Abstract
Fusarium head blight (FHB), caused by Fusarium graminearum, is a devastating disease of small grain cereal crops. FHB causes yield reductions and contamination of grain with trichothecene mycotoxins such as deoxynivalenol (DON). DON inhibits protein synthesis in eukaryotic cells and acts as a virulence factor during fungal pathogenesis, therefore resistance to DON is considered an important component of resistance against FHB. One mechanism of resistance to DON is conversion of DON to DON-3-O-glucoside (D3G). Previous studies showed that expression of the UDP-glucosyltransferase genes HvUGT13248 from barley and AtUGt73C5 (DOGT1) from Arabidopsis thaliana conferred DON resistance to yeast. Over-expression of AtUGt73C5 in Arabidopsis led to increased DON resistance of seedlings but also to dwarfing of transgenic plants due to the formation of brassinosteroid-glucosides. The objectives of this study were to develop transgenic Arabidopsis expressing HvUGT13248, to test for phenotypic changes in growth habit, and the response to DON. Transgenic lines that constitutively expressed the epitope-tagged HvUGT13248 protein exhibited increased resistance to DON in a seed germination assay and converted DON to D3G to a higher extent than the untransformed wild-type. By contrast to the over-expression of DOGT1 in Arabidopsis, which conjugated the brassinosteriod castasterone with a glucoside group resulting in a dwarf phenotype, expression of the barley HvUGT13248 gene did not lead to drastic morphological changes. Consistent with this observation, no castasterone-glucoside formation was detectable in yeast expressing the barley HvUGT13248 gene. This barley UGT is therefore a promising candidate for transgenic approaches aiming to increase DON and Fusarium resistance of crop plants without undesired collateral effects.
Key words: Deoxynivalenol, Fusarium head blight, trichothecenes, UDP-glucosyltransferase
Introduction
Trichothecenes are a diverse family of mycotoxins produced by a complex of Fusarium ssp. (Starkey et al., 2007) including Fusarium graminearum, F. pseudograminearum, and F. culmorum. This complex of Fusarium ssp. causes several major disease problems on wheat and barley including Fusarium head blight (FHB; Leonard and Bushnell, 2003). Depending on the chemotype of the inoculum, the type B trichothecenes deoxynivalenol (DON), nivalenol, 3-acetyldeoxynivalenol (3-ADON), and 15-acetyldeoxynivalenol accumulate in the developing grain (Desjardins et al., 1993). DON is the primary trichothecene found in F. graminearum-infected wheat and barley in Europe and North America (McCormick, 2003). DON contamination of cereals can reach toxicologically relevant levels (Pestka, 2010). In the United States of America, the Food and Drug Administration issued an advisory level of 1 ppm (1mg kg–1) DON in finished wheat products that is believed to provide an adequate level of safety to consumers, whereas the European Commission has binding legislation that prohibits the blending of contaminated grain and enforces maximum tolerated levels of DON (European Commission, 2006) in unprocessed cereals (e.g. 1250 µg kg–1 for wheat intended for human consumption). Nevertheless, based on worldwide intake estimates, the provisional maximum tolerated daily intake of DON (1 µg kg–1 body weight), proposed by the Joint FAO/WHO Expert Committee on Food Additives, seems to be frequently exceeded (Pestka, 2010). A recent urine biomarker-based exposure assessment indicates that about 30% of volunteers exceed this level in Austria (Warth et al., 2010). Thus, a reduction in the contamination of grain with the Fusarium mycotoxin DON is a worldwide goal of plant breeding- and plant biotechnology-based efforts.
Several genetic studies have shown that trichothecenes are virulence factors during FHB development. F. graminearum strains carrying loss-of-function mutations in the TRI5 gene, the first step in the trichothecene biosynthetic pathway encoding trichodiene synthase, result in the lack of trichothecene biosynthesis and reduced virulence on wheat and barley (Proctor et al., 1995; Jansen et al., 2005; Boddu et al., 2007). In susceptible wheat genotypes, the tri5 mutant strains exhibit infection at the site of inoculation but lack symptom spread, indicating that trichothecene accumulation promotes disease spread in wheat (Bai et al., 2001). Quantitative trait locus (QTL) mapping in wheat identified the FHB resistance locus Qfhs.ndsu-3BS (Fhb1) that exhibits the ability to restrict the spread of disease symptoms (Waldron et al., 1999; Liu et al., 2006). In a wheat population segregating for Fhb1, lines containing the Fhb1 resistance allele efficiently conjugate DON to the less toxic DON-3-O-glucoside (D3G; Lemmens et al., 2005). Based on the combined genetic and biochemical evidence, Lemmens et al. (2005) proposed that Fhb1 encodes a UDP-glucosyltransferase (UGT) that conjugates DON to D3G or is a regulator of a UGT. Interestingly, an FHB-susceptible barley genotype exhibiting resistance to disease spread has the capacity to conjugate DON to D3G, indicating that barley exhibits UGT activity that may be responsible for the high resistance to disease spread (Gardiner et al., 2010).
The first UGT capable of detoxifying DON (DOGT1, AtUGT73C5) was identified by the selection of an Arabidopsis cDNA clone that rescued a DON-sensitive yeast strain when plated on DON-containing media (Poppenberger et al., 2003). AtUGT73C5 was shown to catalyse the transfer of glucose to the hydroxyl group at carbon 3 of DON creating D3G (Poppenberger et al., 2003). Over-expression of the Arabidopsis DOGT1 gene in Arabidopsis resulted in increased tolerance to DON (Poppenberger et al., 2003). Yet, these transgenic plants also displayed a dwarf phenotype reminiscent of brassinosteroid deficiency. It was shown that, in AtUGT73C5-overexpressing transgenic plants, the brassinosteroid brassinolide (BR) was converted to the inactive BR-23-O-glucoside (Poppenberger et al., 2006). In addition, the closely related Arabidopsis UGT73C6 can also glycosylate brassinosteroids (Husar et al., 2011). These results point to the need to isolate UGTs from plants and to characterize their trichothecene specificity and activity, but also to investigate possible unwanted activity such as the glycosylation of plant hormones.
UGTs are encoded by a large gene family, with approximately 100–150 members in different plant species (Bowles et al., 2006). Thus, identifying the specific UGT that conjugates DON to D3G is not a trivial task. Candidate UGT genes induced during Fusarium infection were identified in several studies (Hill-Ambroz et al., 2006; Desmond et al., 2008; Walter et al., 2008; Steiner et al., 2009; Lulin et al., 2010). Lulin et al. (2010) identified six DON-induced wheat UGT genes and isolated the TaUGT3 gene and tested the function of the gene via expression in Arabidopsis. Compared with the Arabidopsis DOGT1 transgenic plants, transgenic Arabidopsis lines carrying an over-expressed TaUGT3 gene did not confer clear tolerance against DON (Lulin et al., 2010). Expression of TaUGT3 in yeast did not confer DON resistance (Schweiger et al., 2010). In barley, multiple RNA-profiling studies have been conducted and candidate UGTs have been identified that are up-regulated during F. graminearum infection, trichothecene accumulation, and DON application (Boddu et al., 2006, 2007; Gardiner et al., 2010). Recently, four barley UDP-glucosyltransferases were tested in the DON-sensitive yeast strain and only one UGT, HvUGT13248 was shown to confer DON resistance via the conjugation of DON to D3G (Schweiger et al., 2010).
