Abstract
Stem cell response to a library of scaffolds with varied 3D structures was investigated. Microarray screening revealed that each type of scaffold structure induced a unique gene expression signature in primary human bone marrow stromal cells (hBMSCs). Hierarchical cluster analysis showed that treatments sorted by scaffold structure and not by polymer chemistry suggesting that scaffold structure was more influential than scaffold composition. Further, the effects of scaffold structure on hBMSC function were mediated by cell shape. Of all the scaffolds tested, only scaffolds with a nanofibrous morphology were able to drive the hBMSCs down an osteogenic lineage in the absence of osteogenic supplements. Nanofiber scaffolds forced the hBMSCs to assume an elongated, highly branched morphology. This same morphology was seen in osteogenic controls where hBMSCs were cultured on flat polymer films in the presence of osteogenic supplements (OS). In contrast, hBMSCs cultured on flat polymer films in the absence of OS assumed a more rounded and less-branched morphology. These results indicate that cells are more sensitive to scaffold structure than previously appreciated and suggest that scaffold efficacy can be optimized by tailoring the scaffold structure to force cells into morphologies that direct them to differentiate down the desired lineage.
Keywords: bone tissue engineering, cell morphology, nanotopography, osteogenesis, scaffolds, stem cell
1. Introduction
Use of tissue scaffold properties to direct cell function constitutes a basic tenet in the field of tissue engineering [1]. It is known that surface topography from the micro- to the nanoscale can influence cell behavior [2–4]. For instance, 2D topographical cues can induce osteoprogenitor cell [4,5] and mesenchymal stem cell [6] osteogenesis. Since topography has been reported to direct cell function, the hypothesis that 3D scaffold structure can direct cell differentiation has been tested herein. Many protocols for fabricating polymer scaffolds have been developed that yield a wide variety of structures including salt-leaching (SL) [7], gas foaming (GF) [8], phase-separation (GFPS) [9], electrospinning nanofibers (NF) [10,11] and freeform fabrication (FFF) [12,13]. Scaffolds fabricated by these methods have different physical architectures ranging from porous to fibrous, nanofibrous to microfibrous, and irregular to uniform. However, a systematic study to determine how these different scaffold structures influence cell function has not been performed.
Herein, gene expression signatures and osteogenic differentiation of primary human bone marrow stromal cells (hBMSCs) were assessed during culture on a library of scaffolds with varied structure and chemistry. hBMSCs are adult stem cells that can be isolated from bone marrow and can differentiate into bone, fat and cartilage [14,15]. Scaffolds with varied structure were fabricated from the same material poly(ε-caprolactone) (PCL) so that effects of scaffold chemistry could be decoupled from effects of scaffold structure. A number of controls were also tested including 2D flat samples (spun-coat films) and scaffolds made from a different polymer [PDLLA, poly(D,L-lactic acid)] to control for 3D structure and chemistry, respectively. hBMSC sensitivity to differences in scaffold structure were assessed and a mechanism for their response based on cell shape is proposed.
2. Materials and methods
2.1. PCL Salt-Leached Scaffolds (PCL_SL)
PCL (relative molecular mass 80 000 g/mol, Sigma) solution (30% mass fraction in chloroform) was mixed with sieved NaCl (250 μm to 425 μm) at a (NaCl):(PCL solution) mass ratio of 4:1 to yield a homogenous paste [16]. The paste was pressed into a disk shaped Teflon mold (12 mm diameter, 4 mm thickness). Scaffolds in molds were air dried for 2 h at room temperature, salt-leached 5 d in deionized water, air-dried for 1 d and stored in a desiccator.
2.2. PCL Gas-Foamed Scaffolds (PCL_GF)
PCL solution (30% by mass in chloroform) was mixed with sieved NH4HCO3 (250 μm to 425 μm) at a (NH4HCO3):(PCL solution) mass ratio of 2:1 to yield a homogenous paste [8]. The paste was pressed into a disk shaped Teflon mold (12 mm diameter, 4 mm thickness). Scaffolds in molds were air dried for 2 h at room temperature, removed from molds, incubated in warm d-water (40°C) until no gas bubbles were generated (2 h), further leached in d-water at room temperature for 3 d, air-dried for 1 d and stored in a desiccator.
2.3. PCL Gas-Foamed Phase-Separated Scaffolds (PCL_GFPS)
Gas foamed phase separated scaffolds of PCL were fabricated by combining thermally induced phase-separation technique (TIPS) and a gas-foaming technique [9]. PCL solution (30 % by mass in 7:3 volume ratio chloroform:n-butanol) was mixed with sieved NH4HCO3 (250 μm to 425 μm) at a (NH4HCO3):(PCL solution) mass ratio of 2:1 to yield a homogenous paste. The paste was pressed into a disk-shaped Teflon mold (12 mm diameter, 4 mm thickness) and phase separated at −80 °C for 4 h. Next, the scaffolds were immersed in methanol at −20 °C for 24 h for solvent exchange. Scaffolds were gas-foamed by immersion in warm d-water (40 °C) for 2 h (until no bubbles were released), immersed in d-water at room temperature for 3 d, air-dried for 1 d and stored in a desiccator.
