HMGB1 orchestrates leukocyte recruitment and their induction to secrete inflammatory cytokines by switching between mutually exclusive redox states.
Abstract
Tissue damage causes inflammation, by recruiting leukocytes and activating them to release proinflammatory mediators. We show that high-mobility group box 1 protein (HMGB1) orchestrates both processes by switching among mutually exclusive redox states. Reduced cysteines make HMGB1 a chemoattractant, whereas a disulfide bond makes it a proinflammatory cytokine and further cysteine oxidation to sulfonates by reactive oxygen species abrogates both activities. We show that leukocyte recruitment and activation can be separated. A nonoxidizable HMGB1 mutant in which serines replace all cysteines (3S-HMGB1) does not promote cytokine production, but is more effective than wild-type HMGB1 in recruiting leukocytes in vivo. BoxA, a HMGB1 inhibitor, interferes with leukocyte recruitment but not with activation. We detected the different redox forms of HMGB1 ex vivo within injured muscle. HMGB1 is completely reduced at first and disulfide-bonded later. Thus, HMGB1 orchestrates both key events in sterile inflammation, leukocyte recruitment and their induction to secrete inflammatory cytokines, by adopting mutually exclusive redox states.
Injury can trigger an acute inflammatory response, even in the absence of concomitant infection. “Sterile” inflammation is also associated with several types of cancer. Two events are key for the development of sterile inflammation: the recruitment of leukocytes, especially neutrophils and monocytes, and their activation to release proinflammatory cytokines.
High-mobility group box 1 (HMGB1) is a nuclear protein that signals tissue damage when released into the extracellular medium, and thus works as a damage-associated molecular pattern (DAMP; Bianchi, 2007). Extracellular HMGB1 can act both as a chemoattractant for leukocytes and as a proinflammatory mediator to induce both recruited leukocytes and resident immune cells to release TNF, IL-1, IL-6, and other cytokines. Notably, immune cells secrete HMGB1 when activated by infection or tissue damage (Andersson and Tracey, 2011); mesothelioma and other cancer cells secrete HMGB1 constitutively (Jube et al., 2012).
Recent studies have shown that the proinflammatory cytokine-stimulating activity of HMGB1 depends on the redox state of three cysteines: C23 and C45 must form a disulfide bond within the first HMG-box domain of HMGB1, BoxA, whereas the unpaired C106 within BoxB must be in the thiol state (Yang et al., 2012). Both terminal oxidation of these cysteines to sulfonates (CySO3–) with reactive oxygen species (ROS) and their complete reduction to thiols (CySH) abrogates the cytokine-stimulating activity.
We now find that only the fully reduced form of HMGB1, where all three cysteines are in the thiol state, can recruit motile cells. Because the disulfide and thiol states of cysteines are mutually exclusive, the cytokine-stimulating and chemotactic activities of HMGB1 should also be mutually exclusive, which is what we show experimentally. HMGB1 terminally oxidized to sulfonates has no activity, either as a chemoattractant or in cytokine stimulation. Replacement of all three cysteines with serines makes HMGB1 nonoxidizable, thus preventing both its cytokine-stimulating activity and its eventual inactivation, but preserving the chemoattractant activity.
Our results indicate that mutually exclusive molecular forms of HMGB1 orchestrate both of the key events in sterile inflammation: leukocyte recruitment and activation of cytokine release.