The objectives of this study were to develop transgenic Arabidopsis carrying the barley UDP-glucosyltransferase (HvUGT13248) gene, to evaluate these plants for resistance to DON, to examine the fate of DON, and to examine morphological phenotypes.
Materials and methods
Construction of the plant transformation vector and plant transformation
For the Arabidopsis transformation vector, the open reading frame of the barley UDP-glucosyltransfease (HvUGT13248) gene was amplified using Pfu DNA polymerase with the reverse primer designed to add a carboxy-terminal Flag-tag antibody sequence. The primers used for amplification were HvUGT13248 F: 5'-CACCATGGAGACCACGGTCACCGC-3', and HvUGT13248 R: 5'-TTACTTGTCATCGTCGTCCTTGTAGTCTATTGACGAAT ACTTGGTAGCGA-3' with the Flag-tag site underlined. The PCR products were inserted into the Gateway pENTERTM/D-TOPO vector (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and then inserted into the binary expression plasmid pMDC32 (Curtis and Grossniklaus, 2003) using the Gateway LR recombination reaction. The pMDC32 vector harbours a duplicated CaMV35S promoter driving HvUGT13248 and a hygromycin resistance gene as the selectable marker (Curtis and Grossniklaus, 2003). The pMDC32-HvUGT13248 construct was introduced into Agrobacterium tumefaciens strain GV3101 and Arabidopsis thaliana wild-type (Columbia ecotype Col-0) plants were transformed using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS (Murashige and Skoog, 1962) media containing 25mg l–1 hygromycin and transferred to soil. The seedlings were grown in a growth chamber at 22 °C in a 16/8h light/dark cycle.
Molecular analysis of transgenic Arabidopsis thaliana
For Southern blot analysis of transgenic Arabidopsis carrying the HvUGT13248 gene, genomic DNA (10 µg) was digested with XbaI and BamHI, separated on a 1% agarose gel and transferred onto Hybond N+ membranes (Amersham Biosciences, Piscataway, NJ, USA). The HvUGT13248 gene probe (633bp) was derived from a PCR-amplified product. The forward 5'-CAACTCATTCCGTGACATCG-3' and reverse 5'-CTTTCTCTCCCCATCCATCA-3' primers were used for the HvUGT13248 PCR amplification. The probe sequence was labelled with α-32P dCTP using the Prime-a-Gene labelling system (Promega, Madison, WI, USA), following the manufacturer’s instructions. The radiolabelled HvUGT13248 gene was used as a probe for the hybridization and the subsequent banding patterns were visualized using autoradiography.
For immunodetection of the Flag-tagged HvUGT13248 protein in the transgenic Arabidopsis plants, protein was extracted by grinding rosette leaves in extraction buffer [2% SDS, 60mM TRIS (pH 6.8), 14.4mM β-mercaptoethanol, 10% glycerol, and 0.1% (w/v) bromophenol blue]. Protease inhibitor mixture (1%; Sigma P-9599, St Louis, MO, USA) was added to the extraction buffer/cell debris mixture and the cell debris was removed by micro-centrifugation. Total protein concentration was determined using Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, CA, USA) with bovine serum albumin as a standard. Protein extracts (10 µg) were separated by SDS polyacrylamide electrophoresis (12% acrylamide) and transferred to PVDF transfer membrane (Amersham Biosciences, Piscataway, NJ, USA). A DYKDDDK recognizing the FLAG antibody coupled to HRP (Cell Signaling Technology, Beverly, MA, USA) was used to detect the tagged HvUGT13248 protein at a 1:2000 dilution. The protein was visualized using an ECL Western Blotting Reagent Pack (Amersham Biosciences, Piscataway, NJ, USA).
Germination of transgenic Arabidopsis thaliana on DON-containing media
DON was purified at the Bacterial Foodborne Pathogens and Mycology Research Unit, USDA-ARS, Peoria, IL, USA, and at the IFA-Tulln, Austria and dissolved in 70% ethanol or water (10mg ml–1), respectively. For analysis of DON resistance, the seeds of homozygous T3 lines exhibiting HvUGT13248 expression were surface-sterilized and germinated on Petri dish plates of MS (Murashige and Skoog, 1962) agar media containing different concentrations of DON (0.5, 1, 2 10, 15, 20, 50, and 75mg l–1). Col-0 was used as the non-transgenic control.
DON metabolism in transgenic Arabidopsis thaliana seedlings
Seeds of the parental line Columbia and the transgenic derivatives thereof expressing the HvUGT13248 (line #28) or DOGT1 (Poppenberger et al., 2003) genes were surface-sterilized using sodium hypochlorite (2%) plus 0.01% Triton X solution for 10min, and rinsed twice with sterile water. About 15 seeds per well were distributed into 6-well plates each containing 3ml of liquid half-strength (1% sucrose) MS medium (Murashige and Skoog, 1962). After 48h at 4 °C the plates were transferred to a growth chamber for 12 d (16/8h light/dark conditions at 22 °C). Fresh liquid half-strength MS was added on day six. On day 12, the plates were transferred to continuous light (about 70 µE m–2 s–1) for 3 d. Before DON treatment, the medium was removed by aspiration and substituted with 4ml fresh half-strength MS containing either 0, 50, 75, 100, 150 or 200mg l–1 DON. Immediately after addition (t=0) and after 3, 6, and 24h in continuous light, 300 µl samples of the medium were collected in duplicate, mixed with the same volume of absolute ethanol, and stored at –20 °C until analysis.
The seedlings were recovered at the end-point with a Büchner funnel and rinsed with 20ml absolute ethanol to remove external DON. After the fresh weight of the seedlings was determined they were homogenized with 10ml absolute ethanol using an Ultra-Turrax® T25 mixer (IKA, Staufen, Germany) at 24 000rpm for 1min. The resulting suspensions were centrifuged at 4000rpm for 10min to remove insoluble material and 800 µl of the resulting supernatants were harvested and stored at –20 °C. The culture medium samples and the plant extracts were analysed for DON and D3G using a QTrap-LC-MS/MS system (AB Sciex, Foster City, CA, USA) as previously described by Berthiller et al. (2005).
Kinematic analysis of root growth
Seeds of Col-0 and the transgenic HvUGT13248 expressing line (event #28) were surface-sterilized and plated onto square plates containing half-strength MS agar with 0, 0.5, 1, and 2mg l–1 of DON. Root growth of the seedlings growing on the vertically-positioned plates was traced by marking the position of the root tip on the back of the plate using a scalpel blade every day for 10 d starting 3 d after germination. Root growth dynamics were determined using the program Scion Image (version beta 4.0.3.2; Scion Corp, Frederick, MD, USA). The daily length increase over the final growing period was calculated by adding the measured distances between successive marks along the root axis. The average growth rate was calculated for each day as well as the total root length.
