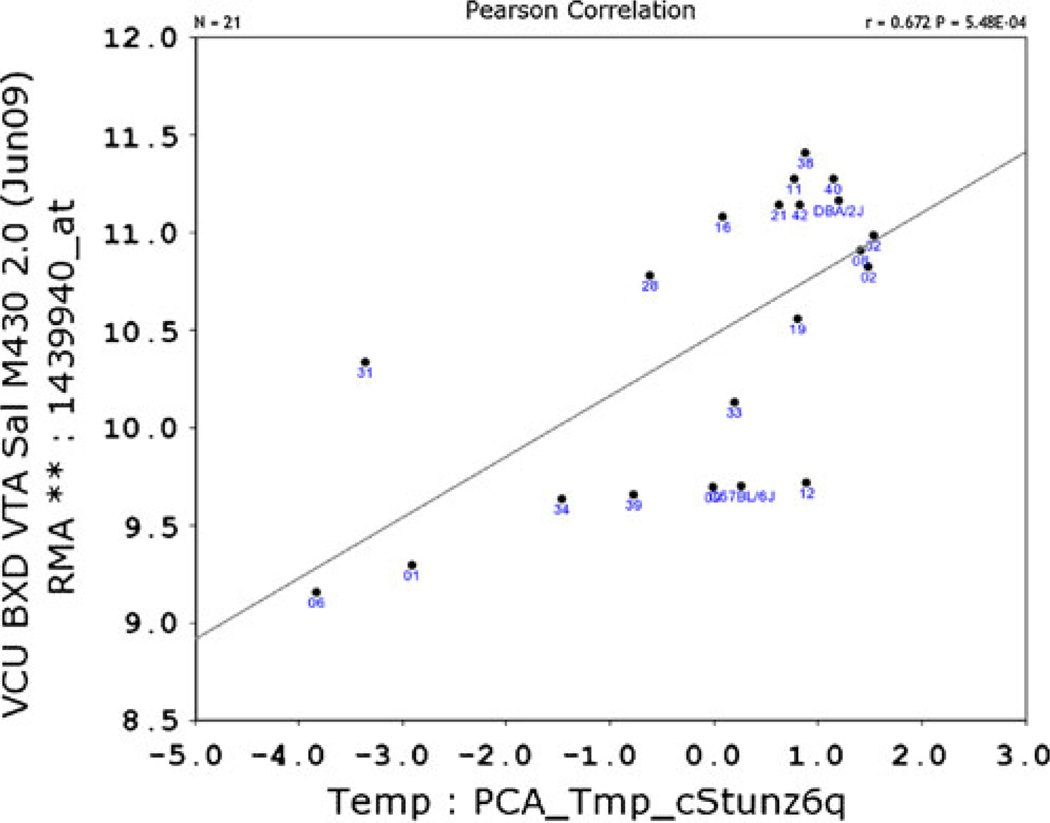
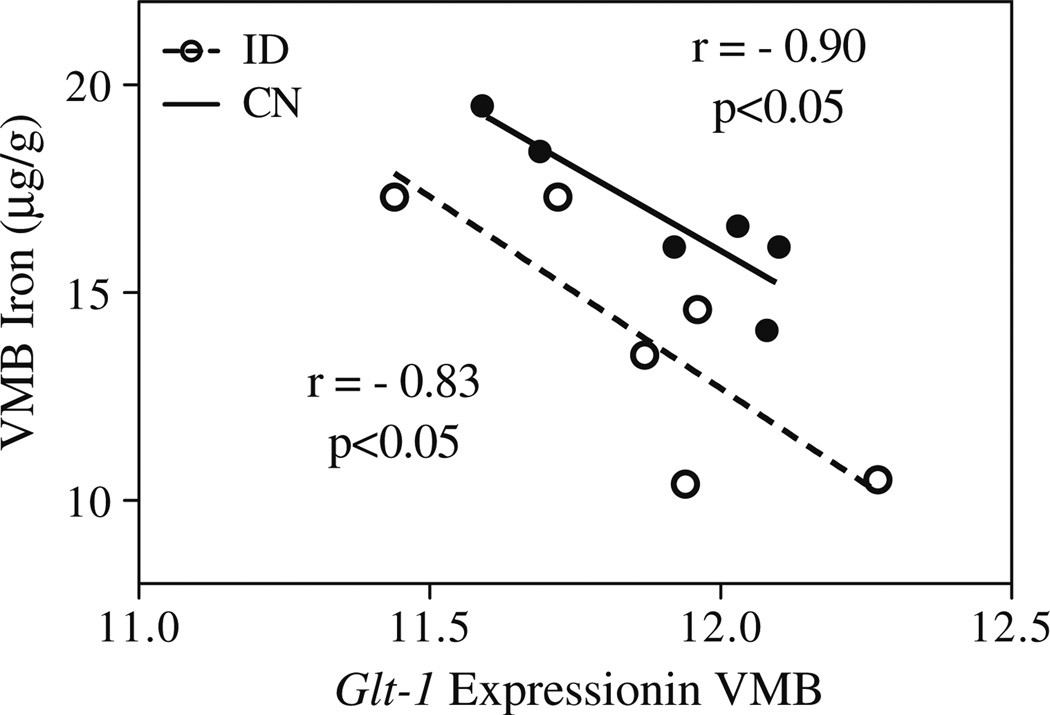
The QTL interval on chromosome 2 spans a number of genes; eQTL mapping using the VTA dataset on GeneNetwork.org highlighted cis-regulated genes that co-map to the VMB iron QTL. A number of cis-regulated genes within the QTL region were identified; of these, six showed expression levels that correlate significantly with previously measured basal VMB iron levels [20]. These include glial high-affinity glutamate transporter (Glt1 (r=−0.574; 102.6 Mb; probe 1439940_at; Figs. 3 and 4), apoptosis inhibitor 5 (Api5; r=−0.498; 94.3 Mb; probe 1415813_at), leucine-rich repeat containing 4C (Lrrc4c; −0.624; 97.3 Mb, probe 1456759_at), pyruvate dehydrogenase complex, component X (Pdhx; r=0.508; 102.9 Mb, probe 1459434), and two yet unclassified genes: C920021A13 (r=0.685; 94.3 Mb, probe 1441471_at) and 2810002D19Rik (r=0.579; 94.3 Mb, probe 1452312_at). Glt1 expression was also correlated with the principal component for general VMB iron regulation (r=0.672, p<0.001; Fig. 4).
Fig. 3.
Chromosome 2 QTL for VMB iron levels co-maps with cis-eQTL for Glt1 expression. a Interval mapping plot for VMB iron levels showing close-up of chromosome 2. b Interval mapping plot for VTA Glt1 expression levels. Blue line represents LOD score, or strength of association between genetic markers and phenotypic trait. Horizontal red dashed line represents significance threshold determined by 1,000 permutations of the data. Vertical orange dashes on x-axis represent SNPs from a high density map; horizontal dashes near top of the image represent gene locations