RESULTS AND DISCUSSION
Recombinant HMGB1 can be reversibly oxidized and reduced
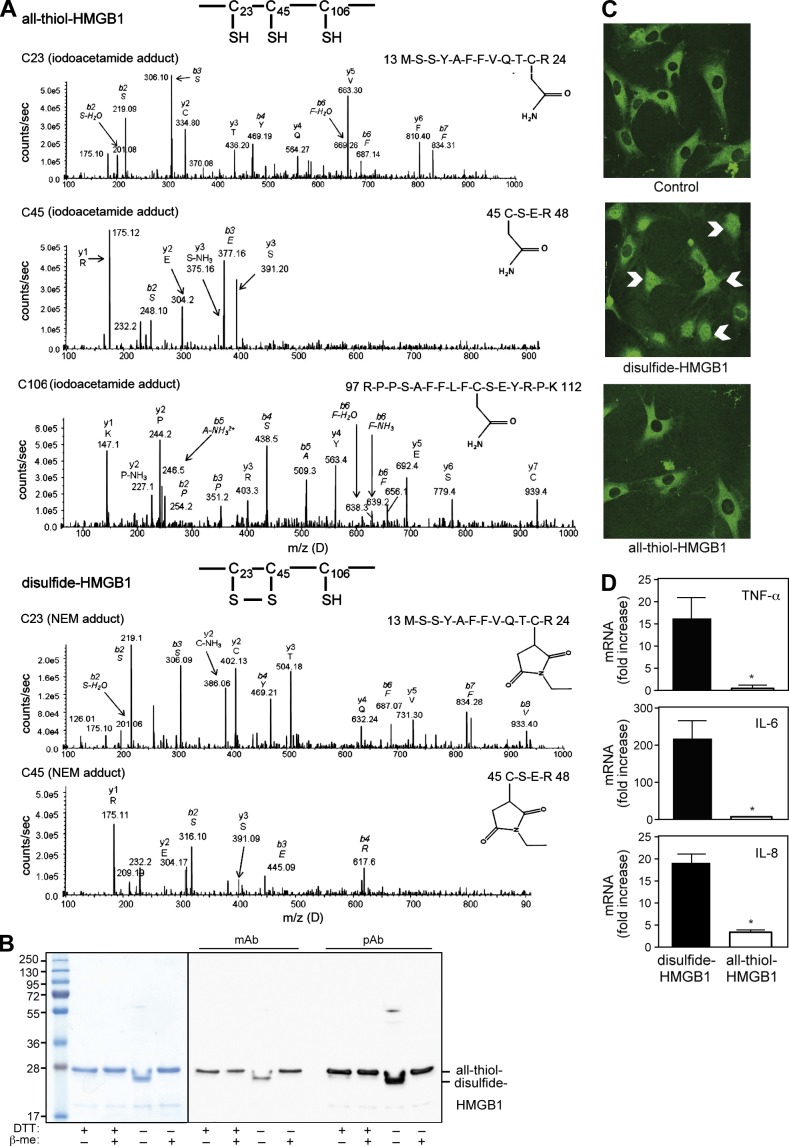
All previous studies by our group relied on bacterially produced full-length HMGB1 protein, which was purified in the presence of dithiothreitol (DTT) added to degassed buffers (Knapp et al., 2004). Because HMGB1 can exist in different redox forms, we compared different batches of HMGB1 purified in the presence or absence of DTT. We specifically excluded any LPS contamination (see Materials and methods). To characterize the redox state of HMGB1, we alkylated reduced cysteines with iodoacetamide, and then reduced disulfide bonds with DTT and reacted them with N-ethylmaleimide (NEM); we then performed tryptic digestion and liquid chromatography tandem mass spectrometric analysis (LC-MS/MS). HMGB1 purified with DTT gave rise to peptides of 1,569.1 (doubly charged ion 784.52+), 622.6 (doubly charged ion 311.32+), and 2,070.0 D (triply charged ion 690.03+). MS/MS revealed an iodoacetamide adduct on all cysteines, which is indicative of a thiol side chain (Fig. 1 A, top); we will henceforth call this form all-thiol-HMGB1. A similar analysis of HMGB1 produced in the absence of DTT revealed NEM adducts on C23 and C45, indicating that they formed a disulfide bond (Fig. 1 A, bottom). C106 was also reduced in this form of HMGB1 (henceforth, disulfide-HMGB1). No SOH, SO2H, or SO3H modification of cysteines was identified within these forms of HMGB1.