Morphological characterization of transgenic plants
Surface-sterilized Arabidopsis seeds from wild-type (Col-0), and transgenic Arabidopsis lines expressing HvUGT13248 (#28, #40, and #42) were sown on MS growth media supplemented with 1% sucrose and 0.2% phytagel (Sigma) and subjected to a 2 d dark treatment at 4 °C to synchronize germination. The seedlings were grown for 2 weeks in a growth chamber at 22 °C on a 16/8h light/dark cycle. The seedlings were then transferred to Metro-Mix 200 growth medium (The Scotts Company, Marysville, OH, USA) in 2.5'' square plastic pots and placed in a growth chamber set at 22 °C on a 16/8h light/ dark cycle. Plants were grown in a randomized complete block design with three replications and eight plants per replication. Height, days to flowering, number of rosette leaves, and number of shoots were measured over a 6-week period.
Determination of brassinosteroid-glucoside formation in yeast
Cultures of the yeast strain YZGA515 transformed with the empty vector or with expression vectors carrying the AtUGT73C5 or HvUGT13248 (Schweiger et al., 2010) were grown in SC-LEU medium. The brassinosteroids, brassinolide and castasterone (Chemiclones Inc., Waterloo Ontaria, Canada) used in pilot experiments were dissolved in ethanol (100 µg ml–1). For the main experiment, castasterone was obtained from OlChemIm (Olomouc, Czech Republic) and dissolved in acetone (stock 500 µg ml–1). Three freshly-prepared independent transformants obtained with the empty vector (pBP910), HvUGT13248 (pWS1921) and AtUGT73C5 (pBP868) were analysed (Schweiger et al., 2010). Logarithmic cultures were concentrated by centrifugation and re-suspended at a density of OD600=7 in fresh medium. At the start of incubation, 2 µl castasterone stock were added to 198 µl yeast cells. After 24h incubation at 180rpm and 30 °C (in 2.2ml plastic tubes with a hole punched in the lid with a hot needle to allow gas exchange) one volume (200 µl) of methanol was added to the tubes to stop the reaction. After vortexing and a 10min centrifugation at 9000rpm in an Eppendorf centrifuge the supernatants were harvested and transferred to glass HPLC vials with inserts for small sample volumes. Determination of brassinolide and castasterone and their respective 23-O-glucosides by HPLC-MS/MS was performed as recently described byHusar et al. (2011).
Results
Development of transgenic Arabidopsis thaliana expressing HvUGT13248
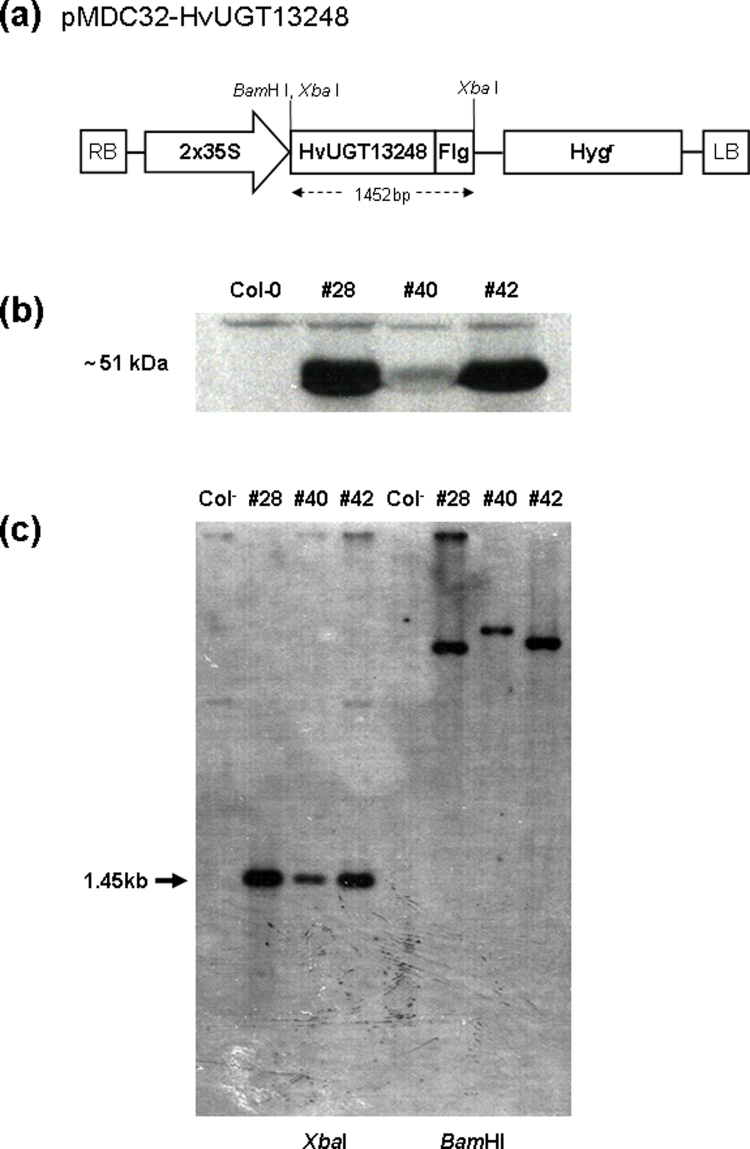
The pMDC32-HvUGT13248 plasmid containing the barley UDP-glucosyltransferase gene HvUGT13248 under the control of a duplicated CaMV 35S promoter (Fig. 1a) was used for Agrobacterium-mediated transformation of Arabidopsis. Initially, 63 T1 plants that grew on MS medium containing hygromycin were selected. To identify plants that express the HvUGT13248 transgene, Western blot analysis utilizing the C-terminal Flag epitope tag was conducted on the T1 plants. Forty-five transgenic Arabidopsis plants accumulating detectable amounts of HvUGT13248 protein were identified. Two events, HvUGT13248 #28 and #42 that showed high levels of recombinant protein, and the HvUGT13248 #40 event that showed a low level of recombinant protein, were chosen for further study (Fig. 1b).
Fig. 1.
Development and molecular characterization of transgenic Arabidopsis thaliana expressing HvUGT13248. (a) The pMDC32-HvUGT13248 plasmid containing the barley UDP-glucosyltransferase (HvUGT13248) gene was used for Arabidopsis transformation. The BamHI and XbaI enzyme sites were used to genomic DNA blot analysis. Hygr, hygromycin resistance was used as a selectable marker; 2x35S, duplicate of the cauliflower mosaic virus 35S promoter, Flg, Flag-epitope tag for western blotting. (b) Western blot analysis of transgenic Arabidopsis plants carrying the barley HvUGT13248 gene. Total protein (10 µg) extracted from leaf tissue of the transgenic lines was subjected to SDS-PAGE analyses. The blot was probed with the HRP conjugate of the Flag antibody. Col-0 was used as a negative control. Two lines (#28 and #42) with high and one line (#40) with low level(s) of Flag-tagged HvUGT13248 protein were identified. Molecular markers indicated that the detected protein has the expected 51kDa. (c) Southern blot analysis of three transgenic Arabidopsis plants carrying the barley HvUGT13248 gene. Genomic DNA from Col-0 and transgenic lines were digested with XbaI and BamHI, and hybridized with a HvUGT13248 gene probe. The arrow indicates the position of the expected 1.45kb hybridizing fragment resulting from an XbaI digestion.
