Abstract
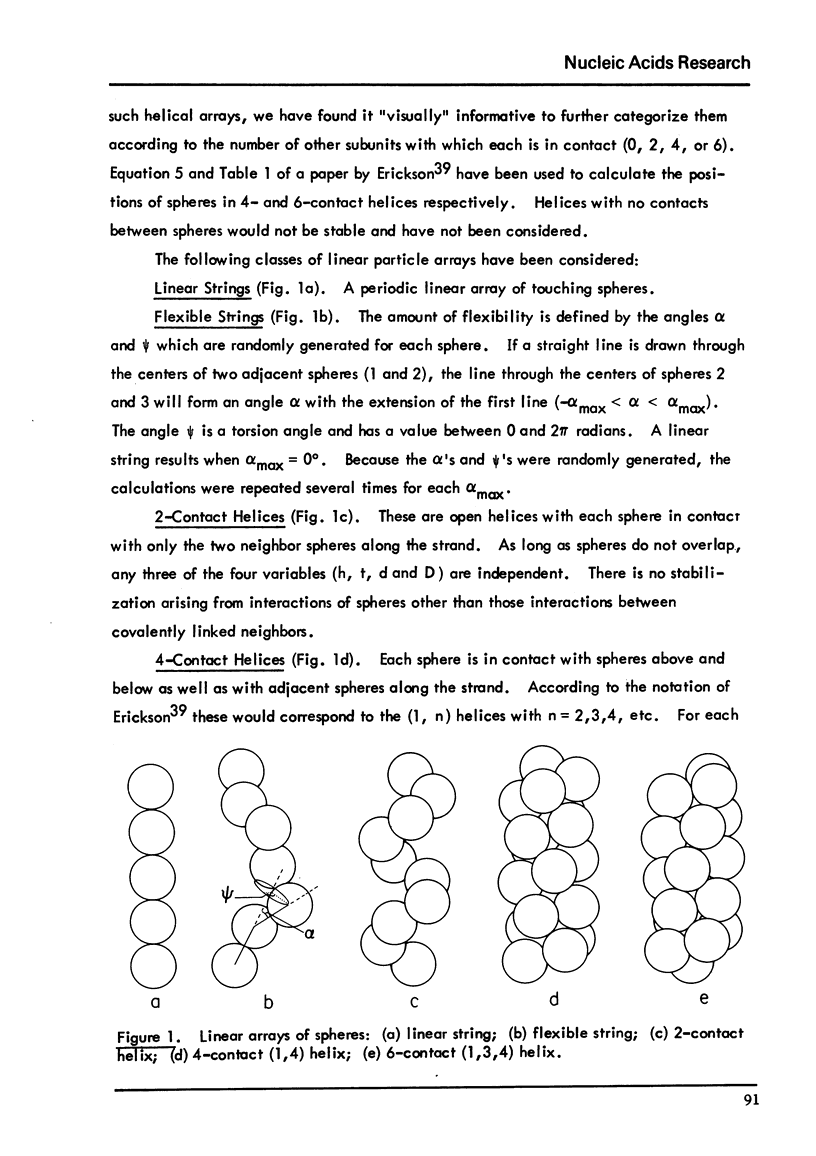
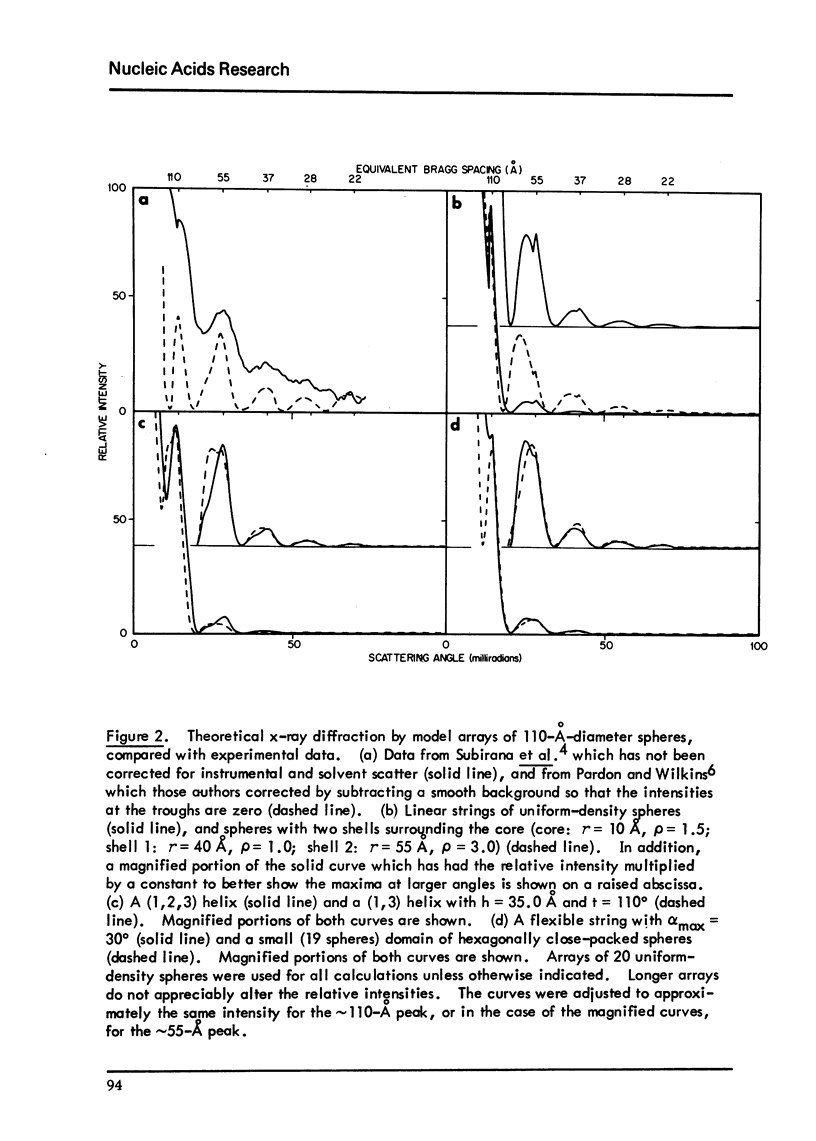
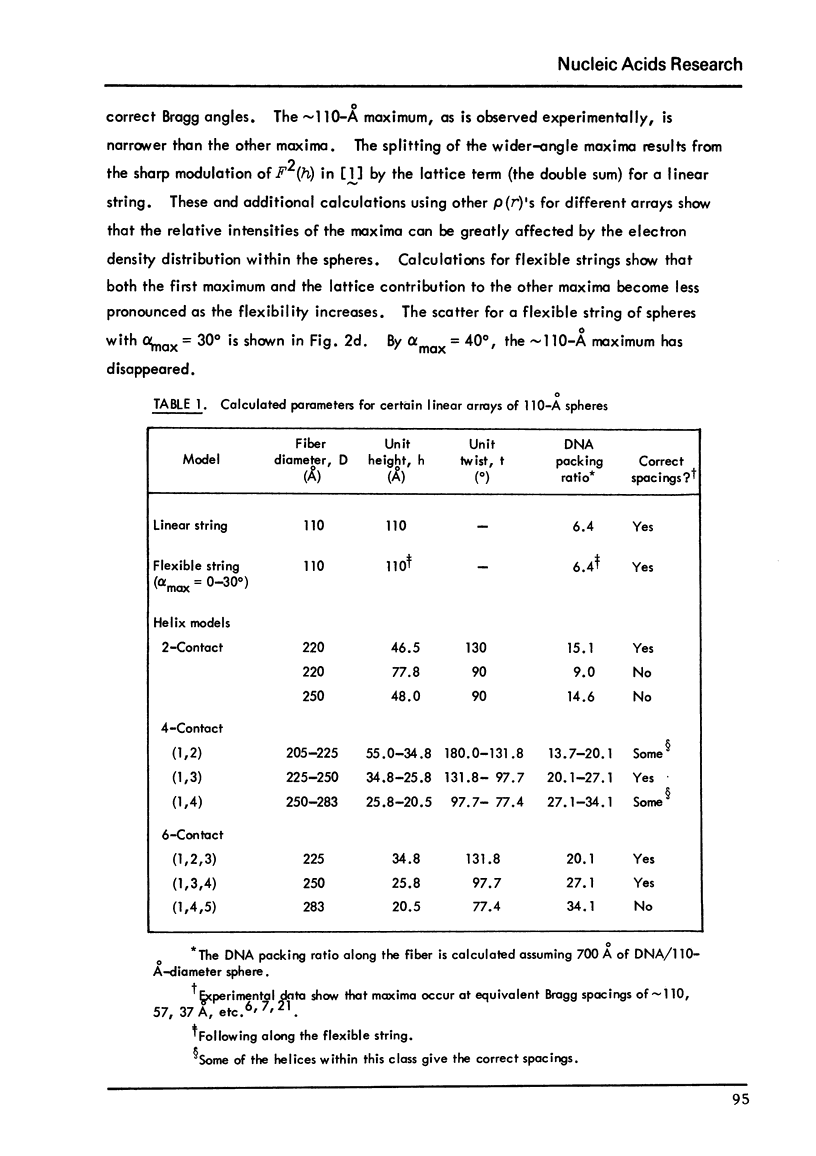
Chromatin fibers consists of globular nucleohistone particles (designated nu bodies) along the length of the chromatin DNA with approximately 6-to7-fold compaction of the DNA within the nu bodies. We have calculated theoretical small-angle x-ray scattering curves and have compared these with experimental data in the literature. Several models predict maxima at the correct angles. The first maximum (approximately 110 degrees A) results from interparticle interference, while both the spatial arrangement and the structure factor the nu bodies can contribute to the additional small-angle maxima. These calculations suggest models which can account for the electron microscopic observation that chromatin is seen as either approximately 100-or approximately 200-to 250 degrees A-diameter fibers, depending on the solvent conditions. They also account for the limited orientability of the x-ray pattern from pulled chromatin fibers.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Bram S., Butler-Browne G., Baudy P., Ibel K. Quaternary structure of chromatin. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1043–1045. doi: 10.1073/pnas.72.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch K., Seligy V. L., Setterfield G. Effects of low salt concentration on structural organization and template activity of chromatin in chicken erythrocyte nuclei. Exp Cell Res. 1971 Mar;65(1):61–72. doi: 10.1016/s0014-4827(71)80050-2. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Hewish D. R., Mobbs J. Mammalian chromatin substructure studies with the calcium-magnesium endonuclease and two-dimensional polyacrylamide-gel electrophoresis. Biochem J. 1974 Oct;143(1):67–72. doi: 10.1042/bj1430067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson R. D., Olins A. L., Olins D. E. Urea denaturation of chromatin periodic structure. Biochemistry. 1975 Jul 15;14(14):3122–3125. doi: 10.1021/bi00685a013. [DOI] [PubMed] [Google Scholar]
- Davies H. G. Electron-microscope observations on the organization of heterochromatin in certain cells. J Cell Sci. 1968 Mar;3(1):129–150. doi: 10.1242/jcs.3.1.129. [DOI] [PubMed] [Google Scholar]
- DuPraw E. J. Macromolecular organization of nuclei and chromosomes: a folded fibre model based on whole-mount electron microscopy. Nature. 1965 Apr 24;206(982):338–343. doi: 10.1038/206338a0. [DOI] [PubMed] [Google Scholar]
- Erickson R. O. Tubular packing of spheres in biological fine structure. Science. 1973 Aug 24;181(4101):705–716. doi: 10.1126/science.181.4101.705. [DOI] [PubMed] [Google Scholar]
- Everid A. C., Small J. V., Davies H. G. Electron-microscope observations on the structure of condensed chromatin: evidence for orderly arrays of unit threads on the surface of chicken erythrocyte nuclei. J Cell Sci. 1970 Jul;7(1):35–48. doi: 10.1242/jcs.7.1.35. [DOI] [PubMed] [Google Scholar]