Fig. 4.
Bioinformatic resources depict correlation between VMB iron levels and Glt1 expression. VMB iron first principal component correlated with expression of Glt1. Correlation with principal component is positive but reflects an inverse relationship between Glt1 expression and VMB iron values. Graph generated using GeneNetwork.org; expression data are from the VTA dataset post-saline injection, with permission from Miles and colleagues
Microarray analysis
None of the six cis-regulated genes within the QTL interval that correlate with VMB iron concentration has a known role in iron homeostasis. To confirm the correlations and determine whether any respond to iron deficiency, we conducted a secondary array analysis of six strains representing the range of iron loss. Of the six genes, only two passed the initial microarray criterion for minimum expression level in the VMB. These were Lrrc4c and Glt1. Of these two, neither showed a change in expression in response to the iron-deficient diet; however, consistent with the correlation we discovered using GeneNetwork.org, Glt1 (ILMN_ 3076439) expression was again negatively correlated with VMB iron concentration, here in both iron-adequate and iron-deficient mice (Fig. 5). In addition, Glt1 expression was significantly higher in the two strains with significant VMB iron loss (strains BXD 38 and 40) than in the four strains that showed nonsignificant iron loss in our iron deficiency study (strain 31, 6, 39, and 19; LR=0.29; Q<0.05).
Fig. 5.
