Abstract
Floral volatile benzenoid/phenylpropanoid (FVBP) biosynthesis is a complex and coordinate cellular process executed by petal limb cells of a Petunia×hybrida cv. ‘Mitchell Diploid’ (MD) plant. In MD flowers, the majority of benzenoid volatile compounds are derived from a core phenylpropanoid pathway intermediate by a coenzyme A (CoA) dependent, β-oxidative scheme. Metabolic flux analysis, reverse genetics, and biochemical characterizations of key enzymes in this pathway have supported this putative concept. However, the theoretical first enzymatic reaction, which leads to the production of cinnamoyl-CoA, has only been physically demonstrated in a select number of bacteria like Streptomyces maritimus through mutagenesis and recombinant protein production. A transcript hasbeen cloned and characterized from MD flowers that shares high homology with an Arabidopsis thaliana transcript ACYL-ACTIVATING ENZYME11 (AtAAE11) and the S. maritimus ACYL-COA:LIGASE (SmEncH). In MD, the PhAAE transcript accumulates in a very similar manner as bona fide FVBP network genes, i.e. high levels in an open flower petal and ethylene regulated. In planta, PhAAE is localized to the peroxisome. Upon reduction of PhAAE transcript through a stable RNAi approach, transgenic flowers emitted a reduced level of all benzenoid volatile compounds. Together, the data suggest that PhAAE may be responsible for the activation of t-cinnamic acid, which would be required for floral volatile benzenoid production in MD.
Key words: AAE, benzenoid, flower, petunia, t-cinnamic acid, volatiles
Introduction
Petunia×hybrida cv. ‘Mitchell Diploid’ (MD) is an excellent model plant system for floral volatile biosynthesis research. MD plants can maintain extensive numbers of large white flowers that emit considerable amounts (>100 µg h–1 g–1 FW) of floral volatile benzenoid/phenylpropanoid (FVBP) compounds from petal limb tissue of an open, unpollinated flower mainly during a dark period (Verdonk et al., 2003, 2005; Underwood et al., 2005; Colquhoun et al., 2010a). Several of the genetic, molecular, biochemical, and physiological aspects of FVBP biosynthesis have been identified and characterized to some extent in MD (reviewed in Dudareva et al., 2006; Schuurink et al., 2006; Colquhoun and Clark, 2011a). Examples include the now obvious network of genes that encompasses the genetic elements of FVBP biosynthesis, the identification of several transcriptional regulators that enable a concerted transcription of the FVBP network genes, the illustration of biological rhythmicity of FVBP emission and gene transcription, but corresponding protein activities fail to demonstrate a correlated rhythm, and the flowers’ physiological response to pollination and/or ethylene exposure resulting in a down-regulation of the FVBP pathway as a whole. However, the entire narrative is far from complete.
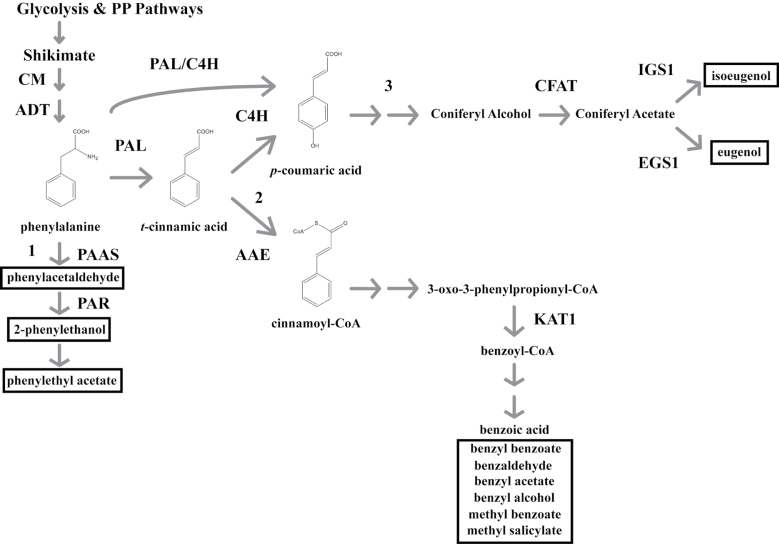
In an attempt to delineate FVBP biosynthesis, the aromatic amino acid phenylalanine serves as the initial, common precursor for FVBP biosynthesis (Fig. 1) with the production of individual FVBP compounds emanating from phenylalanine directly and the core phenylpropanoid pathway metabolites t-cinnamic acid and p-coumaric acid (Boatright et al., 2004; Schuurink et al., 2006). Phenylacetaldehyde, 2-phenylethanol, and 2-phenethyl acetate are enzymatically formed, linearly from phenylalanine; FVBP metabolic branch 1 (Boatright et al., 2004, Guterman et al., 2006; Kaminaga et al., 2006; Tieman et al., 2006, 2007; Chen et al., 2011). The benzenoids benzaldehyde, benzyl alcohol, benzyl benzoate, and methyl benzoate are all thought to be produced indirectly from t-cinnamic acid; FVBP metabolic branch 2 (Boatright et al., 2004; Underwood et al., 2005; Orlova et al., 2006; Long et al., 2009; Van Moerkercke et al., 2009). Isoeugenol and eugenol (phenylpropanoids) are enzymatically derived downstream of t-cinnamic acid, i.e. indirectly from p-coumaric acid; FVBP metabolic branch 3 (Koeduka et al., 2006, 2008, 2009; Dexter et al., 2007; Colquhoun et al., 2011b). Phenethyl benzoate is produced from substrates generated in FVBP branches 1 and 2 (Dexter et al., 2008).
Fig. 1.
A schematic representation of the FVBP pathway in MD. Glycolysis and the pentose phosphate (PP) pathways provide the initial carbon substrates (phosphoenolpyruvate and erythrose-4-phosphate, respectively) for the condensation to a benzene ringed structure through the shikimate pathway. The aromatic amino acid phenylalanine is generated by a succession of two plastid enzymes (CHORISMATE MUTASE, CM; AROGENATE DEHYDRATASE, ADT). PHENYLALANINE AMMONIA-LYASE (PAL) converts phenylalanine to t-cinnamic acid, or PAL can associate with CINNAMTE-4-HYDROXYLASE (C4H) ultimately to form p-coumaric acid. Floral volatile benzenoid\phenylpropanoid (FVBP) metabolic branch 1 emanates from phenylalanine with the PHENYLACETALDEHYDE SYNTHASE (PAAS) enzyme, branch 2 by way of t-cinnamic acid through ACYL-ACTIVATING ENZYME (AAE), and branch 3 from p-coumaric acid. All FVBP compounds are boxed with a black line. Enzymes also depicted: PHENYLACETALDEHYDE REDUCTASE (PAR), CONIFERYL ALCOHOL ACYLTRANSFERASE (CFAT), ISOEUGENOL SYNTHASE 1 (IGS1), EUGENOL SYNTHASE1 (EGS1), and 3-KETOACYL-COA THIOLASE (KAT1).
FVBP metabolic branch 1 begins with a reaction catalyzed by the petunia enzyme PHENYLACETALDEHYDE SYNTHASE (PhPAAS). PhPAAS demonstrates substrate-specific usage of phenylalanine to form phenylacetaldehyde through a decarboxylation-amine oxidation reaction (Kaminaga et al., 2006). Presumably, PhPAAS is competing for substrate with the initial core phenylpropanoid pathway enzyme PHENYLALANINE AMMONIA-LYASE (PAL) (recently reviewed in Vogt, 2010). PAL catalyzes the non-oxidative deamination of phenylalanine to t-cinnamic acid (Koukol and Conn, 1961; Ritter and Schulz, 2004). CINNAMATE-4-HYDROXYLASE (C4H) enzymatically forms p-coumaric acid from t-cinnamic acid (Anterola et al., 2002), which produces substrate for isoeugenol and eugenol biosynthesis and appears to be the initial enzyme of FVBP metabolic branch 3 (Colquhoun et al., 2011b). However, the exact order of biochemical reactions between p-coumaric acid and isoeugenol has not yet been identified in petunia.