2.4. PCL “Big” Nanofiber Scaffolds (PCL_BNF)
This protocol yields large diameter PCL nanofibers (≈900 nm). PCL solution (30 % by mass in 9:1 volume ratio chloroform: methanol) was loaded into a syringe and a syringe pump was used to dispense PCL solution (0.5 mL/h) into a home built electrospinning apparatus [11]. Aluminum foil was used as the target, the distance between needle and target was 15 cm, and voltage was 15 kV. The positive lead from the power supply was fixed to the spinneret which was an 18 gauge needle and the ground lead was fixed to the target (aluminum foil). Non-woven PCL nanofiber mats were collected for 1 h onto tissue culture polystyrene (TCPS) disks (12 mm dia., punched from the bottom of 48-well TCPS plates) placed on the aluminum foil target. TCPS disks were lightly sprayed with 70 % by mass ethanol before electrospinning to help fibers adhere to the disks. TCPS disks were made by using a hot 12 mm punch to punch disks from the bottom of TCPS 48-well plates.
2.5. PCL “Small” Nanofiber Scaffolds (PCL_SNF)
This protocol yields small diameter PCL nanofibers (≈300 nm) and is the same as for PCL-BNF except for the following changes: PCL solution 10 % by mass in 3:1 volume ratio chloroform: methanol, pump rate 2 mL/h and voltage 16.5 kV.
2.6. PCL Freeform Fabricated Scaffolds (PCL_FFF)
Disc-shaped freeform fabricated scaffolds (5 mm dia., 2 mm height, in 96-well plates) made by precision extrusion deposition [12] were purchased from 3D Biotek and used as received.
2.7. 2D Spin-Coated PCL Films (PCL_SC)
Thin films of PCL were then prepared by spin-coating PCL solution (0.8 mL, 10% by mass in glacial acetic acid) at 1000 rpm for 30 s over TCPS disks (100 mm dia.). Films were air-dried at room temperature for 2 h and annealed at 60° C for 30 s to adhere the PCL to the TCPS. Films were punched into 12 mm disks using a hot punch and exposed to sterile PBS to remove residual acetic acid.
2.8. PDLLA “Big” Nanofiber Scaffolds (PDLLA_BNF)
This protocol yields large diameter PDLLA nanofibers (≈900 nm) and is the same as for PCL-BNF except for the following changes: PDLLA (relative molecular mass 100,000 g/mol, Surmodics) solution 15 % by mass in hexafluoroisopropanol (1,1,1,3,3,3-hexafluoro-2-propanol), pump rate 1.5 mL/h and voltage 15 kV.
2.9. 2D Spin-Coated PDLLA Films (PDLLA_SC)
Thin films of PDLLA were then prepared by spin-coating PDLLA solution (0.8 mL, 10% by mass in glacial acetic acid) at 1500 rpm for 30 s over TCPS disks (100 mm dia.). Films were air-dried at room temperature for 2 h and annealed at 60° C for 30 s to adhere the PDLLA to the TCPS. Films were punched into 12 mm disks using a hot punch and exposed to sterile PBS to remove residual acetic acid.
2.10. Scaffold Characterization
PCL_SL, PCL_GF, PCL_GFPS, PCL_BNF, PCL_SNF, PCL_FFF and PDLLA_BNF scaffolds were examined by scanning electron microscopy (SEM). PCL_SL, PCL_GF and PCL_GFPS scaffolds were frozen in liquid nitrogen and fractured with a scalpel to expose interior. PCL_BNF, PCL_SNF, PCL_FFF and PDLLA_BNF scaffolds were examined from the top. Scaffolds were sputter-coated with gold for 90 s and imaged (SEM, 15kV, Hitachi s-4700-II FE-SEM). NF diameter, FFF strut diameter and FFF strut spacing were measured in SEM images using ImageJ software (NIH) (100 measurements of each). Morphology of 2D PCL_SC and TCPS samples were viewed by transmitted white light phase contrast microscopy. Gravimetry was used to measure total porosity of PCL_SL, PCL_GF, PCL_GFPS and PCL_FFF scaffolds using an equation, “Total Porosity = 1 − [(m/d)/v]”, where “m” is mass of the scaffold (g), “d” is PCL density (1.1 g/mL) and “v” is volume of scaffold (mL) (caliper measurements).
2.11. Cell Culture
PCL_SL, PCL_GF, PCL_GFPS, PCL_BNF, PCL_SNF, PCL_SC, PDLLA_BNF and PDLLA_SC specimens were placed in 48-well tissue-culture polystyrene plates for cell culture experiments. Wells without specimens were used as TCPS controls. PCL_FFF were put into polystyrene 96-well plates (non-tissue culture treated) for cell culture experiments. Specimens were sterilized by ethylene oxide (Anderson Products) and degassed for 2 d under house vacuum.
Primary human bone marrow stromal cells (hBMSCs, also known as mesenchymal stem cells, Tulane Center for Gene Therapy) were cultured at 37 °C with 5 % by volume CO2 in α-minimum essential medium (Invitrogen) supplemented with 16.5 % by volume fetal bovine serum (Atlanta Biologicals) and 4 mmole/L L-glutamine [14]. To wet scaffolds fully with culture medium, medium was added to specimens in wells and plates were placed under vacuum for 1 min. The vacuum was released and reapplied two more times. For seeding scaffolds, hBMSCs (80 % confluent) were dissociated with 0.25 % mass fraction trypsin [containing 1 mmole/L ethylenediaminetetraactate (EDTA)] and re-suspended in medium. Passage 4 or 5 cells were used for all experiments. Medium was removed from scaffolds and 10,000 hBMSCs in 0.5 mL of medium was added to each well for 48-well plates (PCL_SL, PCL_GF, PCL_GFPS, PCL_BNF, PCL_SNF, PCL_SC, PDLLA_BNF, PDLLA_SC, TCPS) or 5,000 hBMSCs in 0.2 mL of medium for 96-well plates (PCL_FFF). Medium was changed twice per week and cells cultured for 4 time points (1 d, 7 d, 21 d, 50 d) as indicated in the figures. In some experiments, cells were cultured in medium containing osteogenic supplements (OS): dexamethasone (10 nmole/L), β-glycerophosphate (20 mmol/L) and ascorbic acid (0.05 mmol/L).