Homozygous lines for the three transgenic events (#28, #40, and #42) were identified by examining segregation for hygromycin resistance. Families derived from transgenic events #28, #40, and #42 that were not segregating for hygromycin resistance (100% hygromycin resistant) were classified as homozygous. Homozygous T3 families were used for further testing.
Southern blot analysis was conducted on a single T3 plant from each of the three events. Genomic DNA was digested with XbaI and BamHI and probed with a fragment from the HvUGT13248 gene. The probe did not hybridize to the Col-0 Arabidopsis control; however, each of the lines exhibited a different banding pattern, indicating that the three lines were transgenic and resulted from independent transformation events (Fig. 1c).
Transgenic Arabidopsis thaliana expressing HvUGT13248 exhibit resistance to DON
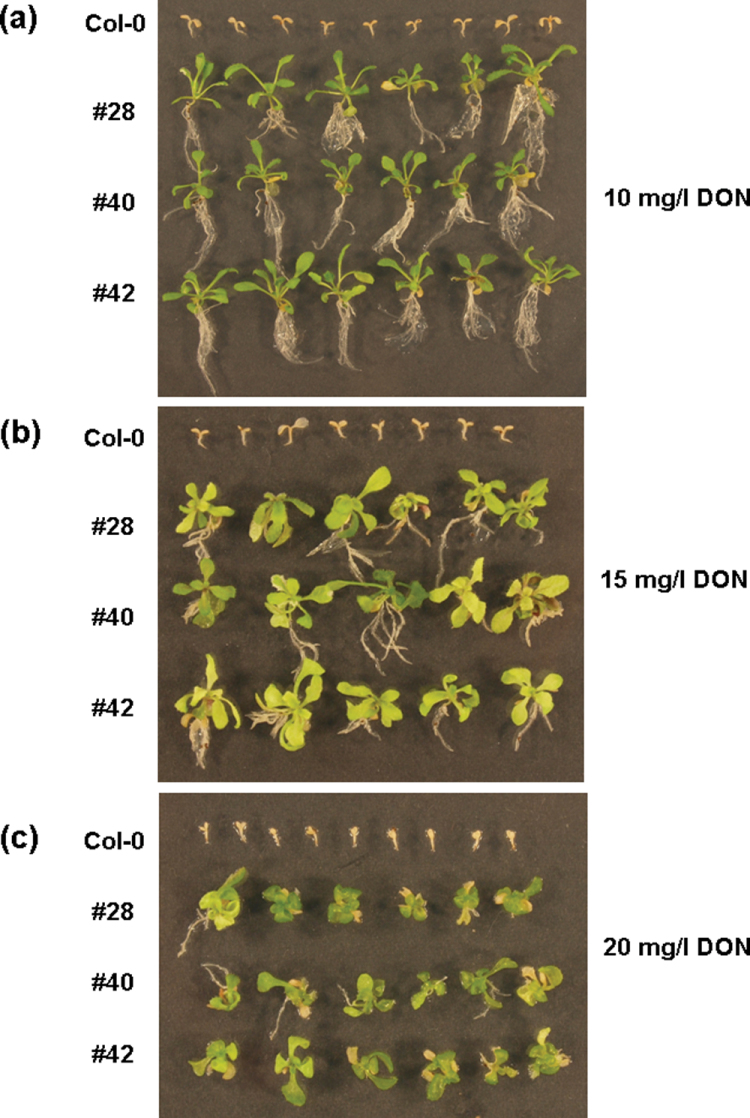
To determine the effect of the expression of the barley HvUGT13248 gene in Arabidopsis, the growth of non-transformed and homozygous transgenic seeds was examined on MS medium supplemented with DON (Fig. 2). T3 seeds from transgenic lines #28, #40, and #42 and the non-transformed control (Col-0) were germinated on MS medium containing 10, 15, and 20mg l–1 DON. At 10–20mg l–1 DON, the non-transformed control exhibited restricted shoot, root, and cotyledon growth, and bleaching before the true leaves could form. After 4 weeks of growth on DON media, the chlorophyll-deficient wild-type ceased growth. By contrast, at 10mg l–1 DON, the transgenic lines exhibited root and shoot growth and stayed green (Fig. 2). Higher concentrations of DON resulted in signs of chlorosis and also the inhibition of root growth in the transgenic seedlings. Only at high DON concentrations was the difference in expression level manifested in a difference in resistance. More seeds of the high expression line #28 than of the low expression line #40 germinated and grew at 50mg l–1and 75mg l–1 DON, respectively, and germination was faster (see Supplementary Fig. S1 at JXB online).
Fig. 4.
Seed germination and growth of transgenic Arabidopsis lines expressing HvUGT13248 on MS medium containing 10–20mg l–1 DON. Three transgenic lines (#28, #40, and #42) and wild-type Col-0 were germinated on MS media containing DON and grown for 4 weeks. The three transgenic Arabidopsis lines exhibit enhanced DON resistance compared to wild-type Col-0.
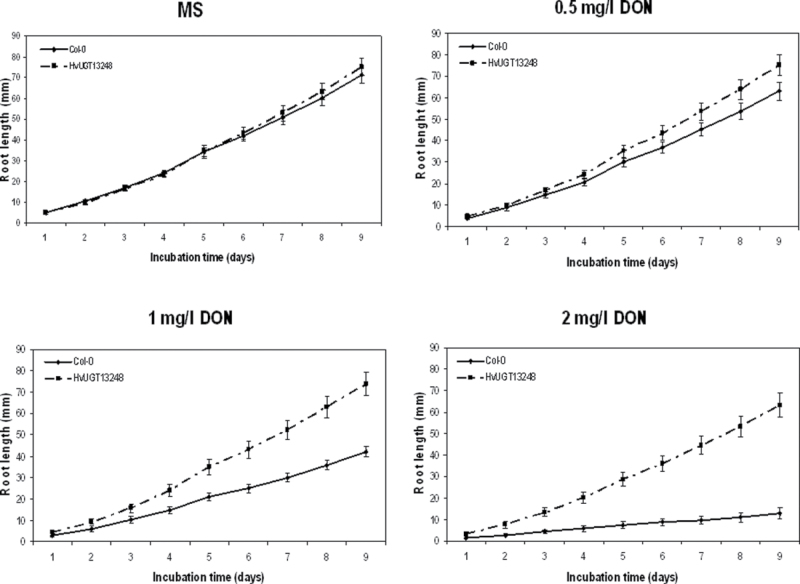
To quantify the impact of DON on root development on the transgenic plants carrying HvUGT13248, root length of the non-transformed control and transgenic line #28 grown on 0–2mg l–1 DON was examined (Fig. 3; see Supplementary Fig. S2 at JXB online). Non-transformed and transgenic plants grown on MS media without DON did not exhibit a difference in root growth from 1–9 d. However, at 0.5mg l–1 DON, the transgenic plants exhibited longer root growth at 7 d. At 1 and 2mg l–1 DON, roots of the transgenic plants were longer than those of the non-transformed plants at 2 ds (Fig. 3; see Supplementary Fig. S2 at JXB online).