Figure 1.
Characterization of HMGB1 redox states. (A) Mass spectrometric characterization of HMGB1 purified with DTT (top) or without DTT (bottom). Characterization of C23: MS-MS trace of the peptide containing amino acids 13–24, with an iodoacetamide adduct indicating reduced C23 (top) or with a NEM adduct after DTT reduction of a disulfide bond (bottom). Characterization of C45: MS-MS trace of the peptide containing amino acids 45–48, with an iodoacetamide adduct indicating reduced C45 (top) or with a NEM adduct after DTT reduction of a disulfide bond (bottom). Characterization of C106: MS-MS trace of the peptide containing amino acids 97–112 with an iodoacetamide adduct indicating reduced C106 (top); the MS-MS trace from disulfide-HMGB1 was identical and is not depicted. (B) Electrophoretic mobility of recombinant HMGB1 prepared in the presence (+DTT) or absence of DTT (−DTT). Samples were heated in the presence (+) or absence (−) of 350 mM β-mercaptoethanol (β-me), loaded onto a 12% SDS-PA gel, and revealed by Coomassie staining (left) or by Western blotting using a mAb or a pAb against HMGB1 (right). (C) Nuclear translocation of p65-GFP (indicated with white arrows) was visualized by confocal microscopy in living mouse embryonic fibroblasts exposed for 30 min to 80 nM disulfide- or all-thiol HMGB1. Data are representative of three independent experiments. (D) Human macrophages were exposed for 4 h to 0.4 µM of disulfide- or all-thiol-HMGB1. The levels of TNF, IL-6, and IL-8 mRNAs were measured by real-time PCR and expressed as fold increase compared with unstimulated macrophages (*, P < 0.05, Student’s t test). Error bars represent standard deviation. Data are representative of three experiments performed with macrophages from unrelated healthy individuals.
The presence of disulfide bonds is often associated with an increased electrophoretic mobility in nonreducing conditions, which is a result of a more compact folding of the polypeptide chain. All-thiol-HMGB1 migrated as a single band with an apparent molecular weight of 28 kD, both in reducing and nonreducing conditions. In contrast, disulfide-HMGB1 migrated in nonreducing conditions as a single band of 26 kD and shifted in reducing conditions to the same 28 kD position as all-thiol-HMGB1 (Fig. 1 B). Monoclonal or polyclonal antibodies against HMGB1 recognized both forms of HMGB1 (Fig. 1 B). Disulfide-HMGB1 was readily shifted to the all-thiol-HMGB1 electrophoretic pattern after 5-min exposure to 5 mM DTT; conversely, we occasionally detected the formation of disulfide-HMGB1 after dilution in air-equilibrated buffers lacking reducing agents. This indicates that all-thiol- and disulfide-HMGB1 forms are readily interconverted in the presence of electron donors (DTT) or acceptors (oxygen).
The cytokine-stimulating and chemoattractant activities of HMGB1 are mutually exclusive
Recently, Yang et al. (2012) showed that disulfide-HMGB1 has cytokine-stimulating activity that is lost after reduction with DTT. We confirmed that disulfide-HMGB1 induces activation of the NF-κB pathway (Fig. 1 C) and cytokine/chemokine expression by macrophages (Fig. 1 D), whereas all-thiol-HMGB1 does not. However, the influence of redox modifications on the chemotactic activity of HMGB1 was not known. We recently showed that HMGB1-induced recruitment of inflammatory cells is dependent on the formation of a HMGB1–CXCL12 heterocomplex that acts exclusively through CXCR4 and not through other HMGB1 receptors (Schiraldi et al., 2012). Using a hybrid ELISA (anti-CXCL12 capture antibody and anti-HMGB1 detection antibody), we found that all-thiol-HMGB1 forms the heterocomplex with CXCL12; no heterocomplex formation could be detected between disulfide-HMGB1 and CXCL12 (Fig. 2 A). All-thiol-HMGB1 synergizes with CXCL12 in inducing human monocyte migration, as expected (Schiraldi et al., 2012), whereas disulfide-HMGB1 does not (Fig. 2 B).