2.12. Alizarin Red Staining
For Alizarin red staining, scaffolds were fixed with 3.7 % formaldehyde for 24 h at 37°C and then stained with Alizarin Red S (10 mg/mL) for 1 h. Scaffolds were washed 5 times with d-water to remove excess stain and air dried. Digital images of stained scaffolds were acquired using a stereomicroscope. All Alizarin red experiments were performed with three specimens (n = 3). Two fields of view at both low and high magnification were randomly selected and captured for each specimen and representative images are shown in the figures.
2.13. Picogreen DNA Assay
The Picogreen DNA assay was used to quantify cell numbers in the scaffolds. Scaffolds were washed with PBS (phosphate-buffered saline) and then incubated with lysis buffer (0.2 mg/mL Proteinase K and 0.02 % by mass sodium dodecyl sulfate) for 24 h at 37°C. After incubation, 100 μL of lysate was transferred to a clean 96-well plate and diluted with 100 μL of Picogreen reagent (Invitrogen, diluted to working concentration as per manufacturer’s protocol). Fluorescence (excitation 485 nm, emission 538 nm) was measured using a plate reader. A DNA standard curve was generated using known DNA concentrations to calibrate readings.
2.14. Fluorescence Staining of Cells
Cells on scaffolds were fixed with 3.7 % formaldehyde (mass/volume in PBS buffer) for 15 min, washed in PBS and permeabilized with 0.2 % by mass Triton X-100 for 5 min. Samples were rinsed with PBS and stained (1 h) with Alexa Fluor 546-phalloidin (20 nmol/L in PBS) and Sytox green (100 nmol/L in PBS buffer) to stain for F-actin and nuclei, respectively (Invitrogen). Cells were washed with PBS.
2.15. Epifluorescence Microscopy
For regular epifluorescence microscopy (non-confocal, Fig. 2 and Fig. S2-S4) to assess cell proliferation, specimens were washed with PBS, then water and air-dried before imaging. Two specimens were prepared for each treatment. Two fields of view at both low and high magnification were randomly selected and captured for green (nuclei) and red channels (actin) for each specimen and representative images are shown in the figures.
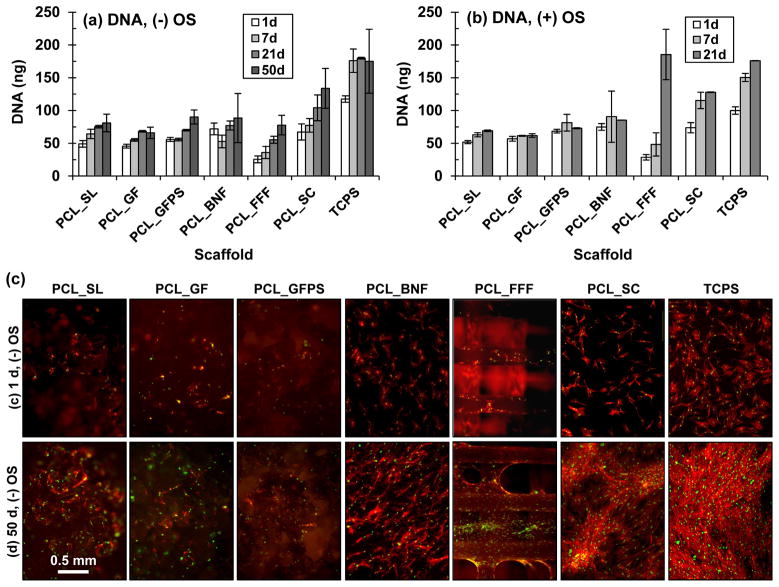
Figure 2.
(a,b) hBMSC cell numbers measured by Picogreen DNA assay after culture (a) without and (b) with OS. Error bars are S.D. (n = 4). (c) Fluorescence micrographs (40×) of hBMSCs cultured on scaffolds without OS for 1 d or 50 d. Nuclei are green (Sytox green) and actin is red (AlexaFluor 546 phalloidin). Scale bar applies to all images in (c).
2.16. Confocal Microscopy
Confocal microscopy (Zeiss LSM-510) was used to measure cell morphology using samples wet with PBS (not air-dried). High resolution z-stack images were captured with a 40×/0.80 water immersion objective (1 μm z-step size) for nuclei (Sytox green) and actin (Alexa Fluor 546 phalloidin). Two specimens were prepared for each treatment and ten cells were imaged and analyzed for each specimen (n = 20 cells).
Sytox green staining of nuclei was used to make sure that cell morphology was assessed for single cells only (only one nucleus per object). Alexa Fluor 546 phalloidin stained actin images were used to assess cell morphology. Confocal Z-stack projections were constructed (to project entire cell surface into two dimensions) and thresholding was performed to result in binary images. ImageJ was used to measure cell area, aspect ratio and roundness. Cell outlines were created and analyzed for branching. Branching was determined by counting the number of projections from the cell body and the number of branch points originating within that projection with a length greater than 5 μm.