Illumina BeadChip analysis corroborates correlation between VMB iron levels and Glt1 expression and extends results across dietary conditions. Glt1 expression was measured in the VMB following 100 days of ID or CN dietary treatment. The six BXD strains assayed were a selected subset of the original 22 strains
Genetic correlations between phenotypes
To identify correlations between VMB iron and other phenotypic traits of plausible biological relevance, we used the BXD phenotypes database on GeneNetwork.org. VMB iron was correlated with several DA-related phenotypes in the BXDs, extending previous findings from animal models of iron deficiency. VMB iron and VMB DA transporter (DAT) density were positively correlated, consistent with the previous report that iron deficiency reduces the protein density of DAT in the rat VMB [15]. VMB iron and several anxiety and depression-related phenotypes were also correlated [29], in line with observations of increased anxiety and depression-related behaviors in iron-deficient rodents and humans [1, 22].
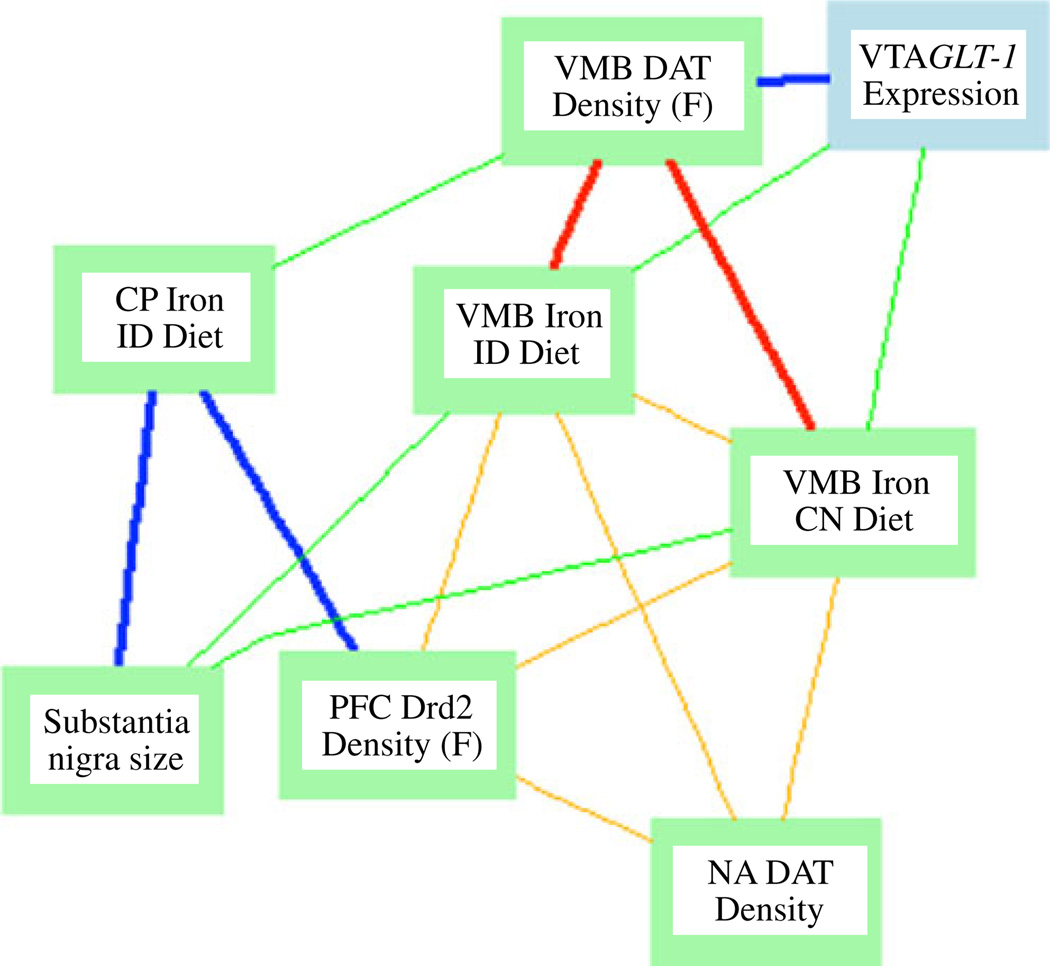
Other phenotypes included hemoglobin (current study) and PFC iron [20]. The direction, magnitude, and significance of these correlations are reported in Table 1. A Ventral Midbrain Iron Glt1 Expression network highlighting several of these correlations and correlations with Glt-1 expression is shown in Fig. 6.