The benzenoid branch of FVBP biosynthesis (branch 2) putatively originates with t-cinnamic acid and has been empirically shown to be dominated by a β-oxidative, coenzyme A (CoA) dependent chemical strategy in petunia (Boatright et al., 2004; Van Moerkercke et al., 2009). In MD, t-cinnamic acid and benzoic acid levels fluctuate in accordance with FVBP production and emission, whereby peak metabolite accumulation coincides with peak FVBP emission (Underwood et al., 2005; Orlova et al., 2006). Benzoic acid is an important intermediate in floral volatile benzenoid biosynthesis (Schuurink et al., 2006), and has been empirically supported to be a direct precursor to methyl benzoate formation in MD (the highest quantity FVBP emitted), benzoyl-CoA formation in Clarkia breweri, Arabidopsis thaliana, and a select number of bacteria, and benzaldehyde formation in Antirrhinum majus (Beuerle and Pichersky, 2002; Negre et al., 2003; Underwood et al., 2005; Long et al., 2009). Two types of transferase enzymes (methyl and benzoyl) have been identified to catalyze reactions that result in the direct formation of FVBP compounds from metabolic branch 2 in MD. BENZOIC ACID/SALICYLIC ACID METHYL TRANSFERASE (PhBSMT1, 2) can utilize benzoic acid or salicylic acid to generate methyl benzoate or methyl salicylate, respectively (Kolosova et al., 2001a; Negre et al., 2003; Underwood et al., 2005). BENZOYL-COA:BENZYL ALCOHOL/PHENYLETHANOL BENZOYLTRANSFERASE (PhBPBT) utilizes benzoyl-CoA as a constant substrate with either benzyl alcohol or 2-phenylethanol to form benzyl benzoate or phenylethyl benzoate, respectively (Boatright et al., 2004; Orlova et al., 2006; Dexter et al., 2008). One more petunia enzyme (a thiolase) has been empirically shown to function in the intermediate steps of FVBP metabolic branch 2. 3-KETOACYL-COA THIOLASE (PhKAT1) can utilize 3-oxo-3-phenylpropionyl-CoA to form benzoyl-CoA, and is localized to peroxisomes in MD petal limb cells (Van Moerkercke et al., 2009).
Despite the ecological and economic importance of plant-derived benzenoid compounds (e.g. as a phytohormone and as a food preservative), relatively little is known about the pathways that lead to the biosynthesis of many of these compounds (reviewed in Wildermuth, 2006). It appears that the exact biosynthetic steps remain unresolved because of the range of different chemical intermediates detected amongst different plant organs and/or plant species.
In contrast to the chemical variations in plants, benzenoids and phenylpropanoids are generally absent in bacterial species, which is mostly attributed to the lack of any recognizable PAL enzyme to initiate the core phenylpropanoid pathway. However, the marine isolate Streptomyces maritimus produces benzoyl-CoA as an intermediate during enterocin (enc) biosynthesis, which is a polyketide bacteriostatic agent (Moore et al., 2002). This bacterium produces benzoyl-CoA indirectly from phenylalanine in a PAL-like mediated manner (SmEncP), and by using t-cinnamic acid as an intermediate, much like what has been observed in plants (Xiang and Moore, 2002). S. maritimus employs a CoA-dependent, β-oxidative chemical scheme to produce benzoyl-CoA with a set of gene products from the enc gene cluster (Piel et al., 2000).
Subsequent to SmEncP (a unique bacterial PAL) function, benzoyl-CoA biosynthesis in S. maritimus includes the activation of t-cinnamic acid to cinnamoyl-CoA by the SmEncH protein, which is a putative acyl-CoA:ligase (Piel et al., 2000; Moore et al., 2002; Xiang and Moore, 2002, 2003). Also included in this gene cluster is an open reading frame encoding a putative β-oxoacyl-CoA thiolase (SmEncJ) enzyme that appears to utilize 3-oxo-3-phenylpropionyl-CoA as a substrate to form benzoyl-CoA directly (Piel et al., 2000; Xiang and Moore, 2003) much like the function of PhKAT1 in MD (Van Moerkercke et al., 2009).
In a continued effort to elucidate FVBP pathway enzymes, a MD transcript has been isolated and characterized that resembles SmEncH in amino acid sequence and function, Petunia×hybrida cv. ‘Mitchell Diploid’ ACYL-ACTIVATING ENZYME (PhAAE). Based on the utilization of several genomic resources, comparative transcript analyses, subcellular localization studies, and reverse genetics approaches, PhAAE appears to be the initial enzyme of FVBP metabolic branch 2 and is located in the peroxisomes of petunia flower petal limb cells.
Materials and methods
Plant material
Inbred Petunia x hybrid cv. ‘Mitchell Diploid’ (MD) plants were used as a ‘wild-type’ control in all experiments (Mitchell et al., 1980). The homozygous ethylene-insensitive CaMV 35S:etr1-1 line 44568, generated in the MD genetic background (Wilkinson et al., 1997), was used as a negative control for ethylene sensitivity. MD, 44568, ir-PhAAE, ir-PhMYB4, and irPhMYB4/ir-PhAAE plants were grown in glass greenhouses as previously described by Dexter et al. (2007).
PhAAE cloning
Partial sequences from the NCBI and Sol Genomics Network petunia EST databases were used as references to obtain full-length cDNAs by 5' and 3' race with the SMART™ RACE cDNA Amplification Kit (Clontech Laboratories, Inc., Mountain View, CA) as per manufacturer’s protocol (see Supplementary Fig. S1 at JXB online). A resulting 1997bp cDNA had a 1536bp coding sequence (GenBank accession number: JQ031717) for a predicted 512 amino acid protein and was termed PhAAE. The PhAAE cDNA sequence was amplified by Phusion™ Hot Start High-Fidelity DNA Polymerase (New England Biolabs, Inc., Ipswich, MA) and cloned into a pGEM®T-EASY vector (Promega Corp., Madison, WI), which were extensively sequenced and checked for errors. These constructs were used as a template to clone the predicted mature PhAAE coding sequences into a pET-32 EK/LIC vector (Novagen, Gibbstown, NJ), the RNAi induction plasmid (Underwood et al., 2005), and the AAE–GFP fusion construct (Van Moerkerker et al., 2009).
RNA isolation, tissue collection, and treatments
In all cases, total RNA was extracted as previously described by by Verdonk et al. (2003) and subjected to TURBO™ DNA-free™ treatment (Ambion Inc., Austin, TX) followed by total RNA purification with RNeasy® Mini protocol for RNA cleanup (Qiagen, Valencia, CA). Total RNA was then quantified on a NanoDrop™ 1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and 50ng µl–1 dilutions were prepared and stored at –20 °C. Generation of cDNA samples used 2 µg of total RNA with SuperScript® Reverse Transcriptase II (Invitrogen Corp., http://invitrogen.com) and it was conducted more than three times for technical replications.
All petunia tissue collections were done as previously described by Colquhoun et al. (2010a). The spatial transcript accumulation analysis consisted of total RNA isolated from petunia root, stem, stigma, anther, leaf, petal tube, petal limb, and sepal tissues from multiple greenhouse-grown MD plants harvested at 16.00h. The developmental transcript accumulation analysis used total RNA isolated from whole petunia flowers (MD and 44568) collected at 11 consecutive stages beginning at small bud to floral senescence from multiple greenhouse-grown plants at 16.00h. For all tissue collections, individual samples consisted of three flowers. All samples were frozen in liquid N2 and stored at –80 °C. Total RNA was then isolated from all samples, with multiple biological replicates included.
Transcript accumulation analysis of PhAAE
All transcript accumulation analyses were conducted multiple times with multiple biological replicates and equivalent results were observed. Semi-quantitative (sq)RT-PCR was performed using a Qiagen One-step RT-PCR kit (Qiagen Co., Valencia, CA, USA) with 50ng total RNA. To visualize RNA loading concentrations, samples were amplified with Ph18S primers and analyzed on an agarose gel. Primers were also designed and utilized for the visualization of the mRNA levels corresponding to PhAAE (see Supplementary Table S1 at JXB online). ∆∆Ct Quantitative (q)RT-PCR was performed and analyzed using a StepOnePlus™ real-time PCR system (Applied Biosystems, Foster City, CA). Power SYBR® Green RNA-to-CtTM 1 and 2-Step kits (Applied Biosystems, Foster City, CA) were used to amplify and detect the products according to the manufacturer’s protocol. The following, qRT-PCR primers were constructed in Primer Express® software v2.0: PhAAE, PhFBP1, PhCM1, PhPAL1, PhPAL2, PhBPBT, PhBSMT, PhC4H1, PhC4H2, PhIGS1, PhMYB4, PhPAAS, PhCFAT, PhKAT1, PhODO1 (see Supplementary Table S1 at JXB online). Optimization of primers was conducted and demonstrated gene specificity during melt curve analysis.
PhAAE RNAi
To directly test the gene function of MD PhAAE, stable RNAi based gene silencing was utilized. A 236 nt sequence at the 5' end of the PhAAE transcript was developed as the RNAi inducing fragment (PhAAE forward primer: 5'-CTTACCAAAATGTGGAGC ATG-3', and reverse primers: 5'-AATGCATTTCGTACGTTGCT-3', 5'-TATTCTCGGGAGCATATTCA-3'). When the 236 nt PhAAE RNAi trigger is fractionated into 22–25 nt fragments, then used as queries for BLAST analysis on the petunia EST databases; the only identical sequences that return belong to AAE petunia sequences. In planta expression of this fragment is driven by a constitutive promoter, pFMV. Fifty independent PhAAE RNAi (ir-PhAAE, inverted repeat) plants were generated in the MD background by leaf disc transformation (Jorgensen et al., 1996). Further details of the technical cloning have been previously described (Underwood et al., 2005; Dexter et al., 2007).
At least 19 T0 plants showing a transcriptional (sqRT-PCR) and physiological phenotype were self-pollinated. The T1 generation was analyzed for a 3:1 segregation based on the presence of the transgene and the transcriptional phenotype. In this manner, two T2 homozygous ir-PhAAE lines (ir-15.15 and ir-24.8) were produced.