Figure 2.
The cytokine-stimulating and chemoattractant activities of HMGB1 are mutually exclusive. (A) HMGB1/CXCL12 heterocomplex detected by hybrid ELISA. All-thiol- or disulfide-HMGB1 (7.5 ng) were preincubated with the indicated amount CXCL12 at 37°C for 15 min. The formation of the heterocomplex was detected by hybrid ELISA (an anti-CXCL12 capture antibody and an anti-HMGB1 detection antibody). Results are expressed as absorbance at 450 nm (*, P < 0.05, ANOVA). (B) Human monocyte migration in response to increasing concentrations of CXCL12 in the presence or absence of 300 nM all-thiol- or disulfide-HMGB1 (*, P < 0.01 vs. CXCL12 alone; 2-way ANOVA). (C) Migration of mouse 3T3 fibroblasts toward disulfide-HMGB1 or all-thiol-HMGB1, or disufide-HMGB1 exposed to 5 mM of DTT for 30 min (newly all-thiol-HMGB1; *, P < 0.05 vs. disulfide-HMGB1, ANOVA). (D) Migration of 3T3 fibroblasts toward all-thiol-HMGB1 in the presence of increasing concentrations of disulfide-HMGB1, and expression of TNF (as fold increase compared with unstimulated macrophages) in human macrophages stimulated for 4 h with disulfide-HMGB1 in the presence of increasing concentrations of all-thiol-HMGB1. The effects of the competing form of HMGB1 are not statistically significant (ANOVA). (E and F) Migration of mouse 3T3 fibroblasts toward WT all-thiol-HMGB1 previously exposed to increasing concentrations of H2O2 for 1 h (E; *, P < 0.05 vs. all-thiol HMGB1 not treated with H2O2, ANOVA), and toward WT all-thiol-HMGB1 or the E106 mutant purified in the presence of DTT (F; *, P < 0.05 vs. untreated control). (G) Human macrophages were stimulated for 4 h with WT disulfide-HMGB1 or the E106 mutant (0.4 µM) prepared in the absence of DTT. Expression of TNF, MIP-2, and IL-8 was measured by real-time PCR and expressed as fold increase compared with unstimulated macrophages (*, P < 0.05 vs. disulfide-HMGB1; Student’s t test). In all panels, data are representative of at least three independent experiments and bars represent the mean ± SD of triplicate samples (when not visible, they fall within symbols).
Fibroblasts, respond to lower concentrations of HMGB1 compared with leukocytes (Palumbo et al., 2007) and support their own migration by secreting CXCL12 both basally and in response to HMGB1 activation of the receptor for advanced glycation end products (RAGE; Schiraldi et al., 2012). Mouse 3T3 fibroblasts migrated in a dose-dependent manner toward all-thiol-HMGB1, but not toward disulfide-HMGB1 (Fig. 2 C). Notably, the addition of DTT to disulfide-HMGB1 (newly all-thiol HMGB1) restored almost completely the chemotactic activity of the protein.
Collectively, our results indicate that the formation of the C23-C45 disulfide bond inhibits the chemoattractant function of HMGB1 (Fig. S1). Thus, the cytokine-stimulating and chemoattractant activities of HMGB1 are mutually exclusive. In addition, disulfide-HMGB1 and all-thiol-HMGB1 do not compete with each other (Fig. 2 D). Indeed, the chemoattractant and cytokine-stimulating functions of HMGB1 require different receptors: CXCR4 and Toll-like receptor 4, respectively (Yang et al., 2010; Schiraldi et al., 2012). The small conformational rearrangements associated with the formation of a single disulfide bond decreased, but did not abrogate, the binding of HMGB1 to DNA (Park and Lippard, 2011); how they can segregate and restrict so effectively the interactions of the all-thiol- and disulfide-HMGB1 to CXCL12 and Toll-like receptor 4, respectively, still needs to be investigated.