Fig. 2.
Root growth of a HvUGT13248-expressing transgenic line and wild-type (Col-0) Arabidopsis seedlings on vertical agar plates containing DON. The transgenic line (#28) and wild-type were germinated on half-strength MS media containing 0, 0.5, 1, and 2mg l–1 DON. Root growth of the transgenic Arabidopsis line exhibited enhanced DON resistance compared with wild-type Col-0 (see Supplementary Fig. S1 at JXB online).
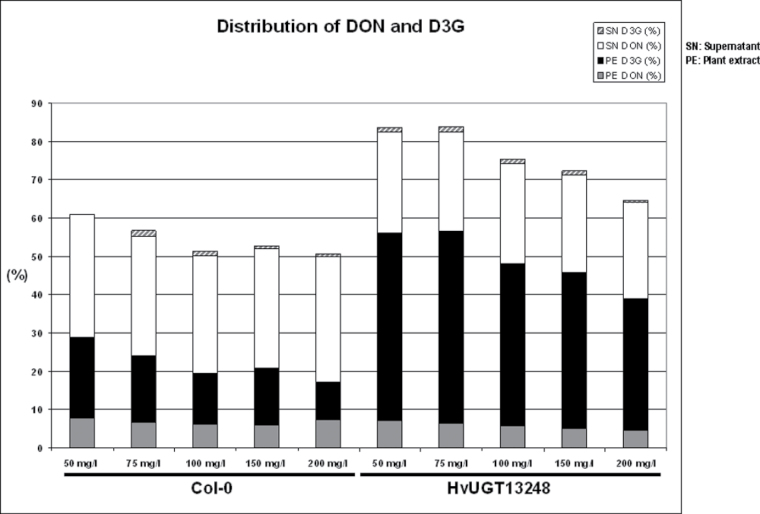
Increased ability to detoxify DON to DON-3-O-glucoside in transgenic Arabidopsis expressing HvUGT13248
To determine whether the HvUGT13248 transgenic plants have a higher capacity to form D3G, seedlings were treated in liquid culture with different concentrations of DON and the concentration of D3G was measured. Arabidopsis has at least two endogenous rapidly DON-inducible UDP-glucosyltransferases (AtUGT73C5, AtUGT73C4) capable of forming D3G (Poppenberger et al., 2003; Schweiger et al., 2010). Therefore, a higher concentration range was used for treatment, which is expected to inhibit protein synthesis in the wild type, thereby preventing expression of DON-induced endogenous gene(s). Our results (Fig. 4) show that the untransformed Col-0 can, nevertheless, form a significant amount of D3G under these conditions. Yet, the introduced constitutively expressed barley HvUGT13248 gene led to a higher concentration of D3G than found in the untransformed Col-0 wild-type. The majority of the D3G was recovered from the plant extract, and only a small portion of the total D3G formed was released to the culture medium. From the total input of DON, about 20% is still present as unchanged DON in the medium, but, depending on the concentration used, the molar fraction of the total added DON converted to D3G is in the range of 34–50% (Fig. 4). In the control line not expressing a heterologous UGT, the unaccounted molar portion (recovered DON+recovered D3G divided by DON input×100) was the highest, indicating that other metabolic reactions leading to the disappearance of DON might exist, which are currently not understood.
Fig. 3.
Metabolism of DON by seedlings in liquid culture. The molar percentage of the deoxynivalenol input recovered as (DON) and DON-3-O-glucoside (D3G) in plant extracts and the medium according to LC-MS/MS analysis is shown. The transgenic Arabidopsis line carrying HvUGT13248 (#28) and the wild-type Col-0 Arabidopsis seedlings were pre-grown in liquid medium and treated with DON (50–200mg l–1) for 24h (see the Materials and methods for details).
Morphology of transgenic Arabidopsis expressing HvUGT13248
To analyse the growth and morphology of the transgenic Arabidopsis lines, seeds of the transgenic lines and the Col-0 non-transgenic control were germinated on agar media. The seedlings were transferred to soil and flowering time, rosette leaf number, plant height, and number of shoots was monitored. All three HvUGT13248 over-expression lines did not show significantly different flowering time, number of rosette leaves, and number of shoots compared with the wild-type control (Table 1). Of the three transgenic lines, only the HvUGT13248 (#40) line was slightly shorter when compared with the non-transgenic control. Thus, expression of the HvUGT13248 gene does not drastically affect Arabidopsis morphology. In addition, the morphological change in transgenic line #40 did not correlate with the level of protein accumulation.
Table 1.
Morphological characterization of transgenic Arabidopsis expressing HvUGT13248
| Genotypes | Days to floweringa | Number of rosette leavesb | Plant height (cm)c | Number of shootsc |
| Control (Col-0) | 26 | 8.7±1.04 | 37.8±3.81 | 6±1.09 |
| #28 | 26 | 8.9±1.14 | 39.5±3.49 | 5.3±1.54 |
| #40 | 26 | 8.2±1.03 | 34.7*±4.74d | 6±1.05 |
| #42 | 26 | 8.4±0.71 | 35.5±4.18 | 5.6±0.93 |
a ±Number of days to flowering.
b The number of the rosette leaves was measured when the plant flowered.
c The number of the shoots and plant height were measured six weeks after planting.
d An asterisk(*) indicates significance at the 0.05 level, respectively compared with the wild-type Col-0 (Student’s t test).