Subcellular localization
To create the CaMV 35S-driven C-terminal GFP fusion construct (35S:AAE-GFP), the AAE CDS was PCR-amplified using forward primer 5'-gcTCTAGAccatggacgagttaccaaaatg-3' and reverse primer 5'-gcTCTAGAacctgccaaatgtacgataatatctg-3' (the XbaI restriction sites are in upper case). The fragment was cloned XbaI/XbaI to replace PhKAT1 into the previously created 35S:PhKAT-GFP shuttle vector, which contains an XbaI site at the 3'-end of the CaMV 35S promoter and is within the Ala-Gly linker between PhKAT1 and GFP (Van Moerkercke et al., 2009). The 35S:AAE-GFP:tnos cassette was sequenced and subsequently cloned into pBIN+ (Vanengelen et al., 1995) and transferred to A. tumefaciens GV3101 (pMP90) as described by Van Moerkercke et al. (2009). The 35S-driven mCherry (Shaner et al., 2004) was cloned into pCAMBIA.
Agro-infiltration of petunia petals was performed as described by Verweij et al. (2008). To enable co-localization, two separate A. tumefaciens GV3101 cultures harbouring the PhAAE–GFP fusion construct or the peroxisomal marker px-rk, which contains mCherry with a C-terminal peroxisomal targeting signal 1 (Nelson et al., 2007), respectively, were mixed 1:1 (v/v) prior to infiltration. For confocal analysis, infiltrated petals were embedded in 80% glycerol to reduce light scattering of the conical epidermal cells. GFP and mCherry were imaged using a Zeiss LSM 510 confocal laser scanning microscope (Vermeer et al., 2008).
Confocal microscopy
Agrobacterium-mediated transient transformation of petunia petals was performed as described by Van Moerkercke et al. (2009). Arabidopsis protoplasts were prepared as described by Dangl et al. (1987) with modifications described by Meskiene (2003). Five µg of plasmids were co-transfected as described by Meskiene et al. (2003) and cultured overnight before analysis. Petunia hybrida cv. Mitchell petals and Arabidopsis protoplasts were observed using a 20× water objective with a Nikon Eclipse T1 microscope coupled to an Nikon A1 confocal scanning head. Green Fluorescent Protein fluorescence was monitored with a 500–550nm band pass emission filter (488nm excitation) and red fluorescent protein was visualized with a 570–620nm band pass emission filter (561nm excitation). The images were processed by ImageJ (http://rsb.info.nih.gov/ij/) and assembled in MS Power Point.
Recombinant protein production and enzyme assay of PhAAE
PhAAE target was amplified with high fidelity Phusion™ DNA polymerase (New England Bio-Labs, Ipswich, MA) using specific adapter primers to pET-32 vector (Novagen, San Diego, CA) at the 5' and 3' ends of the coding sequence (forward primer: 5'-GACGACGACAAGATGGACGAGTTACCAAAATGTGGAG-3', and reverseprimer: 5'-GAGGAGAAGCCCGGTCTCAAATGTACGAT AATATCTGCTTCT-3'). T4 DNA polymerase generated compatible overhangs of the gene sequence with the adapter to anneal the PhAAE target and the Ek/LIC vector, then transformed using NovaBlue GigaSingle Competent Cells (Novagen). Traditional restriction enzyme digests were performed to confirm proper plasmid sequence prior to pET-32 transformation into E. coli BL21(DE3) host strain.
Biologically active pET-32-PhAAE was expressed in E. coli BL21/(DE3) with an induction of 1mM isopentylgalactose (IPTG) overnight at 37 °C. Induction was analyzed from crude cellular extracts on a 10% polyacrylamide, TRIS-HCl Ready Gel (Bio-Rad). Recombinant protein was obtained from induced cells lysed with BugBuster® protein extract reagent (Novagen, San Diego, CA), pET-32-PhAAE localized to the insoluble fraction was inclusion body purified; washed inclusion bodies were solubilized using the Protein Refolding Kit (Novagen, EMD Chemicals). The resulting proteins were separated from any low molecular weight compounds and concentrated with 30 000 NMWL Amicon® Ultra-4 centrifugal filter devices (Millipore, Billerica, MA) and affinity purified with His-Band® resin chromatography (Novagen). Protein refolding dialysis was performed according to the patented procedure (Burgess, 1996) using Slide-A-Lyzer G2 Dialysis Cassettes, 20K MWCO. The recombinant protein concentration was quantified by the Bradford method using BSA as a standard.
Enzyme assays were performed at standard conditions. Assay components included 100mM TRIS (pH 8), 10mM MgCl2, 3mM rATP, 2.5mM DTT, 1mM CoA, 0.5mM t-cinnamic acid, and 10 µg total protein. Spectrophotometric readings at 259nm were used to monitor ATP absorbance, and at 267nm for t-cinnamic acid absorbance.
Volatile emission
For all volatile emission experiments, emitted floral volatiles from excised flowers were collected at 18.00h and quantified as previously described (Underwood et al., 2005; Dexter et al., 2007). All samples consisted of three flowers per sample with at least three biological replicates. Flowers were collected 2 d after the initial opening at 18.00h. Volatile analyses were conducted as previously described by Colquhoun et al. (2010b).
GenBank® accession numbers
Arabidopsis thaliana ACYL-ACTIVATING ENZYME11 (AtAAE11), AY250841; Petunia×hybrida cv. MD PHENYLACETALDEHYDE SYNTHASE (PhPAAS), DQ243784; PHENYLALANINE AMMONIA-LYASE1, 2 (PhPAL1, 2), AY705976 and CO805160; Hordeum vulgare HvPAL, Z49147; Petroselinum cripsum PcPAL, Y07654; CINNAMATE-4HYDROXYLASE (AtC4H), U71081; PhC4H1 and PhC4H2; HM447144 and HM447145, BENZOIC ACID/SALICYLIC ACID METHYL TRANSFERASE (PhBSMT1, 2), AY233465 and AY233466; BENZOYL-COA:BENZYL ALCOHOL/PHENYLETHANOL BENZOYLTRANSFERASE (PhBPBT), AY611496; 3-KETOACYL COA THIOLASE 1 (PhKAT1), FJ657663; Streptomyces maritimus enterocin biosynthetic gene cluster (SmEnc), AF254925; ACYL-ACTIVATING ENZYME (PhAAE), JQ031717; BENZOYLOXYGLUCOSINOLATE1 (AtBZO1), NM_105260; AtAAE12, NM_105261; A. thaliana AMP-DEPENDENT SYNTHASE AND LIGASE –LIKE, AAE34775; SmEncH, AAF81723; CHORISMATE MUTASE1 (PhCM1), EU751616, AROGENATE DEHYDRATASE1 (PhADT1), FJ790412; CONIFERYL ALCOHOL ACYLTRANSFERASE (PhCFAT), DQ767969; ISOEUGENOL SYNTHASE1 (PhIGS1), DQ372813; ODORANT1 (PhODO1), AY705977; MYB4 (PhMYB4), HM447143.
Results
Identification of PhAAE
The characterization of a plant thiolase involved in benzoyl-CoA biosynthesis, and the observation that a β-oxidative chemical strategy is the main contributor to the formation of benzenoid compounds in a petunia flower in the dark period (Van Moerkercke et al., 2009); led us to examine the hypothetical, biochemical steps preceding Petunia×hybrida cv. ‘Mitchell Diploid’ (MD) 3-KETOACYL-COA THIOLASE (PhKAT1). The initial step emanating from the core phenylpropanoid pathway may involve the biochemical activation of t-cinnamic acid resulting in cinnamoyl-CoA (Fig. 1), which would consist of enzyme, t-cinnamic acid, coenzyme A (CoA) molecules, and energy in the form of adenosine triphosphate (ATP). This model is similar to an already identified and somewhat unique biochemical process described in the marine isolate Streptomyces maritimus (Piel et al., 2000). The S. maritimus protein EncH is homologous to long-chain fatty acid-CoA:ligases, contains a putative AMP-dependent synthase/ligase domain (Pfam: PDOC00427, IPR000873), and enzymatically activates t-cinnamic acid to form cinnamoyl-CoA (Xiang and Moore, 2003). The SmEncH amino acid sequence was used to search for petunia transcript sequences from expressed sequence tagged (EST) databases housed at NCBI (http://www.ncbi.nlm.nih.gov/guide/) and Sol Genomics Network (http://solgenomics.net/). The search resulted in many petunia ESTs with homology to 4-coumarate:CoA ligases and acyl-activating enzymes from various plant species. When the resulting ESTs were assayed for similarity using the ContigExpress module of Vector NTI Advanced® 11.0, a single contig was created suggesting that all the ESTs originated from a single transcript that was well-represented in the EST collections (Colquhoun et al., 2011b).