Table 1.
Correlations between ventral midbrain iron and other phenotypes in the BXD panel
| Phenotype | N | Citation | Control diet | ID diet | ||
|---|---|---|---|---|---|---|
| Male | Female | Male | Female | |||
| Dopamine transporter protein density in ventral midbrain (F) | 11 | [18] | 0.77** | 0.78** | ||
| DRD2 protein density in the prefrontal cortex (M) | 12 | [18] | 0.58* | 0.71** | ||
| Anxiety assay, plus maze closed arm entries (M) | 20 | [29] | 0.54* | 0.52* | ||
| Anxiety assay, time in middle of elevated plus maze (M + F) | 21 | [29] | −0.56** | |||
| Depression assay, Porsolt behavioral despair test, time immobile (M) | 19 | [29] | −0.60** | |||
| Ethanol response, locomotion post-injection (2 g/kg ip) | 15 | [30] | −0.64* | −0.83*** | −0.62* | −0.66** |
| Ethanol response, total ethanol intake in two-bottle choice | 13 | [13] | −0.74** | −0.61* | ||
| Max startle to 120 dB (M) | 21 | [29] | 0.61** | 0.69*** | ||
| Iron in the prefrontal cortex | 11 | [20] | 0.64* | 0.68* | ||
| Hemoglobin, control diet (F) | 24 | Jellen et al. current | 0.67*** | |||
The BXD phenotype database available on GeneNetwork.org allows comparison of a trait of interest with other phenotypes previously measured in the BXD panel. Selected correlations with VMB iron are reported here, as Pearson correlation coefficients
N number of strains in the comparison
p<0.05;
p<0.01;
p<0.001
Fig. 6.
Network graph depicting correlations between VMB and CP iron, Glt1 expression, and dopamine-related phenotypes. Nodes represent phenotypes measured in the BXD strains; lines represent correlations. Bold lines −0.7<r<0.7. All other lines: −0.5<r<0.5. Blue and green indicate positive correlations; red and orange indicate negative correlation
Discussion
In this study, we confirmed a wide, genetic-based variation in basal VMB and CP iron concentration [20] and showed a wide variation in susceptibility to iron loss after being fed an iron-deficient diet, both peripherally and in the brain. Our primary interest is in the VMB because it contains the SN, the site for iron-related pathophysiology related to RLS and Parkinson’s disease. We identified a novel QTL for VMB iron on chromosome 2. We used extant expression data to nominate Glt1 as a candidate gene, finding that its expression correlates with VMB iron and is cis-regulated. Moreover, we showed that Glt1 expression is correlated with VMB iron across dietary treatments.
Variation in susceptibility to iron deficiency in the brain
This is the first study to demonstrate large, genetic-based differences in susceptibility to nutritional iron deficiency in the brain. Perhaps more interesting than the VMB iron loss that occurred in some strains was the ability of other strains to maintain baseline iron levels, despite an extremely low iron diet and long duration of treatment, and in some cases despite signs of systemic iron deficiency. Identifying the mechanisms that manage this in humans could prevent DA malfunction and many other neurological complications of nutritional iron deficiency, which affects over 30 % of the world population (http://www.who.int/nutrition/topics/ida/en/index.html). There was also a striking disconnect between the brain and systemic response to iron deficiency. This may be explained in part by the additional regulation that occurs at the blood–brain barrier [25].
Support for Glt1 as candidate
Glt1 (aka Slc1a2) encodes the high-affinity glial glutamate transporter, GLT1.We note several characteristics of Glt1 that support its nomination: it is physically located within our QTL region, its expression shows heritable variation regulated by an eQTL within our QTL region, and its expression is correlated with VMB iron concentration, under both dietary conditions. Glt1 is located on chromosome 2, between 102.498840 and 102.680941 Mb. A search for polymorphic markers between the C57BL/6J and DBA/2J strains (the founders for the BXD RI lines) reveals one nonsynonymous SNP at 102575441 Mb, three indels, and a deletion between 102.505178 and 102.505633 Mb in the DBA/2 strain (Mouse Genomes Project http://www.sanger.ac.uk/cgi-bin/modelorgs/mousegenomes/snps.pl). The extent to which any or all of these polymorphisms affect VMB iron and Glt1 expression is to be determined. Figure 5 presents a gene–phenotype network analysis of direct and indirect associations between Glt1 expression and iron/dopamine phenotypes. Clinically, striatal GLT1 may play a role in Parkinson’s disease, an iron-related, DA-based movement disorder [24] as well as in restless legs syndrome. It has been shown recently that RLS is accompanied by increased glutamate and glutamine levels in the thalamus (RP Allen, personal communication). This could stem from differences in the glutamate transporter, increasing glutamate uptake into astrocytes thereby producing increased glutamatergic activity. This would establish a biological basis for glutamatergic abnormality in RLS.