2.17. Human Genome Microarray
Total RNA from hBMSCs in 3D scaffolds and on 2D surfaces at 1 d and 14 d was collected by homogenization of the cell lysate using the QIAshredder followed by purification using the RNeasy Minikit (Qiagen). Four replicates were performed for all treatments wherein a replicate was prepared by pooling cell lysates from 4 samples (expect for PCL_FFF scaffolds where 8 samples were pooled). Genome-wide transcriptions of hBMSCs were measured by high-throughput screening using the Illumina Human HT-12 v4 Expression BeadChips with 47231 probes for 25130 RefSeq annotated genes (NCBI/NIH) through a contract with Gene Logic (Gaithersburg, MD).
Data analysis was performed using the BRB-Array Tools version 4.1.0 (Biometric Research Branch, National Cancer Institute). The average of the four replicates of TCPS_1d was used as an in silico reference array for median normalization of all of the individual array data. The “significance analysis of microarrays” (SAM) approach [17], with a false discovery rate of 0.05, was chosen to test for significantly changed genes within the osteogenic controls (4 replicates each of TCPS_1d, TCPS_14d, TCPS_OS_1d and TCPS_OS_14d). A total of 9494 genes were identified that were changed significantly between all possible pair-wise comparisons of these treatments. From those 9494 SAM-significant genes, a subset of 831 genes was found to exhibit at least a two-fold change ‘between TCPS_1d and TCPS_OS_1d’ or ‘between TCPS_14d and TCPS_OS_14d’ (this creates “black” regions on the sorted 14d arrays indicating “no change”). Hierarchical cluster analysis (HCA), with centered correlation and average linkage, was performed using all arrays and the 831 two-fold significant genes. A dendrogram of the relationships is shown in Fig. 3a. The log2 ratio of the each gene intensity compared to the reference array (see above) was used to show the relative change in gene expression across the dataset. A heat map for the 831 SAM-significant genes is shown in Fig. 3b. Finally, 29 of those genes were found (January 2011) to be in a Gene Ontology [18] cluster for skeletal development (Fig. 3c).
Figure 3.
Gene expression profiles of hBMSCs on scaffolds and control surfaces at 1d and 14d revealed by microarrays. (a) Dendrogram from hierarchical cluster analysis organizing experiments by similarity. Only three treatments (marked with an asterisk) would need to be relocated to give perfect segregation by treatment in the dendrogram. (b) Heat map of relative gene expression for 831 genes identified to be significantly regulated in osteogenic controls (at least a two-fold change ‘between TCPS 1d and TCPS_OS 1d’ or ‘between TCPS 14d and TCPS_OS 14d’). Data are organized from highest up-regulation to lowest down-regulation for TCPS_OS 14d. (c) From the 831 genes shown in (b), a subset of 29 genes that are involved in skeletal system development was identified by Gene Ontology. Color scale at bottom of figure applies to (b,c) and is log2 of the ratio of fluorescence intensity to the mean value for the four TCPS 1d controls.
2.18. Imaging Fluorescent PDLLA_BNF
In some cases (Fig. 4i-k), PDLLA_BNF were doped with Rhodamine-123 (5 μg/mL in electrospinning solution, Molecular Probes) so that they could be imaged by fluorescence microscopy. For 3D renderings in Fig. 4i-j, confocal microscopy images (Leica TCS SP5 broadband) were collected with a 63x/0.9 water immersion objective (290 nm Z-step size) for nanofibers (Rhodamine-123) and hBMSC actin (Alexa Fluor 633 phalloidin). For the single Z-slice in Fig. 4k, a confocal microscopy image (Zeiss LSM-510) was collected with a 40×/0.80 water immersion objective for nanofibers (Rhodamine-123) and hBMSC actin (Alexa Fluor 546 phalloidin).
Figure 4.
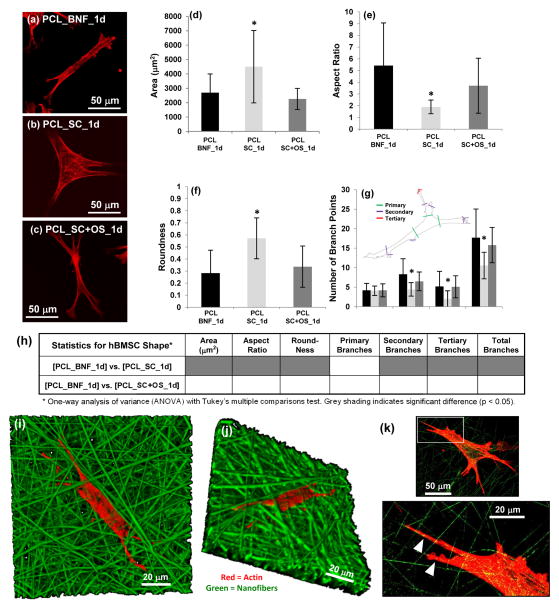
(a–c) hBMSC morphology (400×) after 1 d culture for (a) PCL_BNF, (b) PCL_SC and (c) PCL_SC+OS. Actin is red (AlexaFluor 546 phalloidin) and projections of confocal z-stacks are shown. (d–g) Cell morphology using quantified for (d) area, (e) aspect ratio, (f) roundness and (g) number of primary, secondary, tertiary and total branch points for hBMSCs cultured 1 d on PCL_SNF, PCL_SC and PCL_SC+OS. Confocal z-stack projections of Alexa Fluor 546 phalloidin-stained cells were used for analysis. Error bars are S.D. and 20 cells were analyzed for each treatment (n = 20). Inset in (h) defines branching. Asterisk in (d–g) indicates significantly different from PCL_BNF [p < 0.05, one-way analysis of variance (ANOVA) with Tukey’s test for multiple comparisons]. (h) Statistical analysis for hBMSC morphology. (i–k) Confocal imaging of hBMSCs (red, actin, Alexa Fluor 633 phalloidin) cultured on PDLLA_BNF (green, Rhodamine 123 doping) for 24 h. (i) 3D view of a confocal Z-stack (630×). (j) 3D view of the Z-stack shown in (i) but rotated in 3D space to show how hBMSCs have migrated down into the nanofibers (630×). (k) One Z-slice (400×) shows how hBMSCs extend projections that follow along the nanofibers (white arrowheads). The area outlined by the white box in the top image is enlarged in the bottom image.