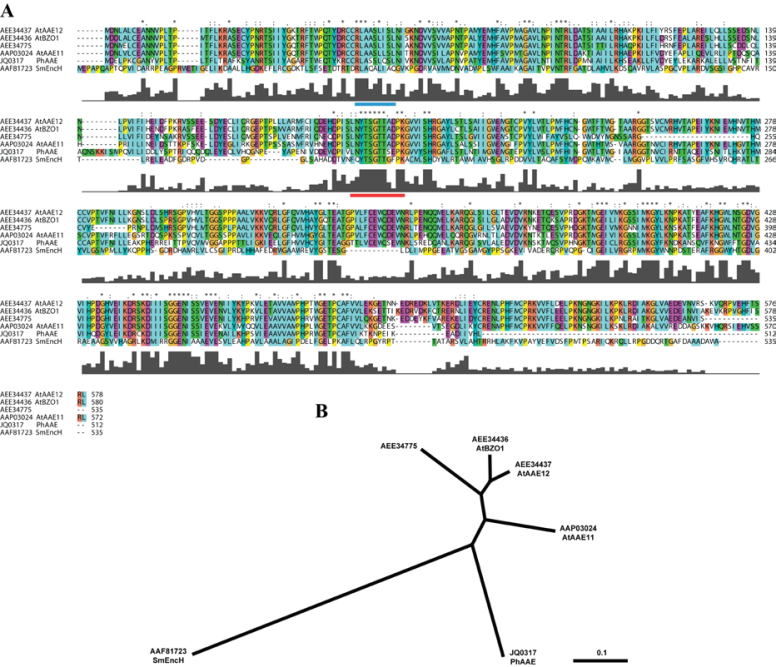
A SMART RACE library constructed from MD flower limb tissue was utilized first to amplify the 5' and 3' segments of the putative MD CoA:ligase with a high fidelity polymerase. These segments enabled the cloning of the full-length transcript (see Supplementary Fig. S1 at JXB online) (GenBank accession no. JQ031717). This transcript was 1997 base pairs (bp) in length and contained a 1536bp coding region for a 512 amino acid product (see Supplementary Fig. S1 at JXB online). Predicted amino acid features include an AMP-dependent synthase/ligase domain (IPR000873) and an AMP-binding, conserved site (IPR020845). The protein sequence was most similar to plant proteins of the acyl-activating enzyme (AAE) family found in Arabidopsis thaliana (Shockey et al., 2003; Shockey and Browse, 2011). An alignment of the predicted petunia protein with three of the most similar members of the A. thaliana AAE family, along with the SmEncH protein, clearly demonstrated the similarities in primary sequence features (Fig. 2A). As expected, the AMP-binding, conserved sequence (LNYTSGTTSEPK) is highly similar throughout the A. thaliana AAE protein family, between the Brassicaceae and Solanaceae plant families, and amid the Plantae and Bacteria kingdoms. Hierarchical association can be readily observed in a phylogenetic tree with the bacterial sequence farthest removed (Fig. 2B).
Fig. 2.
Predicted amino acid sequence alignment and phylogenetic tree. Sequences represented are from Streptomyces maritimus, Arabidopsis thaliana, and Petunia×hybrida. Sequences were aligned using MULTIPLE ALIGNMENT MODE of ClustalX 2.0 program software (Larkin et al., 2007). Above the sequence alignment ‘*’ indicates a fully conserved residue, ‘:’ indicates a fully conserved ‘strong’ group, and ‘.’ indicates a fully conserved ‘weak’ group. Colours are assigned to groups of amino acids as follows: orange for G, P, S, and T; red for H, K, and R; blue for F, W, and Y; and green for I, L, M, and V. Predicted PTS2 (Do-It-Yourself Block Search http://www.peroxisomedb.org/diy_PTS1.html) is represented by a blue bar below residues 51–59 of PhAAE, while the conserved AMP-binding site (InterProScan, http://www.ebi.ac.uk/Tools/pfa/iprscan/) is represented by a red bar below residues 197–208 of PhAAE (A). The Neighbor–Joining tree was derived by ClustalX and visualized in TreeView 1.6.6 as an unrooted tree (B).
Comparative transcript accumulation analysis
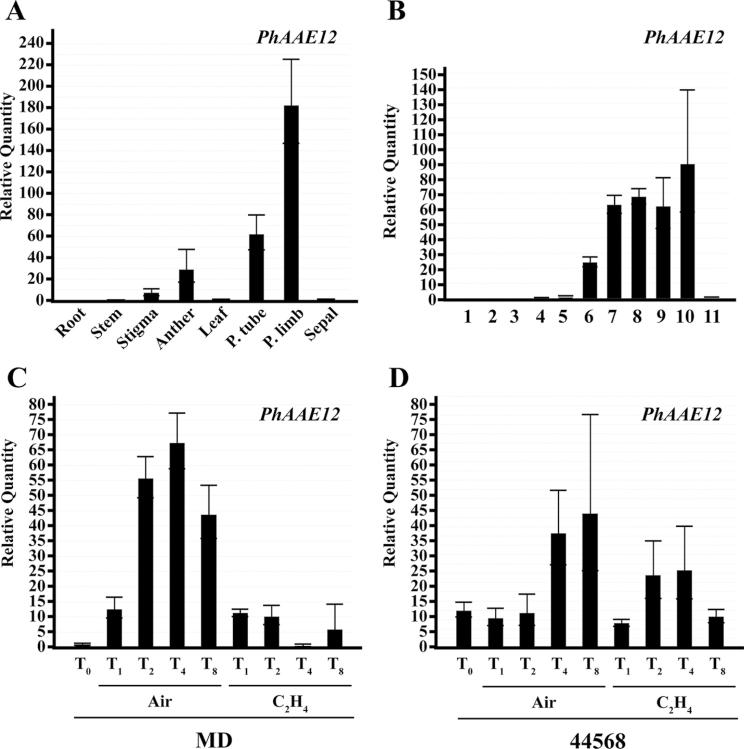
Under the assumption that the petunia homolog identified here and SmEncH share enzymatic function, and this function is required for benzoyl-CoA biosynthesis during FVBP production; it was hypothesized that PhAAE transcript accumulation would be highest in the petal limb much like FVBP genes previously identified. A standard MD spatial series of tissue was used for a ∆∆Ct quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) transcript accumulation assay, which consisted of roots, stem, stigma, anther, leaf, petal tube, petal limb, and sepal tissue. PhAAE transcript was detected at an approximately 185 times higher level in the petal limb compared to leaf tissue (Fig. 3A). Overall transcript accumulation of PhAAE was mainly detected in floral tissues with appreciable detection in stigma, anther, petal tube, and, of course, petal limb tissue.
Fig. 3.
PhAAE transcript accumulation analysis (one-step qRT-PCR). Spatial analysis used root, stem, stigma, anther, leaf, petal tube, petal limb, and sepal tissues of MD harvested at 16.00h (A). Floral developmental analysis used MD flowers from 11 sequential stages collected on one day at 16.00h (B). Ethylene treatment (2 µl l–1 analysis used excised MD and 44 568 whole flowers treated for 0, 1, 2, 4, and 8h (C, D). 50ng total RNA was used per reaction in all cases. Histograms are representative of multiple experiments and multiple biological replicates, and analyzed by the ∆∆Ct method with PhFBP1 and Ph18S as the internal references (mean±SE; n=3).
Since the petal limb tissue used for the spatial analysis was that of an open flower, it was further hypothesized that the PhAAE transcript would accumulate to high levels in an open flower compared to a closed, growing flower bud or senescing flower tissue. Therefore, a flower developmental tissue series from MD and the transgenic, ethylene-insensitive 44568 plants (Wilkinson et al., 1997; Colquhoun et al., 2010a) was used to assay for a developmental transcript accumulation profile of PhAAE. In both MD and 44568, PhAAE transcript was not detected in a closed flower bud (see Supplementary Fig. S2A at JXB online; Fig. 3B). PhAAE was initially detected at considerable levels at stage 6, which is anthesis, and accumulated to high levels throughout the open flower stages of development. In MD, PhAAE transcript accumulation was reduced at stage 11 (Fig. 3B). Stage 11 is the end of the flower life-cycle where the flower progresses through an ethylene-mediated down regulation of the FVBP pathway and a general senescence program (Wilkinson et al., 1997; Underwood et al., 2005; Colquhoun et al., 2010a). By contrast, the PhAAE transcript was not reduced at stage 11 (observably senescing) in flowers of ethylene-insensitive 44568 plants (see Supplementary Fig. S2A at JXB online), suggesting that this transcript is regulated by an ethylene-mediated signal at this stage of flower development.
Based on the developmental disparity of PhAAE transcript accumulation between MD and 44568 flowers at senescence (see Supplementary Fig. S2A at JXB online); the transcript accumulation profile of PhAAE was examined after ethylene treatment in MD and 44568 flowers using both (sq) and qRT-PCR techniques (Fig. 3C, 3D; see Supplementary Fig. S2B, C at JXB online). In MD flowers, PhAAE transcript accumulation was reduced after 2h of ethylene exposure compared with an air-treated flower (Fig. 3C). Comparatively, no detectable to slight change in PhAAE transcript accumulation was observed in 44568 flowers after ethylene treatment (Fig. 3D). Together, these results suggest PhAAE transcript is regulated by ethylene signalling at senescence like that of members of the FVBP gene network.