The involvement of Glt1 in VMB iron regulation requires experimental confirmation. While we hypothesize that differences in its expression modulate iron concentration in this region, its correlation with iron could alternatively reflect linkage between Glt1 and some other polymorphism underlying our trait. One drawback to using expression data to select candidate genes is that the focus is entirely on expression-based QTL, when QTL can also be explained by polymorphisms that act by altering protein sequence. Whether the distance between the eQTL for Glt1 and the first principal component is important remains to be seen. Nonetheless, with careful follow-up work, this approach can be successful. For example, Palmer and colleagues used microarray analysis and mice selectively bred for methamphetamine sensitivity to show that casein kinase 1 epsilon (Csnk1e) expression co-mapped with a QTL for methamphetamine-induced locomotor stimulation [26]. They suggested differences in locomotor stimulation could be explained by differences in Csnk1e expression. In a subsequent study, selective inhibition of Csnk1e indeed attenuated the locomotor response to methamphetamine [7]. In a separate analysis, it was found that a single nucleotide polymorphism in Csnk1e in humans is associated with differences in subjective response to d-amphetamine, demonstrating the translational utility in this approach [38].
Conclusions
Understanding iron-DA mechanisms and what predisposes certain individuals to develop high or low iron in the VMB or other DA-relevant brain regions will aid in developing prevention and treatment strategies for iron-related neurological disease. By combining classical QTL analysis, eQTL analysis, and microarray analysis, we were able to both identify a novel QTL and candidate gene for VMB iron concentration.
Acknowledgments
This work was funded in part by NIA Grant # PO1AG021190, by a grant from the Restless Legs Foundation, by NRSA fellowship 5F31NS060393 to LCJ, and by NIAAA grants U01AA016662 and U01AA016667 to MFM and U01AA13499 to LL and RWW. The authors thank Dr. Kennie Jones for his contributions to database management and Dr. Lisa Tarantino for assistance with identifying polymorphisms within Glt1.
Footnotes
Conflict of interest The authors declare no conflicts of interest in this research.
Contributor Information
Leslie C. Jellen, Intercollege Graduate Degree Program in Neuroscience, The Pennsylvania State University, 315 E. Health and Human Development Building, University Park, PA 16802, USA
Erica L. Unger, Department of Nutritional Sciences, The Pennsylvania State University, University Park, PA, USA
Lu Lu, Department of Anatomy and Neuroscience, University of Tennessee Health Sciences Center, Memphis, TN, USA.
Robert W. Williams, Department of Anatomy and Neuroscience, University of Tennessee Health Sciences Center, Memphis, TN, USA
Sarah Rousseau, Department of Biobehavioral Health, The Pennsylvania State University, University Park, PA, USA.
Xusheng Wang, Department of Anatomy and Neuroscience, University of Tennessee Health Sciences Center, Memphis, TN, USA.
Christopher J. Earley, Department of Neurology and Research Institute, The Johns Hopkins University, Baltimore, MD, USA
Richard P. Allen, Department of Neurology and Research Institute, The Johns Hopkins University, Baltimore, MD, USA
Michael F. Miles, Department of Pharmacology/Toxicology, Virginia Commonwealth University, Richmond, VA, USA Department of Neurology, Virginia Commonwealth University, Richmond, VA, USA.
Byron C. Jones, Email: bcj1@psu.edu, Intercollege Graduate Degree Program in Neuroscience, The Pennsylvania State University, 315 E. Health and Human Development Building, University Park, PA 16802, USA; Department of Biobehavioral Health, The Pennsylvania State University, University Park, PA, USA.