3. Results
In order to test if 3D scaffold architecture affects hBMSC behavior, poly(ε-caprolactone) (PCL) scaffolds with different architectures were fabricated by five different techniques (Table 1): salt-leaching (PCL_SL), gas-foaming (PCL_GF), gas-foaming phase-separation (PCL_GFPS), electrospinning nanofibers (PCL_BNF) and freeform fabrication (PCL_FFF). Spun-coat films of PCL (PCL_SC) were used as a flat 2D control for 3Dness. Tissue culture polystyrene (TCPS) was also used as a control flat surface. Using PCL to fabricate these different scaffold architectures enables contributions from material chemistry to be isolated from topographical effects. The specimens presented a wide variety of morphologies as shown by the range of structural parameters including porosities and fiber diameters (Fig. 1; Table 2).
Table 1.
Scaffold Abbreviations
| Abbreviation | Definition |
|---|---|
| PCL_SL | Poly(ε-Caprolactone) Salt-Leached Scaffold |
| PCL_GF | Poly(ε-Caprolactone) Gas-Foamed Scaffold |
| PCL_GFPS | Poly(ε-Caprolactone) Gas-Foamed Phase-Separated Scaffolds |
| PCL_BNF | Poly(ε-Caprolactone) “Big” Nanofiber Scaffolds |
| PCL_SNF | Poly(ε-Caprolactone) “Small” Nanofiber Scaffolds |
| PCL_FFF | Poly(ε-Caprolactone) Freeform Fabricated Scaffolds |
| PCL_SC | Poly(ε-Caprolactone) Spun-Coat Films |
| PDLLA_BNF | Poly(D,L-Lactic Acid) “Big” Nanofiber Scaffolds |
| PDLLA_SC | Poly(D,L-Lactic Acid) Spun-Coat Films |
| TCPS | Tissue Culture Polystyrene |
| +OS | (+) Osteogenic Supplements |
Figure 1.
(a,b) Structure of scaffolds is shown by SEM for 3D scaffolds PCL_SL, PCL_GF, PCL_GFPS, PCL_BNF, PCL_SNF, PCL_FFF, PDLLA_BNF and by phase contrast microscopy for 2D samples PCL_SC, PDLLA_SC and TCPS. Scale bar in (a) applies to all images in (a) and scale bar in (b) applies to all images in (b). (c) Stereomicrographs of calcium staining (Alizarin red) for hBMSC osteogenesis on scaffolds in the absence of osteogenic supplements (OS) after 50 d culture. The scale bar in TCPS applies to all panels except PCL_FFF. Magnification is 18× for all images except PCL_FFF (PCL_FFF is 32×).
Table 2.
Scaffold Structural Measurements [mean ± S.D. (n)]
| PCL_SL Porositya (%) | 94.3 ± 0.7 (10) |
| PCL_GF Porositya (%) | 92.6 ± 0.8 (10) |
| PCL_GFPS Porositya (%) | 90.3 ± 0.7 (10) |
| PCL_BNF Fiber Diameterb (nm) | 910 ± 526 (100) |
| PCL_SNF Fiber Diameterb (nm) | 326 ± 128 (70) |
| PCL FFF Porositya (%) | 65.6 ± 0.9 (10) |
| PCL_FFF Strut Diameterb (μm) | 288 ± 29 (100) |
| PCL_FFF Strut Spacingb (μm) | 491 ± 39 (100) |
| PDLLA_BNF Fiber Diameterb (nm) | 942 ± 234 (70) |
Porosity measured by gravimetry.
Fiber diameter, strut diameter and strut spacing measured in electron micrographs.
When hBMSCs were cultured on the scaffolds in the absence of osteogenic supplements (OS), the nanofiber scaffolds (PCL_BNF) induced deposition of a bone-like matrix containing calcium (Fig. 1, Fig. S1). hBMSCs did not produce a calcified matrix on the other scaffold types (PCL_SL, PCL_GF, PCL_GFPS, PCL_FFF) or the flat controls (PCL_SC, TCPS) unless OS were added to the culture medium. These results suggest that only the nanofibrous scaffolds can induce hBMSC osteogenesis.
Next, the nanofiber effect was investigated more closely and the effect of nanofiber diameter and chemistry were tested. “Small” diameter 300 nm nanofibers (PCL_SNF) were fabricated and induced hBMSC osteogenesis in the absence of OS. “Big” 900 nm nanofibers made from another polymer, poly(D,L-lactic acid) (PDLLA_BNF), induced osteogenesis but control flat PDLLA films (PDLLA_SC) did not. When control PCL_BNF were incubated in complete medium without hBMSCs, they did not induce non-specific calcium deposition from medium (Fig. S1). A summary of all experiments is given in Fig. S8. These results indicate that nanofiber scaffolds of different sizes (300 nm and 900 nm) and different chemistries (PCL and PDLLA) induced hBMSC osteogenesis without OS, while flat films of the same polymers required OS to support osteogenic differentiation of hBMSCs.