PhAAE recombinant enzyme assay
In an attempt to test the biochemical role of PhAAE more directly, its coding sequence was expressed in E. coli BL21/(DE3) cells and the recombinant protein was isolated and purified. Unfortunately, the recombinant protein was sequestered to inclusion bodies in every attempt to induce protein production (see Supplementary Fig. S3 at JXB online). Therefore, purified protein (from inclusion bodies) was chemically refolded and utilized for enzymatic assays. Recombinant PhAAE protein showed activity with t-cinnamic acid, CoA, and ATP for minutes only (data not shown) and, therefore, was not conclusive for identifying a specific biochemical function. Any future studies may benefit from a eukaryotic expression system of some sort.
Subcellular localization
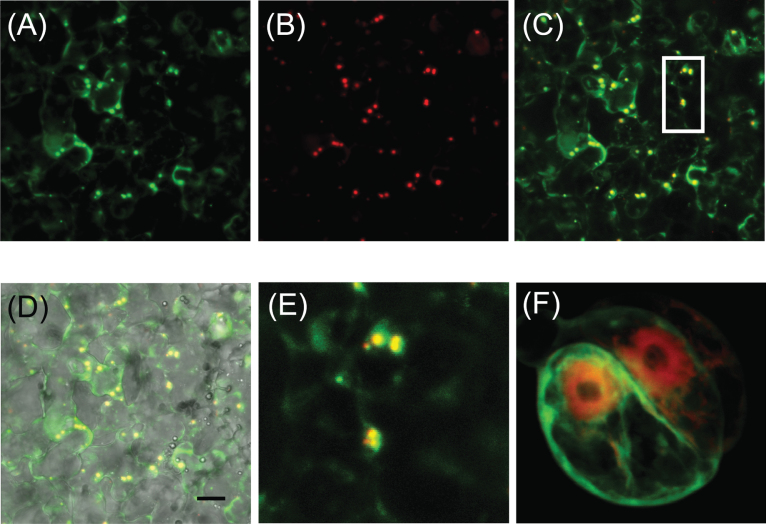
PhAAE is predicted to encode a protein of 512 amino acids containing a well-conserved AMP-binding site (IPR020845) and a putative, weak N-terminal PTS2 (peroxisomal targeting sequence II) site (Fig. 2). To verify whether PhAAE is localized to peroxisomes in planta, a PhAAE-GFP fusion vector was constructed and expressed in MD petals by way of an Agrobacterium-mediated transient transformation (Shang et al., 2007; Van Moerkercke et al., 2009). PhAAE-GFP co-localizes with the mCherry-peroxisomal marker px-rk (Nelson et al., 2007) (Fig. 4), which indicates peroxisomal targeting of PhAAE in planta. By contrast, PhAAE does not co-localize with mCherry in Arabidopsis protoplasts (Fig. 4F) or Nicotiana tobacum leaves (data not shown).
Fig. 4.
In vivo subcellular localization of PhAAE in MD petals and Arabidopsis protoplasts. (A) 35S:PhAAE-GFP and (B) the mCherry peroxisomal marker px-rk (Nelson et al., 2007) were co-infiltrated using A. tumefaciens in MD petal tissue. (C) The merged image of(A) and (B). (D) A merge of (C) and a brightfield image. (E) Magnification of the boxed area in (C). (F) Co-expression of a 35S:mCherry and 35S:PhAAE:GFP in Arabidopsis protoplasts. Representative images are shown. The scale bar in (D) represents 10 µm.
Functional characterization of PhAAE
PhAAE is very similar in its primary amino acid sequence to CoA:ligase family proteins from many other plant species (Fig. 2), and shares considerable similarity to a CoA:ligase that utilizes t-cinnamic acid from the bacterium S. maritimus (Piel et al., 2000). PhAAE transcript accumulates to peak levels in tissue producing large amounts of volatile organic compounds derived from t-cinnamic acid through a β-oxidative pathway. Therefore, it was hypothesized that, upon reduction of endogenous PhAAE transcript, a concomitant reduction in FVBPs derived from t-cinnamic acid would result.
A 5' segment of the PhAAE transcript (see Supplementary Fig. S4 at JXB online) was used to create approximately 50 independent T0 ir-PhAAE (stable RNAi, inverted repeat) plants. These transgenic plants were generated in the MD genetic background by leaf-disc transformation (Jorgensen et al., 1996; Underwood et al., 2005; Dexter et al., 2007; Colquhoun et al., 2010b). Comparison (BLAST analysis in multiple databases: NCBI, SGN, and Petunia inflata genome) of the PhAAE 5' segment used for the RNAi component of the T-DNA resulted in nucleotide sequences matching PhAAE only (i.e. the RNAi induction sequence used for PhAAE seems unique to itself). All T0 plants exhibiting a reduction of PhAAE transcript in stage eight flowers compared to MD flowers (see Supplementary Fig. S5 at JXB online) were self-pollinated and the subsequent T1 generation was screened for the presences of a genomic element (the T-DNA itself) and a molecular component (reduction of PhAAE transcript). Two independent lines (ir-PhAAE 15.15 and ir-PhAAE 24.8) demonstrated a Mendelian segregation ratio in the T1 generation with a lack of segregation in the T2 generation, which is highly suggestive of a homozygous transgenic line (methodology further detailed in Colquhoun et al., 2011b).
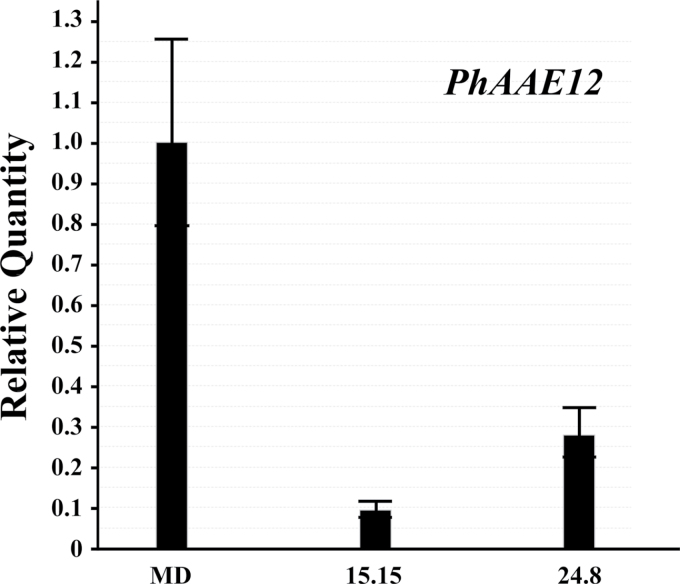
PhAAE transcript was reduced in the independent, homozygous ir-PhAAE 15.15 and 24.8 lines by approximately 93% and 75%, compared with MD (Fig. 5). As expected from wild-type transcript accumulation profiles of PhAAE (mainly in the flower tissues; Fig. 3); from germination to flower senescence, no visual phenotypes were readily observed when compared with MD plants at similar developmental stages.
Fig. 5.
PhAAE comparative transcript accumulation analysis between MD and two independent, homozygous T2 ir-PhAAE lines (15.15 and 24.8). 50ng total RNA was used per reaction in all cases for one-step qRT-PCR with RNA isolated from stage 8 flowers at 16.00h. Histograms are representative of multiple experiments and multiple biological replicates, and analyzed by the ∆∆Ct method with PhFBP1 and Ph18S as the internal references. The individual petunia transcript analyzed is PhAAE (mean±SE; n=3).
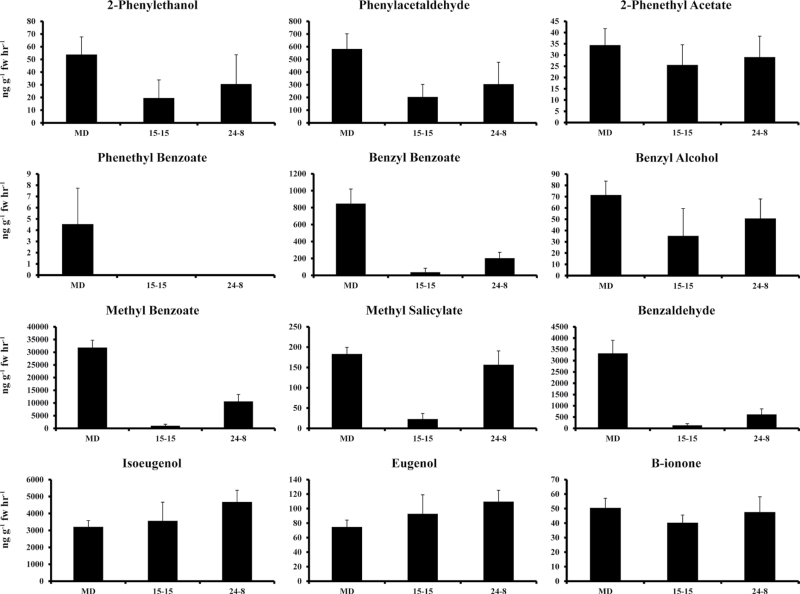
When FVBP emission from the ir-PhAAE lines was compared with MD, the original hypothesis was supported with results illustrating a reduction in FVBPs derived from t-cinnamic acid (Fig. 6). The most abundant FVBP, methyl benzoate (produced from benzoic acid by two isoforms of a methyl transferase, PhBSMT1 and 2 (Negre et al., 2003; Underwood et al., 2005)) was reduced by approximately 96% in ir-PhAAE 15.15 and 67% in ir-PhAAE 24.8 (Fig. 6), which was similar in magnitude to the reduction of PhAAE transcript in the respective, transgenic plant lines (Fig. 5). Two FVBPs directly derived from benzoyl-CoA by the benzoyl transferase, PhBPBT (Dexter et al., 2007) were also severely reduced in the ir-PhAAE lines compared with MD (Fig. 6). Benzaldehyde emission was reduced in the ir-PhAAE 15.15 line by 96%, which closely mirrors the percentage reduction observed for methyl benzoate emission. Benzyl alcohol emission was more variable between collections (a relatively low emitted FVBP in MD; Colquhoun et al., 2010a), but emission of this FVBP was reduced by approximately 50% and 29% in ir-PhAAE 15.15 and 24.8, respectively (Fig. 6).