ROS abrogate the proinflammatory activity of HMGB1 by terminally oxidizing its cysteines to sulfonates (Kazama et al., 2008; Yang et al., 2012). We show that HMGB1 exposed to H2O2 has no chemotactic activity (Fig. 2 E). Thus, terminal oxidation makes HMGB1 inactive, both as proinflammatory and chemotactic factor (Fig. S1). To mimic irreversible oxidation to sulfonate, C106 was replaced by a glutamic acid. The electrophoretic mobility of mutant E106 in reducing and nonreducing conditions was identical to that of WT HMGB1 (unpublished data). Whereas WT all-thiol-HMGB1 has optimal chemotactic activity on fibroblasts at 1 nM, the E106 mutant, whether exposed to DTT or not, was ineffective up to 3 nM (Fig. 2 F). It also failed to induce cytokine/chemokine expression by macrophages (Fig. 2 G). Moreover, the E106 mutant cannot compete with WT all-thiol- and disulfide-HMGB1 in migration and inflammation assays, respectively (unpublished data), suggesting that the E106 mutant cannot bind to HMGB1 receptors.
Overall, our results demonstrate that the activities of HMGB1 are redox-dependent. After the original description of HMGB1-induced cytokine release by macrophages (Andersson et al., 2000), several studies could not reproduce these results, leading us to conclude that HMGB1 can only act in synergy with other inflammatory mediators (Bianchi, 2009). Our study differs in that we conclude that inconsistent results were obtained by using inconsistently defined forms of HMGB1. To understand pathogenesis, it will be important to consider which HMGB1 is present in each specific condition and locale in vivo.
HMGB1 inhibitors BoxA and monoclonal antibody DPH1.1 prevent cell migration but not cytokine production
Several inhibitors of HMGB1 have been developed, among which the most commonly used are monoclonal antibodies and BoxA. We investigated whether they block HMGB1’s chemoattractant and/or cytokine-inducing activities. BoxA inhibited in a dose-dependent manner the migration of fibroblasts in response to HMGB1, but failed to prevent HMGB1-induced TNF expression in macrophages (Fig. 3, A and B). We also tested the activity of the monoclonal antibody DPH1.1, directed against an epitope between BoxB and the acidic tail of HMGB1 (Sitia et al., 2011). We prepared F(ab’)2 fragments to avoid the unspecific activation of macrophages: they inhibit HMGB1-induced cell migration but not TNF expression (Fig. 3, C and D). Previous studies have demonstrated that a different anti-HMGB1 monoclonal antibody (2G7) inhibits HMGB1-induced cytokine formation in macrophages (Yang et al., 2010).
Figure 3.
BoxA and monoclonal antibody DPH1.1 prevent HMGB1-induced cell migration but not cytokine expression. (A and C) Migration of mouse 3T3 fibroblasts toward WT all-thiol-HMGB1 in the presence or not of BoxA (A) or F(ab’)2 fragments from the DPH1.1 anti-HMGB1 monoclonal antibody (C). (B and D) Human macrophages were stimulated for 4 h with disulfide-HMGB1 (0.4 µM) in the presence of BoxA (B) or DPH1.1 F(ab’)2 fragments (D). Expression of TNF was measured by real-time PCR and expressed as fold increase compared with unstimulated macrophages. In all panels, data are representative of at least three independent experiments and bars represent the mean ± SD of triplicate samples (*, P < 0.05 vs. control; ANOVA).
Thus, it is possible to neutralize the chemoattractant activity of HMGB1 without interfering with the cytokine-inducing activity. In fact, the known therapeutic effects of BoxA in models of hepatitis (Sitia et al., 2007), peritonitis (Orlova et al., 2007), ischemia/reperfusion of the heart and the brain (Andrassy et al., 2008; Muhammad et al., 2008), and other ailments, may be entirely attributable to the reduced recruitment of inflammatory cells in the injured tissue.