The effect of scaffold architecture on hBMSC proliferation was tested using Picogreen DNA assay, fluorescence microscopy and stereomicroscopy (Fig. 2; Fig. S2-S5). hBMSCs adhered and proliferated on all specimens tested (PCL_SL, PCL_GF, PCL_GFPS, PCL_BNF, PCL_FFF, PCL_SC, TCPS) both in the presence and absence of osteogenic supplements. For the DNA assay, hBMSC numbers increased the fastest on PCL_FFF scaffolds (both without and with OS) and the highest amount of DNA extracted from any specimen was for PCL_FFF in the presence of OS after 21 d culture (Fig. 2). This was observed despite the fact that that PCL_SL, PCL_GF, PCL_GFPS, PCL_BNF, PCL_FFF, PCL_SC and TCPS were seeded with more hBMSCs (10,000) and were cultured in larger wells (48-well) than the PCL_FFF (PCL_FFF were seeded with 5000 cells in 96-well plates). The hBMSCs formed a tissue-like matrix at the strut joints on the PCL_FFF scaffolds by 21 d as observed in fluorescence micrographs and stereomicrographs (both without and with OS) (Fig. 2; Fig. S1, S2, S4). By 50 d without OS, a tissue-like matrix formed a continuous sheet between some struts on the PCL_FFF scaffolds (Fig. S1c, S2). These data indicate that hBMSCs adhered and proliferated on all substrates tested (PCL_SL, PCL_GF, PCL_GFPS, PCL_BNF, PCL_FFF, PCL_SC, TCPS) but that PCL_FFF scaffolds supported the most hBMSC proliferation.
The effect of scaffold architecture on hBMSC gene expression was assessed using microarrays (mRNA). A dendrogram resulting from cluster analysis (Fig. 3a) plus heat maps of the expression level for significantly regulated genes (Fig. 3b) are shown. Heat maps of the significantly regulated genes involved in skeletal development are also given (Fig. 3c, Table S1). Cluster analysis indicated that hBMSC gene expression at 14 d on the nanofiber specimens correlated most closely with the osteogenic control treatments. Thus, Node A (TCPS+OS 14d: hBMSCs cultured 14 d in OS on TCPS) and Node B (14d nanofiber specimens: PCL_BNF, PCL_SNF, PDLLA_BNF) grouped together under Node C. These data indicate that nanofiber scaffolds induced a gene expression signature that was similar to that induced by OS, further supporting the observation that culture of hBMSCs on nanofibrous scaffolds induces hBMSC osteogenesis.
It is also astonishing that for the different treatments, the 4 replicates group together almost exclusively (Fig. 3a). Indeed, only three treatments (marked with an asterisk in Fig. 3) would need to be relocated to give perfect segregation by treatment in the dendrogram. Also, all 14 d flat specimens (TCPS, PCL_SC, PDLLA_SC) group together under node D, indicating that the hBMSCs can sense the 2-dimensionality of these substrates. For these experiments, specimen architecture was more important than surface chemistry, since PCL_SC grouped with the other 2D specimens (PDLLA_SC, TCPS) instead of with other PCL specimens. Likewise, PDLLA_SC grouped with the other 2D specimens (PCL_SC, TCPS) instead of with the other PDLLA specimen (PDLLA_BNF). These results indicate that hBMSCS are exquisitely sensitive to scaffold structure resulting in a unique gene expression profile for each type of scaffold tested. The results also suggest that scaffold structure has a bigger influence on stem cell behavior than does polymer chemistry.
The ability of nanofibers to drive osteogenic differentiation of hBMSCs was investigated further to understand the mechanism. Fluorescence micrographs suggested that cell shape could be driving osteogenesis on nanofiber scaffolds. hBMSCs assumed a spindly, highly branched morphology on PCL_BNF which was very different than the well spread morphology observed for flat films (PCL_SC) (Fig. S2–S4). Since cell morphology is tightly linked to cell function [19,20], this possibility was investigated further using high resolution confocal microscopy to quantify and compare cell morphology on PCL nanofibers (PCL_BNF) and films (PCL_SC) after 1 d culture (Fig. 4, Fig. S7). hBMSCs cultured on PCL_BNF had smaller cell area, higher aspect ratio, lower roundness and more branching than did hBMSCs on PCL_SC. Strikingly, hBMSCs cultured on PCL_SC in the presence of OS also took on a spindly, branched morphology that was similar to hBMSCs on PCL_BNF (Fig. 4). Thus, elongated, highly branched hBMSC morphologies induced osteogenesis regardless of whether morphological changes were driven by scaffold architecture (nanofiber scaffolds) or by biochemical supplements (OS).
Confocal 3D renderings of hBMSCs on fluorescent nanofiber scaffolds show that the hBMSCs penetrate into the nanofiber scaffolds (Fig. 4i–j). In addition, the hBMSCs extended projections that follow along the individual nanofibers resulting in the elongated, branched hBMSC morphology (Fig. 4k). The hBMSCs cannot attach to the aqueous medium in the voids between the nanofibers and have no choice but to adhere to the nanofibers, which forces them into and elongated and highly branched morphology. Taken together, these results suggest that the intrinsic structure of the nanofiber scaffolds forces the hBMSCs into an osteogenic morphology that drives their osteogenic differentiation.