Fig. 6.
FVBP emission analysis of representative plants from two independent, homozygous T2 ir-PhAAE lines (15.15 and 24.8) and MD. Developmentally staged flowers (stage 8) were used to collect FVBP emission at 18:00h (mean±SE; n=3). Twelve major FVBP compounds were identified and quantified with all measurements ng g–1 FW h–1.
Emission of FVBPs derived more directly from phenylalanine and p-coumaric acid was less altered in the transgenic lines. Specifically, emission of phenylacetaldehyde and 2-phenylethanol were slightly reduced in the ir-PhAAE lines, while isoeugenol and eugenol were slightly elevated in the ir-PhAAE lines, all compared with MD (Fig. 6).
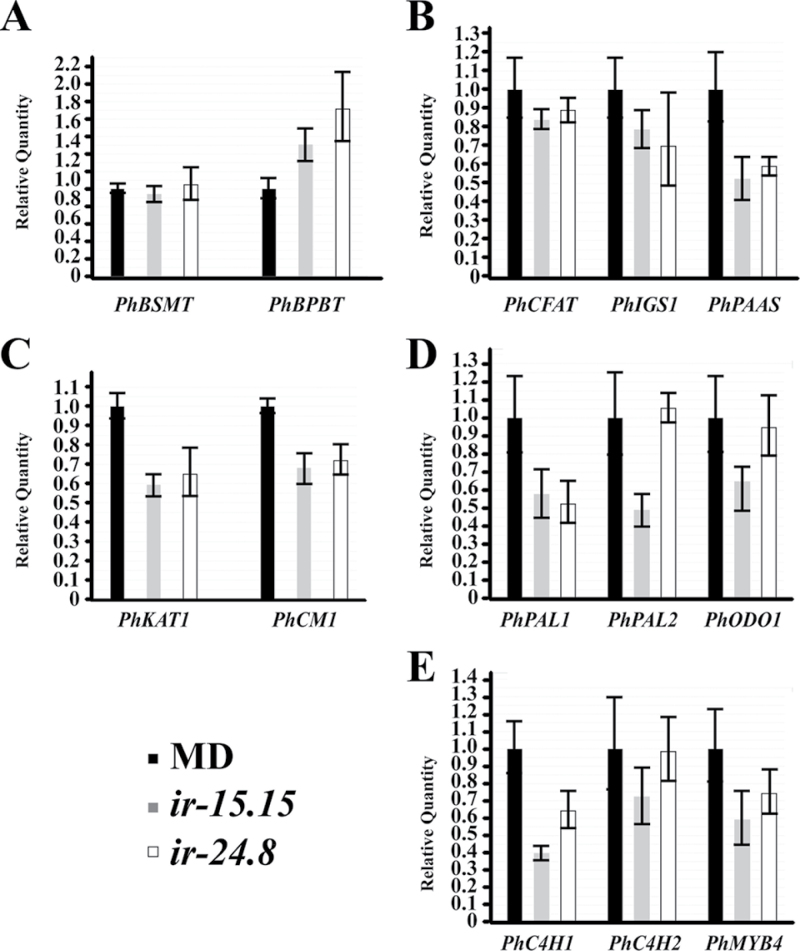
The FVBP metabolic branch phenotype of the RNAi lines (Fig. 6), the subcellular localization of PhAAE (Fig. 4), and the observation that cinnamic acid can have a feedback regulatory effect on the phenylpropanoid pathway; led us to ask whether there were any effects to the known FVBP ‘transcriptome’. A comparative transcript accumulation analysis between MD and ir-PhAAE lines illustrated a perturbation to the FVBP gene network system (Fig. 7). PhBSMT transcript accumulation was unchanged from MD in the RNAi lines, but PhBPBT transcript was elevated in ir-PhAAE 15.15 and 24.8 (Fig. 7A). The FVBP branch 3 transcripts PhCFAT and PhIGS1 were not significantly changed compared with MD (Fig. 7B). PhPAAS (FVBP branch 1) transcript level was between 50 to 60% compared with MD transcript accumulation (Fig. 7B), which coincides with the reduction of emitted phenylacetaldehyde from the ir-PhAAE lines (Fig. 6). The only other empirically demonstrated FVBP protein to be localized in the peroxisomes of MD petal cells, PhKAT1 transcript was approximately 60% of MD levels, and transcript level of the plastid localized CHORISMATE MUTASE1 (PhCM1) protein was reduced to about 70% of MD transcript levels (Fig. 7C). The core phenylpropanoid transcripts for PhPAL1, PhPAL2, and PhC4H1 were all reduced compared with MD (Fig. 7D, 7E), except for PhPAL2 in ir-PhAAE 24.8 were transcript was similar to MD (Fig. 7D). Noteworthy here was a petunia R2R3-MYB (PhODO1) transcript level in ir-PhAAE 24.8 where it was similar to MD, while ir-PhAAE 15.15 it was reduced (Fig. 7D). Another petunia R2R3-MYB transcript PhMYB4 was slightly reduced compared with MD (Fig. 7E), which demonstrated that the reduction of PhC4H is not due to an increase of its negative regulator PhMYB4 (Colquhoun et al., 2011b).
Fig. 7.
PhAAE comparative transcript accumulation analysis between MD and two independent, homozygous T2 ir-PhAAE lines (15.15 and 24.8). 50ng total RNA was used per reaction in all cases for one-step qRT-PCR with RNA isolated from stage 8 flowers at 16.00h. Histograms are representative of multiple experiments and multiple biological replicates, and analyzed by the ∆∆Ct method with PhFBP1 and Ph18S as the internal references. The individual petunia transcripts analyzed are PhBSMT, PhBPBT, PhCFAT, PhIGS1, PhPAAS, PhKAT1, PhCM1, PhPAL1, PhPAL2, PhODO1, PhC4H1, PhC4H2, and PhMYB4 (mean±SE; n=3).
Many of the previously mentioned genes, transcripts, and/or protein functions have been illustrated with reverse genetic approaches, which have produced transgenic plant lines with very specific FVBP phenotypes. These plants and phenotypes can be utilized to examine the metabolic pathway hypothesis further. One example, the transcriptional regulator PhMYB4 negatively regulates PhC4H genes and, upon reduction of PhMYB4 through stable RNAi approaches, PhC4H transcript levels are increased resulting in an increased isoeugenol and eugenol emission from the petunia flower (Colquhoun et al., 2011b). In a condition were PhC4H levels and overall activity are increased (ir-PhMYB4 lines), if levels of t-cinnamic acid were increased (presumably in the ir-PhAAE lines), a further increase of isoeugenol and eugenol could be produced. In short, it was hypothesized that FVBP metabolic branch 3 is not at a biochemical limit even in the ir-PhMYB4 lines. In an attempt to address this hypothesis, homozygous ir-PhMYB4 lines and homozygous ir-PhAAE lines were crossed several times, the progeny grown, and then assayed for an additive FVBP phenotype. However, in every case, an additive volatile phenotype was not observed (see Supplementary Fig. S6 at JXB online).
Discussion
Benzenoid biosynthesis appears to begin with t-cinnamic acid and PhAAE in the peroxisome of petunia petal limb cells. This is not surprising since benzenoid formation in petunia follows a β-oxidative chemical strategy much like fatty acid degradation (Boatright et al., 2004; Van Moerkercke et al., 2009), which occurs in the peroxisomes of many plants (Goepfert and Poirier, 2007). In addition, the compartmentalization of t-cinnamic acid into peroxisomes has been suggested over a decade ago, and may be a regulatory mechanism to prevent t-cinnamic acid accumulation in the cytoplasm where there is a high probability of it being involved in a feedback regulatory mechanism of phenylpropanoid biosynthesis (Blount et al., 2000).