References
- 1.Beard JL, Erikson KM, Jones BC. Neurobehavioral analysis of developmental iron deficiency in rats. Behav Brain Res. 2002;134:517–524. doi: 10.1016/s0166-4328(02)00092-x. [DOI] [PubMed] [Google Scholar]
- 2.Beard JL. Iron deficiency alters brain development and functioning. J Nutr. 2003;133:1468S–1472S. doi: 10.1093/jn/133.5.1468S. [DOI] [PubMed] [Google Scholar]
- 3.Belknap JK. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet. 1998;28:29–38. doi: 10.1023/a:1021404714631. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 5.Ben-Shachar D, Youdim MBH. Intranigral iron injection induces behavioral and biochemical ‘Parkinsonism’ in rats. J Neurochem. 1991;57:2133–2135. doi: 10.1111/j.1471-4159.1991.tb06432.x. [DOI] [PubMed] [Google Scholar]
- 6.Boone EM, Cook MN, Hou X, Jones BC. Sex and strain influence the effects of ethanol on central monoamines. J Stud Alcohol. 1997;58:590–599. doi: 10.15288/jsa.1997.58.590. [DOI] [PubMed] [Google Scholar]
- 7.Bryant CD, Graham ME, Distler MG, Munoz MB, Li D, Vezina P, Sokoloff G, Palmer AA. A role for casein kinase 1 epsilon in the locomotor stimulant response to methamphetamine. Psychopharmacology (Berl) 2009;203:703–711. doi: 10.1007/s00213-008-1417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Connor JR, Beard JL. Alterations of brain iron, transferrin, and ferritin concentrations in iron deficient developing rats. J Nutr. 2000;125:1529–1535. doi: 10.1093/jn/125.6.1529. [DOI] [PubMed] [Google Scholar]
- 9.Chesler EJ, Lu L, Wang J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7:485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- 10.Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 11.Connor JR, Boyer PJ, Menzies SL, Dellinger B, Allen RP, Ondo WG, Earley CJ. Neuropathological examination suggests impaired brain iron acquisition in restless legs syndrome. Neurology. 2003;61:304–309. doi: 10.1212/01.wnl.0000078887.16593.12. [DOI] [PubMed] [Google Scholar]
- 12.Connor JR, Wand XS, Allen RP, Beard JL, Wiesinger JA, Felt BT, Earley CJ. Altered dopaminergic profile in the putamen and SN in restless legs syndrome. Brain. 2009;132:2403–2412. doi: 10.1093/brain/awp125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crabbe JC, Kosobud A, Young ER, Janowsky JS. Polygenic and single-gene determination of responses to ethanol in BXD/Ty recombinant inbred mouse strains. Neurobehav Toxicol Teratol. 1983;5:181–187. [PubMed] [Google Scholar]
- 14.Erikson KM, Pinero DJ, Connor JR, Beard JL. Regional brain iron, ferritin and transferrin concentrations during iron deficiency and iron repletion in developing rats. J Nutr. 1997;127:2030–2038. doi: 10.1093/jn/127.10.2030. [DOI] [PubMed] [Google Scholar]
- 15.Erikson KM, Jones BC, Beard JL. Iron deficiency alters dopamine transporter functioning in rat striatum. J Nutr. 2000;130:2831–2837. doi: 10.1093/jn/130.11.2831. [DOI] [PubMed] [Google Scholar]
- 16.Hitzemann R, Malmanger B, Reed C, et al. A strategy for the integration of QTL, gene expression, and sequence analyses. Mamm Genome. 2003;14:733–747. doi: 10.1007/s00335-003-2277-9. [DOI] [PubMed] [Google Scholar]
- 17.Jellen LC, Beard JL, Jones BC. Systems genetics analysis of iron regulation in the brain. Biochimie. 2009;91:1255–1259. doi: 10.1016/j.biochi.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones BC, Tarantino LM, Rodriguez LA, Reed CL, McClearn GE, Plomin R, Erwin VG. Quantitative-trait loci analysis of cocaine-related behaviours and neurochemistry. Pharmacogenetics. 1999;9:607–617. [PubMed] [Google Scholar]
- 19.Jones BC, Reed CL, Hitzemann R, Wiesinger JA, McCarthy KA, Buwen JP, Beard JL. Quantitative genetic analysis of ventral midbrain and liver iron in BXD recombinant inbred mice. Nutr Neurosci. 2003;6:369–377. doi: 10.1080/10284150310001624192. [DOI] [PubMed] [Google Scholar]