4. Discussion
The cluster analysis dendrogram (Fig. 3) is powerful because it provides an unbiased approach of sorting scaffold treatments according to gene expression patterns. The analysis considers all genes and does not focus on pre-selected markers. In addition, this approach provides a unique way to analyze cell-material interactions where the sorting of the treatments provides insight into which material properties are influencing cell fate. In Fig. 3a, the different scaffold treatments sort by scaffold structure over scaffold composition, suggesting that scaffold structure has a greater ability to control cell fate than does composition. Combining the microarray cluster analysis with the scaffold library approach, where many types of scaffold architectures are systematically compared side-by-side, robustly demonstrates that cells are more sensitive to scaffold structure than previously appreciated.
Although the advantages of nanofiber scaffolds for tissue engineering are well-documented and their ability to promote osteogenesis has been demonstrated [21–23], the mechanism for these effects has not been defined. The current scaffold library results further emphasize the ability of nanofibers to induce osteogenesis by systematically comparing multiple scaffold architectures side by side. A nanofibrous topography was key since only the nanofiber scaffolds, of all the scaffold morphologies that were tested (SL, GF, GFPS, NF, FFF), induced osteogenic differentiation. In addition, the current results provide a mechanistic basis for the effects of nanofibers. Nanofibers provided fibrous adhesion sites for the hBMSCs that caused the hBMSCs to take on an elongated, highly branched morphology which drove them down and osteogenic lineage (Fig. 4). The current experiments suggest that the osteoinductive effects of nanofibers come from their ability to force cells into an osteogenic morphology.
Decellularized extracellular matrices (dECM) have been very successful as tissue engineering scaffolds and collagen nanofibers are the dominant structure of dECM [24–27]. Synthetic polymer nanofiber scaffolds such as PCL_BNF mimic the structure of the collagen nanofibers present in dECM [11]. The current results suggest that the dECM scaffolds may be effective because their nanofibrous structure drives cells into morphologies that enhance their differentiation. It will be interesting to see if this holds true in future work.
It is well-established that cell morphology and cell function are strongly linked [19,20] and that this principle applies to hBMSC osteogenesis [28–30]. McBeath et al. demonstrated that modulation of hBMSC shape with surface patterning could be used to switch hBMSC differentiation between osteogenic or adipogenic [28]. In addition, Rodriguez et al. observed that osteogenic supplements caused changes in hBMSC morphology [29]. In vivo, hBMSCs progress to osteoblasts and then to mature osteocytes. While osteoblasts are cuboidal, osteocytes are highly branched, residing in lacunae and radiating multiple extensions down canaliculi [31]. Thus, nanofiber scaffolds may force the hBMSCs into a highly branched “osteocyte-like” morphology, which pushes the hBMSCs towards an osteogenic lineage.
The data suggest that scaffolds should be designed to drive cells into morphologies that will induce the desired cell function (proliferation, migration, differentiation, etc.). Models for cell spreading could be adapted to predict cell shape on different 3D structures. Alternatively, more extensive libraries of 3D structures could be probed for their effect on cell shape and function to create a catalog of scaffold structures versus cell shape/behavior. Coupling scaffold structure to cell shape could yield new targets for scaffold design that could enhance the inherent regenerative capabilities of scaffolds.
5. Conclusions
A systematic comparison of multiple scaffold types highlights the unexpected sensitivity that cells have for scaffold structure. hBMSCs took on a unique gene expression profile on each type of scaffold structure tested demonstrating that hBMSCs are highly responsive to scaffold architecture. In addition, the effect of scaffold structure on stem cell behavior was mediated by cell shape. Nanofibers scaffolds induced osteogenic differentiation of hBMSCs by forcing the cells into an elongated, highly-branched, osteogenic morphology. These results advance our understanding of how scaffold architecture can control cell fates. The data suggest a new paradigm for improving 3D tissue scaffold efficacy where the structure of the scaffold can be designed to drive cells into morphologies that direct their differentiation down a desired lineage.
Supplementary Material
Acknowledgments
G.K. and K.C. were supported by an NIH-NIBIB/NIST NRC Research Associateship. C.K.T. was supported by a NIST NRC Research Associateship. This work was supported by NIST and the Intramural Program of the NIH/NIDCR (National Institute of Dental and Craniofacial Research). Some of the materials employed in this work were provided by the Tulane Center for Gene Therapy through a grant from NCRR of the NIH P40RR017447. The “standard deviation” (S.D.) is the same as the “combined standard uncertainty of the mean” for the purposes of this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH, NIBIB, NIDCR or NIST. This article, a contribution of NIST, is not subject to US copyright. Certain equipment and instruments or materials are identified in the paper to adequately specify the experimental details. Such identification does not imply recommendation by NIST, nor does it imply the materials are necessarily the best available for the purpose.