PhAAE is the latest member added to the growing list of the floral volatile benzenoid/phenylpropanoid (FVBP) gene network in MD. Transcript accumulation of PhAAE (Fig. 3) is very similar to what has been identified for all the FVBP genes, whereby, transcript accumulates to high levels in open flower limb tissue and is negatively regulated by ethylene (Van Moerkercke et al., 2009; Colquhoun et al., 2010a, b, 2011b; Maeda et al., 2010). PhAAE is localized in the same subcellular compartment as PhKAT1, an already characterized protein involved in the same branch of FVBP biosynthesis (Van Moerkercke et al., 2009). The predicted PhAAE protein contains an AMP-dependent ligase domain and an extremely conserved AMP-binding site (Fig. 2), and recombinant PhAAE protein showed weak signs of utilizing t-cinnamic acid in an ATP-dependent manner. This is all very similar to SmEncH, a protein from a benzoyl-CoA producing species of Streptomyces that catalyzes a reaction of t-cinnamic acid, CoA, and ATP to form cinnamoyl-CoA (Xiang and Moore, 2003). Upon a reduction of endogenous PhAAE transcript through a reverse genetic approach, all emitted benzenoid volatile compounds derived from t-cinnamic acid are reduced (Figs 5, 6). In summary, PhAAE is most likely a new member to the FVBP gene network, and functions in a concerted manner ultimately to produce the specific floral volatile bouquet emitted by MD plants.
The characterization of PhAAE has brought another important aspect of cellular regulation to the forefront, compartmentalization of metabolites and proteins. Tracking the carbon flow of benzenoid biosynthesis starting in the plastid from the shikimate pathway, the benzene ring would initially be biosynthesized as chorismic acid and then enzymatically manipulated to phenylalanine (Rippert et al., 2009; Colquhoun et al., 2010b; Maeda et al., 2010). Soluble PAL enzyme then utilizes phenylalanine to form t-cinnamic acid, but soluble PAL proteins are located in the cytosol (Achnine et al., 2004), so phenylalanine should somehow be exported from the plastid. Subsequent to t-cinnamic acid formation in the cytosol, t-cinnamic acid must then be imported (somehow) into the peroxisome where PhAAE could generate cinnamoyl-CoA, which initiates an enzymatic cascade that results in a two carbon reduction from the side chain of t-cinnamic acid ultimately producing benzoic acid. Of particular future interests are the possible existence of t-cinnamic acid transport and partitioning mechanisms this study and previous studies have suggested exist. Also of importance will be further comparisons between MD and Arabidopsis with respect to PhAAE peroxisomal import (Fig. 4), since PhAAE was not localized to peroxisomes in Arabidopsis.
PhAAE and PhKAT1 are localized to the peroxisome and are involved in floral volatile benzenoid formation (Figs 4, 6) (Van Moerkercke et al., 2009). MD flower petal limb cells maintain a CoA dependent/β-oxidative pathway to generate volatile benzenoid compounds (Boatright et al., 2004; Van Moerkercke et al., 2009). β-oxidative chemical strategies incorporating various metabolites have been described in peroxisomes of plants (Goepfert and Poirier, 2007). All of this may suggest a benzoic acid pathway and/or ‘cycle’ (including t-cinnamic acid, cinnamoyl-CoA, 3-oxo-3-phenylpropionyl-CoA, benzoyl-CoA, benzyl benzoate, and benzoic acid; http://metacyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-6443) is localized to the peroxisome organelles in petunia flowers. Since benzyl benzoate could be a key metabolite of this cycle and an emitted volatile compound from MD flowers, it would be of real interest to assay for PhBPBT localization with the hypothesis that PhBPBT is localized to the peroxisome. Interestingly, PhBPBT transcript levels were elevated in the ir-PhAAE flowers, but PhBSMT transcript accumulation mirrored that of MD flowers (Fig. 7). Assuming benzoic acid is ultimately formed in the peroxisome; is benzoic acid then transported from the peroxisome to support methyl benzoate biosynthesis since PhBSMT is most likely located in the general cytosolic compartment (Kolosova et al., 2001b). Elucidating these questions may very well generate a new mechanism of regulation imparted upon FVBP biosynthesis at numerous metabolite and protein interactions.
This study identified, isolated, and characterized a new member of the FVBP gene network in MD. Through this work, PhAAE has been empirically supported to function in the biosynthesis of benzenoid compounds in peroxisomal cellular compartments of flower petal limb cells. The compartmentalization of PhAAE in MD and the suspected substrate of PhAAE introduce the rational probability that t-cinnamic acid is imported into peroxisomes. In addition, the results of this study suggest that benzoyl-CoA formation in MD flowers occurs much like that found in a select number of bacterial species (i.e. S. maritimus).
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Transcript specific primers used for transcript accumulation analyses (sqRT-PCR and qRT-PCR) throughout this study.
Supplementary Fig. S1. A schematic of the PhAAE cloning strategy.
Supplementary Fig. S2. A comparative PhAAE transcript accumulation analysis with MD and 44568 (sqRT-PCR).
Supplementary Fig. S3. PhAAE recombinant protein expression in E. coli BL21/(DE3) cells.
Supplementary Fig. S4. Schematic representation of the full-length PhAAE transcript model as shown in Vector NTI Advance™ 11.
Supplementary Fig. S5. Comparative sqRT-PCR transcript accumulation analysis.
Supplementary Fig. S6. FVBP emission analysis of representative plants from MD, ir-PhMYB4, ir-PhAAE, and resulting progeny from crossing ir-PhMYB4 and ir-PhAAE.
Supplementary Material
Acknowledgements
This work was supported by grants from the USDA Nursery and Floral Crops Initiative, the Fred C. Gloeckner Foundation, and the Florida Agricultural Experiment Station. We thank Carlos Galván-Ampudia for help with confocal microscopy and Anna Pietraszewska for the 35S:mCherry binary construct. Microscopy was performed at the Advanced Centre for Microscopy of the University of Amsterdam.
References
- Achnine L, Blancaflor EB, Rasmussen S, Dixon RA. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. The Plant Cell. 2004;16:3098–3109. doi: 10.1105/tpc.104.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anterola AM, Jeon JH, Davin LB, Lewis NG. Transcriptional control of monolignol biosynthesis in Pinus taeda: factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. Journal of Biological Chemistry. 2002;277:18272–18280. doi: 10.1074/jbc.M112051200. [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E. Purification and characterization of benzoate:coenzyme A ligase from Clarkia breweri . Archives of Biochemistry and Biochemistry. 2002;400:258–264. doi: 10.1016/S0003-9861(02)00026-7. [DOI] [PubMed] [Google Scholar]
- Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA. Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiology. 2000;122:107–116. doi: 10.1104/pp.122.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright J, Negre F, Chen X, Kish CM, Wood B, Peel G, Orlova I, Gang D, Rhodes D, Dudareva N. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiology. 2004;135:1993–2011. doi: 10.1104/pp.104.045468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RR. Purification of overproduced Escherichia coli RNA polymerase sigma factors by solubilizing inclusion bodies and refolding from Sarkosyl. Methods in Enzymology. 1996;273:145–149. doi: 10.1016/s0076-6879(96)73014-8. [DOI] [PubMed] [Google Scholar]
- Chen X-M, Kobayashi H, Sakai M, Hirata H, Asai T, Ohnishi T, Baldermann S, Watanabe N. Functional characterization of rose phenylacetaldehyde reductase (PAR), an enzyme involved in the biosynthesis of the scent compound 2-phenylethanol. Journal of Plant Physiology. 2011;168:88–95. doi: 10.1016/j.jplph.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Colquhoun TA, Clark DG. Unraveling the regulation of floral fragrance biosynthesis. Plant Signaling and Behavior. 2011a;6:378–381. doi: 10.4161/psb.6.3.14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, Schuurink RC, Clark DG. PhMYB4 fine-tunes the floral volatile signature of Petunia×hybrida through PhC4H. Journal of Experimental Botany. 2011b;62:1133–1143. doi: 10.1093/jxb/erq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Schimmel BC, Kim JY, Reinhardt D, Cline K, Clark DG. A petunia chorismate mutase specialized for the production of floral volatiles. The Plant Journal. 2010b;61:145–155. doi: 10.1111/j.1365-313X.2009.04042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Verdonk JC, Schimmel BC, Tieman DM, Underwood BA, Clark DG. Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry. 2010a;71:158–167. doi: 10.1016/j.phytochem.2009.09.036. [DOI] [PubMed] [Google Scholar]
- Dangl JF, Hauffe KD, Lipphardt S, Hahlbrock K, Scheel D. Parsley protoplasts retain differential responsiveness to UV light and fungal elicitor. EMBO Journal. 1987;6:2551–2556. doi: 10.1002/j.1460-2075.1987.tb02543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter R, Qualley A, Kish CM, Ma CJ, Koeduka T, Nagegowda DA, Dudareva N, Pichersky E, Clark D. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. The Plant Journal. 2007;49:265–275. doi: 10.1111/j.1365-313X.2006.02954.x. [DOI] [PubMed] [Google Scholar]