The three HMGB1 cysteine residues are required for the cytokine-stimulating activity but not for the chemoattractant activity
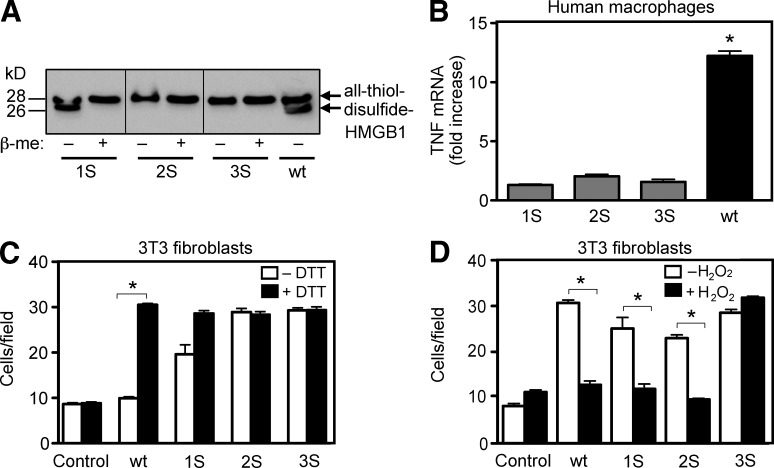
To study the involvement of individual cysteines in the activities of HMGB1, we generated mutants 1S-HMGB1 (where C106 is replaced by a serine), 2S-HMGB1 (where C23 and C45 are replaced), and 3S-HMGB1 (where all three cysteines are replaced). All mutants were purified without DTT, and were reduced where indicated by exposure to 5 mM DTT. Electrophoretic mobility showed the presence of the C23-C45 disulfide bond in 1S-HMGB1, but not in 2S-HMGB1 and 3S-HMGB1 (Fig. 4 A). All mutants failed to induce TNF expression in macrophages, but they all induced fibroblast migration (Fig. 4, B and C). As expected, 2S- and 3S-HMGB1 had the same chemotactic activity whether exposed to DTT or not, as there is no disulfide bond that can be reduced. Thus, all cysteines are required for the cytokine-stimulating activity, but none for the chemoattractant activity of HMGB1.
Figure 4.
HMGB1 cysteines are essential to promote cytokine/chemokine production, but not for chemotaxis. (A) Electrophoretic mobility of recombinant WT HMGB1 and mutants prepared in the absence of DTT, determined as described in the legend of Fig. 1 B. (B) Human macrophages were stimulated for 4 h with mutants or WT HMGB1 prepared in the absence of DTT (0.4 µM). TNF expression was measured by real-time PCR (*, P < 0.05 vs. control, ANOVA). (C and D) 3T3 fibroblast migration toward 1 nM HMGB1 mutants, exposed or not for 1 h to 5 mM DTT (C) or to 100 mM H2O2 (D). Bars represent the mean ± SD of triplicate samples (*, P < 0.05 vs. control, ANOVA).
We next investigated if the chemotactic activity of the mutants was abrogated by ROS. As expected, treatment with H2O2 abrogated the chemotactic activity of 1S- and 2S-HMGB1, but not of 3S-HMGB1, which has no cysteine that can be terminally oxidized (Fig. 4 D).
Cysteine oxidation modulates the different functions of HMGB1 in vivo
Many studies have shown that during inflammation, ROS production supports an extracellular oxidative environment, causing the formation of disulfide bonds between thiols (Carta et al., 2009; Rubartelli and Sitia, 2009). High ROS levels produced during apoptosis cause terminal oxidation of HMGB1, redirecting its ability to activate acquired immune responses toward tolerance (Kazama et al., 2008). A previous study suggested that HMGB1 released by necrotic cells also gets oxidized (Urbonaviciute et al., 2009). Finally, HMGB1 has been shown to be a redox sensor in the context of autophagy (Kang et al., 2011). Thus, we investigated whether the redox state of HMGB1 depends on its source.