- Golomb H. M., Bahr G. F. Electron microscopy of human interphase nuclei. Determination of total dry mass and DNA-packing ratio. Chromosoma. 1974;46(3):233–245. doi: 10.1007/BF00284879. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Henley C. Chromatin condensation involving lamellar strands in spermiogenesis of Goniobasis proxima. Chromosoma. 1973;42(2):163–174. doi: 10.1007/BF00320938. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hill W. E., Rossetti G. P., Van Holde K. E. Physical studies of ribosomes from Escherichia coli. J Mol Biol. 1969 Sep 14;44(2):263–277. doi: 10.1016/0022-2836(69)90174-0. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Olins D. E. Visualization of chromatin substructure: upsilon bodies. J Cell Biol. 1975 Mar;64(3):528–537. doi: 10.1083/jcb.64.3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oosterhof D. K., Hozier J. C., Rill R. L. Nucleas action on chromatin: evidence for discrete, repeated nucleoprotein units along chromatin fibrils. Proc Natl Acad Sci U S A. 1975 Feb;72(2):633–637. doi: 10.1073/pnas.72.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H. A super-coil model for nucleohistone. J Mol Biol. 1972 Jul 14;68(1):115–124. doi: 10.1016/0022-2836(72)90267-7. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H., Richards B. M. Super-helical model for nucleohistone. Nature. 1967 Jul 29;215(5100):508–509. doi: 10.1038/215508a0. [DOI] [PubMed] [Google Scholar]
- Pooley A. S., Pardon J. F., Richards B. M. The relation between the unit thread of chromosomes and isolated nucleohistone. J Mol Biol. 1974 Jan 5;85(4):533–549. doi: 10.1016/0022-2836(74)90314-3. [DOI] [PubMed] [Google Scholar]