References
- 1.Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310(5751):1135–8. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 2.Dalby MJ, Riehle MO, Johnstone HJ, Affrossman S, Curtis AS. Polymer-demixed nanotopography: control of fibroblast spreading and proliferation. Tissue Eng. 2002;8(6):1099–108. doi: 10.1089/107632702320934191. [DOI] [PubMed] [Google Scholar]
- 3.Thapa A, Miller DC, Webster TJ, Haberstroh KM. Nano-structured polymers enhance bladder smooth muscle cell function. Biomaterials. 2003;24(17):2915–26. doi: 10.1016/s0142-9612(03)00123-6. [DOI] [PubMed] [Google Scholar]
- 4.Lovmand J, Justesen E, Foss M, Lauridsen RH, Lovmand M, Modin C, et al. The use of combinatorial topographical libraries for the screening of enhanced osteogenic expression and mineralization. Biomaterials. 2009;30(11):2015–22. doi: 10.1016/j.biomaterials.2008.12.081. [DOI] [PubMed] [Google Scholar]
- 5.Zhao G, Zinger O, Schwartz Z, Wieland M, Landolt D, Boyan BD. Osteoblast-like cells are sensitive to submicron-scale surface structure. Clin Oral Implants Res. 2006;17(3):258–64. doi: 10.1111/j.1600-0501.2005.01195.x. [DOI] [PubMed] [Google Scholar]
- 6.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007;6(12):997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 7.Shea LD, Wang D, Franceschi RT, Mooney DJ. Engineered bone development from a pre-osteoblast cell line on three-dimensional scaffolds. Tissue Engineering. 2000;6(6):605–17. doi: 10.1089/10763270050199550. [DOI] [PubMed] [Google Scholar]
- 8.Nam YS, Yoon JJ, Park TG. A novel fabrication method of macroporous biodegradable polymer scaffolds using gas foaming salt as a porogen additive. J Biomed Mater Res. 2000;53(1):1–7. doi: 10.1002/(sici)1097-4636(2000)53:1<1::aid-jbm1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 9.Whang K, Thomas CH, Healy KE, Nuber G. A Novel Method to Fabricate Bioabsorbable Scaffolds. Polymer. 1995;36(4):837–42. [Google Scholar]
- 10.Ma PX, Zhang R. Synthetic nano-scale fibrous extracellular matrix. J Biomed Mater Res. 1999;46(1):60–72. doi: 10.1002/(sici)1097-4636(199907)46:1<60::aid-jbm7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 11.Li WJ, Laurencin CT, Caterson EJ, Tuan RS, Ko FK. Electrospun nanofibrous structure: a novel scaffold for tissue engineering. J Biomed Mater Res. 2002;60(4):613–21. doi: 10.1002/jbm.10167. [DOI] [PubMed] [Google Scholar]
- 12.Darling AL, Sun W. 3D microtomographic characterization of precision extruded poly-epsilon-caprolactone scaffolds. J Biomed Mater Res B Appl Biomater. 2004;70(2):311–7. doi: 10.1002/jbm.b.30050. [DOI] [PubMed] [Google Scholar]
- 13.Kim K, Lee CH, Kim BK, Mao JJ. Anatomically shaped tooth and periodontal regeneration by cell homing. J Dent Res. 2010;89(8):842–7. doi: 10.1177/0022034510370803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominici M, Le BK, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 15.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–9. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorsey SM, Lin-Gibson S, Simon CG., Jr X-ray microcomputed tomography for the measurement of cell adhesionand proliferation in polymer scaffolds. Biomaterials. 2009;30(16):2967–74. doi: 10.1016/j.biomaterials.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273(5661):345–9. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 20.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 21.Smith LA, Liu X, Hu J, Ma PX. The influence of three-dimensional nanofibrous scaffolds on the osteogenic differentiation of embryonic stem cells. Biomaterials. 2009;30(13):2516–22. doi: 10.1016/j.biomaterials.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith LA, Liu X, Hu J, Wang P, Ma PX. Enhancing osteogenic differentiation of mouse embryonic stem cells by nanofibers. Tissue Eng Part A. 2009;15(7):1855–64. doi: 10.1089/ten.tea.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruckh TT, Kumar K, Kipper MJ, Popat KC. Osteogenic differentiation of bone marrow stromal cells on poly(epsilon-caprolactone) nanofiber scaffolds. Acta Biomater. 2010;6(8):2949–59. doi: 10.1016/j.actbio.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Abrams GA, Goodman SL, Nealey PF, Franco M, Murphy CJ. Nanoscale topography of the basement membrane underlying the corneal epithelium of the rhesus macaque. Cell Tissue Res. 2000;299(1):39–46. doi: 10.1007/s004419900074. [DOI] [PubMed] [Google Scholar]
- 25.Atala A, Bauer SB, Soker S, Yoo JJ, Retik AB. Tissue-engineered autologous bladders for patients needing cystoplasty. Lancet. 2006;367(9518):1241–6. doi: 10.1016/S0140-6736(06)68438-9. [DOI] [PubMed] [Google Scholar]
- 26.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008 Feb;14(2):213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 27.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329(5991):538–41. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–95. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez JP, Gonzalez M, Rios S, Cambiazo V. Cytoskeletal organization of human mesenchymal stem cells (MSC) changes during their osteogenic differentiation. J Cell Biochem. 2004;93(4):721–31. doi: 10.1002/jcb.20234. [DOI] [PubMed] [Google Scholar]
- 30.Treiser MD, Yang EH, Gordonov S, Cohen DM, Androulakis IP, Kohn J, et al. Cytoskeleton-based forecasting of stem cell lineage fates. Proc Natl Acad Sci U S A. 2010;107(2):610–5. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marks SC, Jr, Hermey DC. The structure and development of bone. In: Bilezikian JP, Raisz LG, Rodan GA, editors. Principles of Bone Biology. 1. New York: Academic Press; 1996. pp. 3–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.