HMGB1 is released passively into the extracellular medium during necrosis and is secreted actively by monocytes and macrophages after stimulation with LPS and ATP (Gardella et al., 2002). We characterized by LC-MS/MS the redox state of HMGB1 before and after release by THP-1 cells (Fig. S2). The HMGB1 contained inside the cell (nucleus or cytosol) is completely reduced, also when THP-1 cells are exposed to LPS. Notably, the supernatants from THP-1 cells mechanically necrotized with freeze–thaw cycles or from cells secreting HMGB1 after LPS exposure contained both all-thiol- and disulfide-HMGB1. Thus, we identified monocytic cells as a source of disulfide-HMGB1. Activated monocytes may participate in the inflammatory response by producing the cytokine-stimulating HMGB1.
We next investigated the redox state of HMGB1 in vivo during muscle injury and the subsequent sterile inflammation, using electrophoretic mobility as an assay. Tibialis anterior muscles of mice were damaged or not by cardiotoxin (CTX) injection, which causes muscle cell necrosis (Ownby et al., 1993). Muscles were harvested 2, 6, 24, or 72 h after CTX injection and incubated in PBS with Collagenase D (Fig. 5 A). HMGB1 was barely detectable in the medium bathing healthy muscles, but was abundant in the medium bathing CTX-injured muscles. At early time points (2 h) all-thiol-HMGB1 was detected, but after 6 h disulfide-HMGB1 appeared. It is possible that infiltrating inflammatory cells secrete disulfide-containing HMGB1, as we observed in vitro with monocytic cells (Fig. S2). Indeed, leukocytes are recruited into damaged muscle already by 3 h after CTX injection (Schiraldi et al., 2012). A nonmutually exclusive explanation is that the infiltrating cells, a well-known source of ROS, may induce the oxidation of HMGB1 released by the damaged tissue. Moreover, disulfide-HMGB1 is not present in the healthy muscle as demonstrated by the lysate control. Thus, disulfide-HMGB1 can be considered as a marker of tissue damage.
Figure 5.
Oxidation modulates the activities of HMGB1 in vivo. (A) Electrophoretic mobility of HMGB1 from tibialis anterior muscles harvested at the indicated times after CTX injection. Samples were heated in the presence (+) or absence (–) of 350 mM β-mercaptoethanol (β-me), loaded onto a 12% SDS-PAGE gel and revealed by Western blotting using a polyclonal antibody against HMGB1. Total lysate of tibialis anterior muscle was added as a control (L). (B–D) 3S-HMGB1 induces leukocyte recruitment in vivo. An air pouch was created in mice by the dorsal subcutaneous injection of air. At day 6, the air pouches were injected with 200 µl of PBS containing 10 pmol CXCL12, 300 pmol HMGB1 (WT or 3S), or both. (C) Alternatively, air pouches were injected with 200 µl of PBS containing or not containing 1 or 10 nmol HMGB1 (WT or 3S; C) or with 1 nmol HMGB1 (WT or 3S) in absence or presence of N-acetylcysteine (NAC; 100 nmol/g; D). After 6 h, cells were collected from the air pouches, stained with anti-Ly6C and anti-CD11b antibodies, and analyzed by flow cytometry (WBCs, white blood cells; *, P < 0.05; **, P < 0.01; ***, P < 0.001, ANOVA plus Dunnett’s posttest).