- Richards B. M., Pardon J. F. The molecular structure of nucleohistone (DNH). Exp Cell Res. 1970 Sep;62(1):184–196. doi: 10.1016/0014-4827(79)90519-6. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- Ris H., Kubai D. F. Chromosome structure. Annu Rev Genet. 1970;4:263–294. doi: 10.1146/annurev.ge.04.120170.001403. [DOI] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Senior M. B., Olins A. L., Olins D. E. Chromatin fragments resembling v bodies. Science. 1975 Jan 17;187(4172):173–175. doi: 10.1126/science.1111096. [DOI] [PubMed] [Google Scholar]
- Subirana J. A., Puigjaner L. C. X-ray diffraction studies of nucleohistone: a polyhelical model of chromosome organization. Proc Natl Acad Sci U S A. 1974 May;71(5):1672–1676. doi: 10.1073/pnas.71.5.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. A model for particulate structure in chromatin. Nucleic Acids Res. 1974 Nov;1(11):1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKINS M. H. Physical studies of the molecular structure of deoxyribose nucleic acid and nucleoprotein. Cold Spring Harb Symp Quant Biol. 1956;21:75–90. doi: 10.1101/sqb.1956.021.01.007. [DOI] [PubMed] [Google Scholar]
- Wolfe S. L., Grim J. N. The relationship of isolated chromosome fibers to the fibers of the embedded nucleus. J Ultrastruct Res. 1967 Aug;19(3):382–397. doi: 10.1016/s0022-5320(67)80226-0. [DOI] [PubMed] [Google Scholar]
- van Bruggen E. F., Arnberg A. C., van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. Electron microscopy of chromatin subunit particles. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1365–1370. doi: 10.1016/0006-291x(74)90348-9. [DOI] [PubMed] [Google Scholar]