Brassinosteroid glycosylation in yeast
Several members of the large UGT gene family of plants are capable of altering the activity of plant hormones by the formation of the respective glucosides resulting in an impact on the morphology and stress physiology of plants over-expressing such genes. Previously, Poppenberger et al. (2005) showed that Arabidopsis seedlings over-expressing the DON-inactivating AtUGT73C5 displayed phenotypes resembling brassinosteroid deficiency. An increased ability to inactivate exogenously added brassinosteroids into the respective glucosides was demonstrated for transgenic plants over-expressing AtUGT73C5 and AtUGT73C6 (Husar et al., 2011). Since brassinosteroids are expensive and the effects of gene over-expression in plants is confounded by the presence of numerous endogenous UGTs, it was tested whether it is possible to analyse brassinosteroid glycosylation by candidate genes in yeast. The Saccharomyces cerevisiae genome, with the exception of a specialized sterol-glucosyltransferase, is devoid of small molecule-conjugating UGTs. In initial small-scale experiments, the brassinosteroids brassinolide and castasterone were tested at a concentration of 5mg l–1. This concentration is probably very low for yeast, which efficiently prevents uptake of various substances by active efflux systems. On the other hand, the concentration cannot be practically increased, due to the low solubility of the apolar steroids in water. With brassinolide, the formation of small concentrations of brassinolide-glucoside (1–2 µg l–1) were only observed in the medium of the AtUGT73C5 transformed yeast after 19h of incubation. Seemingly, the uptake of castasterone into yeast is higher, since castasterone-glucoside was found in the medium of the AtUGT73C5 transformed yeast after 19h in a range of 9–20 µg l–1, but was undetectable in the medium of the HvUGT13248-expressing yeast and yeast containing the empty vector. In a further experiment, with three independent yeast transformants of each plasmid with two independent LC-MS/MS determinations, 58.6±11.7 and 50.9±9.2 µg l–1 castasterone-glucoside in the media of AtUGT73C5 transformants was observed after 24h. By contrast, no castasterone-glucoside was detectable in the media of the transformants containing the empty vector or HvUGT13248.
Discussion
Transgenic approaches to develop trichothecene-resistant plants
DON acts as a virulence factor and increases the aggressiveness of F. graminearum during infection of wheat. Presumably the toxin, acting as an inhibitor of eukaryotic protein biosynthesis, could interfere with the expression of pathogen-induced defence transcripts of host plants, thereby suppressing or delaying plant defence responses. Thus, identifying genes conferring DON resistance could become important for protecting wheat, and potentially other crops, against F. graminearum infection.
Several strategies aimed at antagonizing the Fusarium virulence factor DON using transgenic approaches have been described. The first identified toxin-resistance mechanism, reduced toxin uptake due to active efflux mediated by the yeast ABC transporter protein PDR5, does not seem to be effective in transgenic crops, despite initial encouraging reports (Dahleen et al., 2001). In addition, attempts to modify the trichothecene ribosomal target by transformation with variants of the ribosomal protein L3 (RPL3), which contain amino acid changes conferring toxin resistance in yeast, had little success due to competition with the endogenous gene products (Mitterbauer et al., 2004). However, expression of a truncated form of yeast ribosomal protein L3 in transgenic wheat appeared to result in improved resistance to F. graminearum in greenhouse and field trials and in slightly reduced DON contamination (Di et al., 2010). The F. sporotrichiodes Tri101 gene encodes a 3-OH trichothecene acetyltransferase that converts DON to an acetylated form, 3-ADON. Transgenic barley and wheat with the Tri101 gene have been developed (Okabara et al., 2002; Manoharan et al., 2006). Although these transgenic plants exhibited reduced FHB severity and DON accumulation in greenhouse studies, in field trials these transgenic plants did not provide increased resistance. According to Alexander (2008), Syngenta has, through modifications of the Tri101 gene sequence and expression levels, generated lines with good field resistance to FHB and agronomic performance. Yet, it is unclear whether DON is shifted to 3-ADON. The acetylation of the C3-OH of DON clearly reduces the toxicity at the level of the ribosome (Kimura et al., 1998), however, the oral toxicity of 3-ADON is even higher than that of DON (mouse oral LD50 for DON=78mg kg–1 and for 3-ADON=34mg kg–1; Yoshizawa and Morooka, 1977). This apparent contradiction might be caused by a higher uptake of 3-ADON than DON and a high level deacetylation in mammalian cells (Wu et al., 2010). In the 2010 FAO/WHO Joint Expert Committee meeting, a group level provisional maximum tolerable daily intake for DON and its acetylated derivatives was established.
In this paper, it has been demonstrated that transgenic Arabidopsis expressing HvUGT13248 shows increased resistance to DON and increased capability to convert DON into DON-3-O-glucoside. The phenotype of the transformants with a high expression level is very clear (Fig. 2), in obvious contrast to Arabidopsis seedlings transformed with the candidate gene TaUGT3, that were previously claimed to display increased resistance (Lulin et al., 2010). The results are comparable with the resistance phenotype previously reported for the Arabidopsis gene AtUGT73C5 (Poppenberger et al., 2003), with the difference that the Arabidopsis gene inactivates barassinosteroids. The conversion of DON into D3G is a detoxification reaction from the plant perspective, yet it may primarily lead to the formation of masked DON, as a currently unknown portion of the consumed D3G might be converted back to the parental toxin by the glucosidases of intestinal bacteria (Berthiller et al., 2011). It remains to be tested whether antagonizing the fungal virulence factor leads to reduced fungal spread and reduced fungal biomass in infected wheat and, consequently, to a lower total DON content (DON+D3G) in wheat. Preliminary results with transgenic wheat constitutively expressing HvUGT13248 are very encouraging, indicating that resistance to Fusarium spread can be engineered (Shin et al., 2011).
Role of UGTs in hormone homeostasis
Glycosyltransferases are enzymes that transfer sugars to a wide range of acceptors including plant hormones, plant secondary metabolites, microbial toxins, and man-made xenobiotics in the environment (Bowles et al., 2006). UGTs are encoded by a very large gene family in plants with more than 100 genes in Arabidopsis (Bowles et al., 2006) and 166 Pfam-database (Pf00201) hits in Brachypodium (Schweiger et al., unpublished results). Individual gene products may have a broad range of substrates, and over-expression therefore may impact other pathways in unanticipated ways and cause unwanted side-effects. Various plant hormones and signalling molecules having a strong impact on plant morphology and physiology and are inactivated by the formation of glucosides. For instance, over-expression of AtUGT84B1 in Arabidopsis, which acts as indole-3-acetic acid (IAA, auxin) glucosyltransferase, caused growth inhibition of Arabidopsis shoots (Jackson et al., 2002). Also, the activity of cytokinin and abscissic acid is modulated by glycosylation and affected by UGT gene overexpression (Priest et al., 2006; Pineda Rodo et al., 2008). Similarly, the defence signalling molecule salicylic acid is inactive as a glucoside, and over-expression of AtSGT1 led to increased susceptibility to a bacterial pathogen in Arabidopsis (Song et al., 2008).
Brassinosteroids play a major role in regulating plant growth and development and also play an important role in a broad range of disease resistance in tobacco and rice (Nakashita et al., 2003). Poppenberger et al. (2005) reported that over-expression of DOGT1 in transgenic Arabidopsis resulted in a dwarf phenotype. These authors also showed that DOGT1 glucosylates the brassinosteroids typhasterol, 6-deoxocastasterone, and castasterone, resulting in a reduction of BR activity and dwarfism. Based on these results, the impact of HvUGT13248 expression in transgenic Arabidopsis on plant growth and development was examined. Only transgenic line #40 exhibited a slightly significant statistical difference compared with the non-transgenic control. However, transgenic line #40 did not show the dwarfism displayed by the DOGT1 over-expression plants. So effects on hormones other than brassinosteroids (see below) cannot be ruled out. Yet, the lack of correlation with protein levels of the transgene product could mean that transformation-induced changes related to the insertion site or other transformation-induced stresses (hygromycin selection) might have caused the observed differences (Ziemienowicz, 2010).