- 20.Jones BC, Beard JL, Gibson JN, Unger EL, Allen RP, McCarthy KA, Earley CJ. Systems genetic analysis of peripheral iron parameters in the mouse. Am J Physiol Regul Integr Comp Physiol. 2007;293:R116–R124. doi: 10.1152/ajpregu.00608.2006. [DOI] [PubMed] [Google Scholar]
- 21.Lozoff B, Jimenez E, Hagen J, Mollen E, Wolf AW. Poorer behavioral and developmental outcome more than 10 years after treatment for iron deficiency in infancy. Pediatrics. 2000;105:E51. doi: 10.1542/peds.105.4.e51. [DOI] [PubMed] [Google Scholar]
- 22.Lu L, Wei L, Peirce JL, Wang X, Zhou J, Homayouni R, Williams RW, Airey DC. Using gene expression databases for classical QTL candidate gene discovery in the BXD recombinant inbred genetic reference population: mouse forebrain weight. BMC Genomics. 2008;9:444. doi: 10.1186/1471-2164-9-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massie A, Goursaud S, Schallier A, Vermoesen K, Meshul CK, Hermans E, Michotte Y. Time-dependent changes in GLT1 functioning in striatum of hemi-Parkinson rats. Neurochem Int. 2010;57:572–578. doi: 10.1016/j.neuint.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Moos T. Brain iron homeostasis. Dan Med Bull. 2002;49:279–301. [PubMed] [Google Scholar]
- 25.Palmer AA, Verbitsky M, Suresh R, Kamens HM, Reed CL, Li N, Burkhar-Kasch S, McKinnon CS, Belknap JK, Gilliam TC, Phillips TJ. Gene expression differences in mice divergently selected for methamphetamine sensitivity. Mamm Genome. 2005;16:291–305. doi: 10.1007/s00335-004-2451-8. [DOI] [PubMed] [Google Scholar]
- 26.Pelz CR, Kulesz-Martin M, Bagby G, Sears RC. Global rank-invariant set normalization (GRSN) to reduce systematic distortions in microarray data. BMC Bioinformatics. 2008;9:520. doi: 10.1186/1471-2105-9-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinero DJ, Jones B, Beard JL. Alterations in brain iron metabolism in response to dietary iron changes. J Nutr. 2000;130:254–263. doi: 10.1093/jn/130.2.254. [DOI] [PubMed] [Google Scholar]
- 28.Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips TJ, Huson M, Gwiazdon C, Burkhart-Kasch S, Shen EH. Effects of acute and repeated ethanol exposures on the locomotor activity of BXD recombinant inbred mice. Alcohol Clin Exp Res. 1995;19:269–278. doi: 10.1111/j.1530-0277.1995.tb01502.x. [DOI] [PubMed] [Google Scholar]
- 30.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder AM, Connor JR. Iron, the SN and related neurological disorders. Biochim Biophys Acta. 2009;1790:606–614. doi: 10.1016/j.bbagen.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Sofic E, Paulus W, Jellinger K, Riederer P, Youdim MBH. Selective increase of iron in SN zona compacta of Parkinsonian brains. J Neurochem. 1991;56:978–982. doi: 10.1111/j.1471-4159.1991.tb02017.x. [DOI] [PubMed] [Google Scholar]
- 33.Stefansson H, Rye DB, Hicks A, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–647. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- 35.Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Gen Biol 2 RESEARCH0046. 2001 doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkelmann J, Schormair B, Lichtner P, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 37.Veenstra-VanderWeele J, Qaadir A, Palmer AA, Cook EH, Jr, de Wit H. Association between the casein kinase 1 epsilon gene region and subjective response to D-amphetamine. Neuropsychopharmacology. 2006;5:1056–1063. doi: 10.1038/sj.npp.1300936. [DOI] [PubMed] [Google Scholar]
- 38.Yin L, Unger EL, Jellen LC, et al. Systems genetic analysis of multivariate response to iron deficiency in mice. Am J Physiol Regul Integr Comp Physiol. 2012 doi: 10.1152/ajpregu.00634.2011. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]