- Dexter RJ, Verdonk JC, Underwood BA, Shibuya K, Schmelz EA, Clark DG. Tissue-specific PhBPBT expression is differentially regulated in response to endogenous ethylene. Journal of Experimental Botany. 2008;59:609–618. doi: 10.1093/jxb/erm337. [DOI] [PubMed] [Google Scholar]
- Dudareva N, Negre F, Nagegowda D, Orlova I. Plant volatiles: recent advances and future perspectives. Critical Reviews in Plant Science. 2006;25:417–440. [Google Scholar]
- Goepfert S, Poirier Y. β-oxidation in fatty acid degradation and beyond. Current Opinion in Plant Biology. 2007;10:245–251. doi: 10.1016/j.pbi.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Guterman I, Masci T, Chen X, Negre F, Pichersky E, Dudareva N, Weiss D, Vainstein A. Generation of phenylpropanoid pathway-derived volatiles in transgenic plants: rose alcohol acetyltransferase produces phenylethyl acetate and benzyl acetate in petunia flowers. Plant Molecular Biology. 2006;60:555–563. doi: 10.1007/s11103-005-4924-x. [DOI] [PubMed] [Google Scholar]
- Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Molecular Biology. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- Kaminaga Y, Schnepp J, Peel G, et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. Journal of Biological Chemistry. 2006;281:23357–23366. doi: 10.1074/jbc.M602708200. [DOI] [PubMed] [Google Scholar]
- Koeduka T, Fridman E, Gang DR, et al.2006Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester Proceedings of the National Academy of Sciences, USA 10310128–10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T, Louie GV, Orlova I, et al.2008The multiple phenylpropene synthases in both Clarkia breweri and Petunia hybrida represent two distinct protein lineages The Plant Journal 54362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T, Orlova I, Baiga TJ, Noel JP, Dudareva N, Pichersky E. The lack of floral synthesis and emission of isoeugenol in Petunia axillaris subsp. parodii is due to a mutation in the isoeugenol synthase gene. The Plant Journal. 2009;58:961–969. doi: 10.1111/j.1365-313X.2009.03834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N, Gorenstein N, Kish CM, Dudareva N. Regulation of circadian methyl benzoate emission in diurnally and nocturnally emitting plants. The Plant Cell. 2001a;13:2333–2347. doi: 10.1105/tpc.010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N, Sherman D, Karlson D, Dudareva N. Cellular and subcellular localization of S-adenosyl-L-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methylbenzoate in snapdragon flowers. Plant Physiology. 2001b;126:956–964. doi: 10.1104/pp.126.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukol J, Conn EE. Metabolism of aromatic compounds in higher plants. IV. Purification and properties of phenylalanine deaminase of Hordeum vulgare . Journal of Biological Chemistry. 1961;236:2692–2698. [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al.2007Clustal W and Clustal X version 2.0 Bioinformatics 232947–2948. [DOI] [PubMed] [Google Scholar]
- Long MC, Nagegowda DA, Kaminaga Y, Ho KK, Kish CM, Schnepp J, Sherman D, Weiner H, Rhodes D, Dudareva N. Involvement of snapdragon benzaldehyde dehydrogenase in benzoic acid biosynthesis. The Plant Journal. 2009;59:256–265. doi: 10.1111/j.1365-313X.2009.03864.x. [DOI] [PubMed] [Google Scholar]
- Maeda H, Shasany AK, Schnepp J, Orlova I, Taguchi G, Cooper BR, Rhodes D, Pichersky E, Dudareva N. RNAi suppression of arogenate dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. The Plant Cell. 2010;22:832–849. doi: 10.1105/tpc.109.073247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriquez PL, Jelinek H, Hirt H. Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. Journal of Biological Chemistry. 2003;278:18945–18952. doi: 10.1074/jbc.M300878200. [DOI] [PubMed] [Google Scholar]
- Mitchell AZ, Hanson MR, Skvirsky RC, Ausubel FM. Anther culture of petunia - genotypes with high-frequency of callus, root, or plantlet formation. Zeitschrift für Pflanzenphysiologie. 1980;100:131–146. [Google Scholar]
- Moore BS, Hertweck C, Hopke JN, et al. Plant-like biosynthetic pathways in bacteria: from benzoic acid to chalcone. Journal of Natural Products. 2002;65:1956–1962. doi: 10.1021/np020230m. [DOI] [PubMed] [Google Scholar]
- Negre F, Kish CM, Boatright J, Underwood B, Shibuya K, Wagner C, Clark DG, Dudareva N. Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. The Plant Cell. 2003;15:2992–3006. doi: 10.1105/tpc.016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Orlova I, Marshall-Colon A, Schnepp J, et al.2006Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport The Plant Cell 183458–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J, Hertweck C, Shipley PR, Hunt DM, Newman MS, Moore BS. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster form the marine isolate ‘Streptomyces maritimus’: evidence for the derailment of an aromatic polyketide synthase. Chemistry and Biology. 2000;7:943–955. doi: 10.1016/s1074-5521(00)00044-2. [DOI] [PubMed] [Google Scholar]
- Rippert P, Puyaubert J, Grisollet D, Derrier L, Matringe M. Tyrosine and phenylalanine are synthesized within the plastids in Arabidopsis. Plant Physiology. 2009;149:1251–1260. doi: 10.1104/pp.108.130070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter H, Schulz GE. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. The Plant Cell. 2004;16:3426–3436. doi: 10.1105/tpc.104.025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurink RC, Haring MA, Clark DG. Regulation of volatile benzenoid biosynthesis in petunia flowers. Trends inn Plant Science. 2006;11:20–25. doi: 10.1016/j.tplants.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature. 2004;12:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shang Y, Schwinn KE, Bennett MJ, Hunter DA, Waugh TL, Pathirana NN, Brummell DA, Jameson PE, Davies KM. Methods for transient assay of gene function in floral tissues. Plant Methods. 2007;3:1–12. doi: 10.1186/1746-4811-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Browse J. Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. The Plant Journal. 2011;66:143–160. doi: 10.1111/j.1365-313X.2011.04512.x. [DOI] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse J. Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme A synthetases. Plant Physiology. 2003;132:1065–1076. doi: 10.1104/pp.103.020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Taylor M, Schaurer N, Fernie AR, Hanson AD, Klee HJ. Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proceedings of the National Academy of Sciences, USA. 2006;103:8287–8292. doi: 10.1073/pnas.0602469103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Lucas HM, Kim JY, Clark DG, Klee HJ. Tomato phenylacetaldehyde reductase catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochemistry. 2007;68:2660–2669. doi: 10.1016/j.phytochem.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Underwood BA, Tieman DM, Shibuya K, Dexter RJ, Loucas HM, Simkin AJ, Sims CA, Schmelz EA, Klee HJ, Clark DG. Ethylene-regulated floral volatile synthesis in petunia corollas. Plant Physiology. 2005;138:255–266. doi: 10.1104/pp.104.051144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Moerkercke A, Schauvinhold I, Pichersky E, Haring MA, Schuurink RC. A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. The Plant Journal. 2009;60:292–302. doi: 10.1111/j.1365-313X.2009.03953.x. [DOI] [PubMed] [Google Scholar]
- Vanengelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Research. 1995;4:288–290. doi: 10.1007/BF01969123. [DOI] [PubMed] [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC. ODORANT1 regulates fragrance biosynthesis in petunia flowers. The Plant Cell. 2005;17:1612–1624. doi: 10.1105/tpc.104.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk JC, Ric de Vos CH, Verhoeven HA, Haring MA, van Tunen AJ, Schuurink RC. Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry. 2003;62:997–1008. doi: 10.1016/s0031-9422(02)00707-0. [DOI] [PubMed] [Google Scholar]
- Vermeer JE, Thole JM, Goedhart J, Nielsen E, Munnik T, Gadella TW., Jr Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. The Plant Journal. 2008;57:356–372. doi: 10.1111/j.1365-313X.2008.03679.x. [DOI] [PubMed] [Google Scholar]
- Verweij W, Spelt C, Di Sansebastiano GP, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F. An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nature Cell Biology. 2008;10:1456–1462. doi: 10.1038/ncb1805. [DOI] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Molecular Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC. Variations on a theme: synthesis and modification of plant benzoic acids. Current Opinion in Plant Biology. 2006;9:288–296. doi: 10.1016/j.pbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ. A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nature Biotechnology. 1997;15:444–447. doi: 10.1038/nbt0597-444. [DOI] [PubMed] [Google Scholar]
- Xiang L, Moore BS. 2002. Inactivation, complementation, and heterologous expression of encP, a novel bacterial phenylalanine ammonis-lyase gene Journal of Biology and Chemistry 277 32505-32509 [DOI] [PubMed] [Google Scholar]
- Xiang L, Moore BS. Characterization of benzoyl coenzyme A biosynthesis genes in the enterocin-producing bacterium ‘Streptomyces maritimus’. Journal of Bacteriology. 2003;185:399–404. doi: 10.1128/JB.185.2.399-404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.