To get more direct evidence whether HvUGT13248 like AtUGT73C5 can inactivate brassinosteroids, a small-scale assay was developed based on yeast expressing individual UGTs. In previous tests (Husar et al., 2011), Arabidopsis seedlings in 30ml medium were treated with a final concentration of 1 µg ml–1. In the yeast test, only 200 µl of 5 µg ml–1 castasterone were used, corresponding to a 30-fold reduction of the expensive brassinosteroid needed. The limit of detection of the LC-MS/MS method is below 1 µg l–1. The finding that the empty vector transformants did not show detectable levels of castasterone-glucoside indicates that the yeast sterol-glucosyltransferase UGT51/YLR189C is not significantly active with castasterone. The yeast transformants containing the positive control gene AtUGT73C5 clearly produced castasterone-glucoside (about 50 µg l–1), while none was detected in case of HvUGT13248. If one assumes similar protein levels, the barley UGT would be at least 50-fold less active with castasterone as AtUGT73C5. No attempt was made to determine protein levels in the small-scale brassinosteroid-treated samples. Yet, based on other experiments (Schweiger et al., 2010), the barley gene shows a higher expression level in yeast than AtUGT73C5, so this is a very conservative estimate. In summary, severe side-effects due to brassinosteroid conjugation seem to be an unlikely problem with HvUGT13248.
This gene is the first crop plant UGT that confers a clear DON-resistance phenotype and represents an attractive candidate gene for developing transgenic wheat with higher Fusarium resistance.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Seeds from transgenic lines #28 and #40 and the non transgenic control Col-0 were germinated and grown on MS, 50 and 75mg l–1 of deoxynivalenol (DON).
Supplementary Fig. S2. Growth of HvUGT13248 expressing Arabidopsis thaliana (#28) and control (Col-0) plants on MS medium containing different concentrations of DON.
Supplementary Material
Acknowledgements
We would like to thank Dr Fumiaki Katagiri at the University of Minnesota for providing the pMDC32 plasmid. We are indebted to Abigail Cole and Anthony Jakubiak for excellent technical assistance. This project is supported by funds from the USDA-ARS US Wheat and Barley Scab Initiative and the Minnesota Small Grains Initiative to GJM. GA, ML, and FB received funding from the Austrian Science fund (SFB Fusarium, F3701, F3706, F3708 and L475-B09) and FB from the Federal Ministry of Economy, Family, and Youth as well as from the National Foundation for Research, Technology, and Development. Sanghyun Shin was supported by a postdoctoral fellowship programme of the National Institute of Crop Science (NICS), Rural Development Administration (RDA), Republic of Korea, and a grant from the Next-Generation BioGreen21 program (No.PJ00800602), Rural Development Administration, Republic of Korea.
Glossary
Abbreviations:
- FHB
Fusarium head blight;
- DON
deoxynivalenol
- D3G
DON-3-O-glucoside
- UGT
UDP-glucosyltransferase
References
- Alexander NJ. 2008. The TRI101 story: engineering wheat and barley to resist Fusarium head blight. World Mycotoxin Journal 1 31–37 [Google Scholar]
- Bai GH, Desjardins AE, Plattner RD. 2001. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes Mycopathologia 153 91–98 [DOI] [PubMed] [Google Scholar]
- Berthiller F, Dall’Asta C, Schuhmacher R, Lemmens M, Adam G, Krska R. 2005. Masked mycotoxins: determination of a deoxynivalenol glucoside in artificially and naturally contaminated wheat by liquid chromatography-tandem mass spectrometry. Journal of Agricultural and Food Chemistry 53 3421–3425 [DOI] [PubMed] [Google Scholar]
- Berthiller F, Krska R, Domig KJ, Kneifel W, Juge N, Schuhmacher R, Adam G. 2011. Hydrolytic fate of deoxynivalenol-3-glucoside during digestion. Toxicology Letters 206 264–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddu J, Cho S, Kruger WM, Muehlbauer GJ. 2006. Transcriptome analysis of the barley–Fusarium graminearuminteraction Molecular Plant–Microbe Interactions 19 407–417 [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Muehlbauer GJ. 2007. Transcriptome analysis of Trichothecene-induced gene expression in barley. Molecular Plant–Microbe Interactions 20 1364–1375 [DOI] [PubMed] [Google Scholar]
- Bowles D, Lim EK, Poppenberger B, Vaistij FE. 2006. Glycosyltransferases of lipophilic small molecules Annual Review of Plant Biology 57 567–597 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana The Plant Journal 16 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta Plant Physiology 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahleen LS, Okubara PA, Blechl AE. 2001. Transgenic approaches to combat Fusarium head blight in wheat and barley Crop Science 41 628–637 [Google Scholar]
- Desjardins AE, Hohn TM, McCormick SP. 1993. Trichothecene biosynthesis in Fusariumspecies: chemistry, genetics, and significance Microbiology and Molecular Biology Reviews 57 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond OJ, Manners JM, Schenk PM, Maclean DJ, Kazan K. 2008. Gene expression analysis of the wheat response to infection by Fusarium pseudograminearum Physiological and Molecular Plant Pathology 73 40–47 [Google Scholar]
- Di R, Blechl A, Dill-Macky R, Tortora A, Tumer N. 2010. Expression of a truncated form of yeast ribosomal protein L3 in transgenic wheat improves resistance to Fusarium head blight Plant Science 178 374–380 [Google Scholar]
- Gardiner SA, Boddu J, Berthiller F, Hametner C, Stupar R, Adam G, Muehlbauer GJ. 2010. Transcriptome analysis of the barley–deoxynivalenol interaction: evidence for a role of glutathione in deoxynivalenol detoxification. Molecular Plant–Microbe Interactions 23 962–976 [DOI] [PubMed] [Google Scholar]
- European Commission 2006. Commission recommendation (EC) no. 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs Official Journal European Union Legislation 364 5–24 [Google Scholar]
- Hill-Ambroz K, Webb CA, Matthews AR, Li W, Gill BS, Fellers JP. 2006. Expression analysis and physical mapping of a cDNA library of Fusarium head blight infected wheat spikes Crop Science 46 S15–S26 [Google Scholar]
- Husar S, Berthiller F, Fujioka S, et al. 2011. Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana BMC Plant Biology 11, 51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ. 2002. Over-expression of an Arabidopsisgene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterization of transgenic lines The Plant Journal 32 573–583 [DOI] [PubMed] [Google Scholar]
- Jansen C, Wettstein DV, Schäfer W, Kogel KH, Felk A, Maier FJ. 2005. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum Proceedings of the National Academy of Sciences, USA 102 16892–16897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kaneko I, Komiyama M, Takatsuki A, Koshino H, Yoneyama K, Yamaguchi I. 1998. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. Journal of Biological Chemistry 273 1654–1661 [DOI] [PubMed] [Google Scholar]
- Lemmens M, Scholz U, Berthiller F, et al. 2005. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat Molecular Plant–Microbe Interactions 18 1318–1324 [DOI] [PubMed] [Google Scholar]
- Leonard KJ, Bushnell WR. . eds. 2003. Fusarium head blight of wheat and barley St Paul, MN: APS Press; [Google Scholar]
- Liu S, Zhang X, Pumphrey MO, Stack RW, Gill BS, Anderson JA. 2006. Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat Functional and Integrative Genomics 6 83–89 [DOI] [PubMed] [Google Scholar]
- Lulin M, Yi S, Aizhong C, Zengjun Q, Liping X, Peidu C, Dajun L, Xiu-e W. 2010. Molecular cloning and characterization of an up-regulated UDP-glucosyltransferase gene induced by DON from Triticum aestivumL. cv. Wangshuibai Molecular Biology Reports37, 785–795 [DOI] [PubMed] [Google Scholar]
- Manoharan M, Dahleen LS, Hohn TM, Neate SM, Yu XH, Alexander NJ, McCormick SP, Bregitzer P, Schwarz PB, Horsley RD. 2006. Expression of 3-OH trichothecene acetyltransferase in barley (Hordeum vulgare L.) and effects on deoxynivalenol Plant Science 171 699–706 [Google Scholar]
- McCormick S. 2003. The role of DON in pathogenicity. In: Leonhard KJ, Bushnell WR, eds. Fusarium head blight of wheat and barley St. Paul, MN, USA: APS Press; 165–183 [Google Scholar]
- Mitterbauer R, Poppenberger B, Raditschnig A, Lucyshyn D, Lemmens M, Glössl J, Adam G. 2004. Toxin-dependent utilization of engineered ribosomal protein L3 limits trichothecene resistance in transgenic plants Plant Biotechnology Journal 2 329–340 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures Physiologia Plantarum15, 473–479 [Google Scholar]
- Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K, Takatsuto S, Yamaguchi I, Yoshida S. 2003. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice The Plant Journal 33 887–898 [DOI] [PubMed] [Google Scholar]
- Okubara PA, Blechl AE, McCormick SP, Alexander NJ, Dill-Macky R, Hohn TM. 2002. Engineering deoxynivalenol metabolism in wheat through the expression of a fungal trichothecene acetyltransferase gene Theoretical and Applied Genetics 106 74–83 [DOI] [PubMed] [Google Scholar]
- Pestka JJ. 2010. Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Archives of Toxicology 84 663–679 [DOI] [PubMed] [Google Scholar]
- Pineda Rodo A, Brugière N, Vankova R, Malbeck J, Olson JM, Haines SC, Martin RC, Habben JE, Mok DW, Mok MC. 2008. Over-expression of a zeatin O-glucosylation gene in maize leads to growth retardation and tasselseed formation Journal of Experimental Botany 59 2673–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B, Berthiller F, Bachmann H, Lucyshyn D, Peterbauer C, Mitterbauer R, Schuhmacher R, Krska R, Glössl J, Adam G. 2006. Heterologous expression of ArabidopsisUDP-glucosyltransferases in Saccharomyces cerevisiaefor production of zearalenone-4-O-glucoside Applied and Environmental Microbiology 72 4404–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B Berthiller F Lucyshyn D Sieberer T Schuhmacher R Krska R Kuchler K Glössl J Luschnig C Adam G. 2003. Detoxification of the Fusariummycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana Journal of Biological Chemistry 278 47905–47914 [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Fujioka S, Soeno K, et al. 2005. The UGT73C5 of Arabidopsis thalianaglucosylates brassinosteroids. Proceedings of the National Academy of Sciences, USA 102 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest DM, Ambrose SJ, Vaistij FE, Elias L, Higgins GS, Ross AR, Abrams SR, Bowles DJ. 2006. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. The Plant Journal 46 492–502 [DOI] [PubMed] [Google Scholar]
- Proctor RH, Hohn TM, McCormick SP. 1995. Reduced virulence of Gibberella zeaecaused by disruption of a trichothecene toxin biosynthetic gene. Molecular Plant–Microbe Interactions 8 593–601 [DOI] [PubMed] [Google Scholar]
- Schweiger W, Boddu J, Shin S, Poppenberger B, Berthiller F, Lemmens M, Muehlbauer GJ, Adam G. 2010. Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Molecular Plant–Microbe Interactions 23 977–986 [DOI] [PubMed] [Google Scholar]
- Shin S, Torres-Acosta A, Berthiller F, Schwiger W, Adam G, McCormick S, Clemente T, Muehlbauer GJ. 2011. Identifying and characterizing barley genes that protect against trichothecene mycotoxins Proceedings of the National Fusarium Head Blight Forum.4–6DecemberSt Louis, MO, USA, page 96 [Google Scholar]
- Song JT, Koo YJ, Seo HS, Kim MC, Choi YD, Kim JH. 2008. Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase, leads to increased susceptibility to Pseudomonas syringae Phytochemistry 69 1128–1134 [DOI] [PubMed] [Google Scholar]
- Starkey DE, Ward TJ, Aoki T, Gale LR, Kistler C, Geiser DM, Suga H, Toth B, Varga J, O’Donnell K. 2007. Global molecular surveillance reveals novel Fusariumhead blight species and trichothecene toxin diversity Fungal Genetics Biology 44 1191–1204 [DOI] [PubMed] [Google Scholar]
- Steiner B, Kurz H, Lemmens M, Buerstmayr H. 2009. Differential gene expression of related wheat lines with contrasting levels of head blight resistance after Fusarium graminearuminoculation Theoretical and Applied Genetics 118 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron BL, Moreno-Sevilla B, Anderson JA, Stack RW, Frohberg RC. 1999. RFLP mapping of QTL for Fusarium head blight resistance in wheat Crop Science 805–811 [Google Scholar]
- Walter S, Brennan JM, Arunachalam C, et al. 2008. Components of the gene network associated with genotype-dependent response of wheat to the Fusarium mycotoxin deoxynivalenol Functional and Integrative Genomics 8 421–427 [DOI] [PubMed] [Google Scholar]
- Warth B, Sulyok M, Fruhmann P, Berthiller F, Schuhmacher R, Hametner C, Adam G, Fröhlich J, Krska R. 2010. Assessment of human deoxynivalenol exposure using an LC-MS/MS based biomarker method Toxicology Letters [Epub ahead of print] PMID: 22429874 [DOI] [PubMed] [Google Scholar]
- Wu Q, Dohnal V, Huang L, Kuča K, Yuan Z. 2010. Metabolic pathways of trichothecenes Drug Metabolism Reviews 42 250–267 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T, Morooka N. 1977. Trichothecenes from mold infested cereals in Japan. In: Rodricks JV, Hesseltine CW, Mehlman MA, eds. Mycotoxins in human and animal health Park Forest South, IL, USA: Pathotox Publishers, 309–321 [Google Scholar]
- Ziemienowicz A. 2010. Plant transgenesis Methods in Molecular Biology 631 253–268 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.