Because 3S-HMGB1 is resistant to oxidation, we hypothesized that its activity in vivo should not be modified by ROS production. We previously showed (Schiraldi et al., 2012) that the HMGB1–CXCL12 heterocomplex induces a massive influx of leukocytes into air pouches created by the injection of air in the dorsal derma of mice; such air pouches provide a cavity into which drugs can be administered and from which recruited cells can be recovered. We injected into air-pouches WT or 3S-HMGB1 (300 pmol) together with CXCL12 (10 pmol). HMGB1 (WT or 3S) or CXCL12 alone failed to induce leukocyte recruitment, but both WT and 3S-HMGB1 in association with CXCL12 induced a massive influx of leukocytes (Fig. 5 B). Notably, the number of recruited leukocytes was increased in response to 3S-HMGB1–CXCL12 compared with WT HMGB1–CXCL12. Because a low amount of CXCL12 is always present in the extracellular fluids, and HMGB1 can induce the secretion of additional CXCL12 (Schiraldi et al., 2012), we also performed the air-pouch experiments using high concentrations of HMGB1 alone. The injection into the air pouch of 1 nmol 3S-HMGB1 induced leukocyte recruitment, but the equivalent amount of all-thiol WT HMGB1 had no effect (Fig. 5 C). However, higher doses of all-thiol WT or 3S-HMGB1 (10 nmol) both recruited leukocytes, with no significant difference. This is compatible with stoichiometric inactivation of HMGB1 by ROS produced in situ. To confirm this hypothesis, we performed the same experiment in the presence or not of N-acetylcysteine (NAC), an antioxidant (Fig. 5 D). We observed that 1 nmol WT protein induced leukocyte recruitment as effectively as 3S-HMGB1 in the presence of NAC, demonstrating that WT HMGB1 gets inactivated in vivo by ROS-induced oxidation.
Collectively, in vitro and in vivo experiments show that 3S-HMGB1 can induce leukocyte recruitment without inducing cytokine/chemokine production and is also resistant to terminal oxidation by ROS. HMGB1 has been shown to promote regeneration in several models of tissue damage, and all such experiments were performed with the reduced form of recombinant WT HMGB1 (Biscetti et al., 2011). However, the redox states of HMGB1 administered in vivo may interconvert among each other. Future investigations of inflammation and tissue regeneration will require the identification of the HMGB1 redox form involved; for example, no studies so far have shown which is the redox state of HMGB1 required for neo-angiogenesis, stem cell recruitment, and cell proliferation. 3S-HMGB1 may help solve these questions, and may hold promise as a therapeutic intervention to favor tissue regeneration.
In conclusion, we show that the DAMP activities of HMGB1—recruitment of leukocytes and their activation to secrete proinflammatory cytokines—depend on different and mutually exclusive redox states of the same polypeptide, which can be modified within the injured tissues after its release. In the model of muscle injury, all-thiol-HMGB1 is prevalent in the extracellular environment immediately after damage, and disulfide-HMGB1 appears a few hours later; we suggest that all-thiol-HMGB1 is released first to recruit leukocytes, which in turn produce disulfide-HMGB1 directly by secretion and/or indirectly by partially oxidizing extracellular HMGB1 with ROS. Finally, sustained ROS production eventually induces the terminal oxidation of HMGB1, which gets inactivated during the resolution of inflammation. Because HMGB1 can be readily modified by electron addition and removal, the limelight shifts onto the cells, the molecules, and the biochemical pathways that control electron flow to HMGB1 both within and outside of the cell.
MATERIALS AND METHODS
Reagents.
Human macrophage colony-stimulating-factor (M-CSF) and granulocyte macrophage colony-stimulating-factor (GM-CSF) were purchased from R&D Systems. DME, X-Vivo medium, PBS, glutamine and penicillin/streptomycin were obtained from Lonza. FBS was obtained from Invitrogen, and hydrogen peroxide (30%) was purchased from BDH Chemicals Ltd. Polyclonal rabbit anti–human HMGB1 antibody was purchased from Abcam, and the monoclonal mouse anti-HMGB1 DPH1.1 antibody was from HMGBiotech Srl. F(ab’)2 fragments were generated from the DPH1.1 antibody using the mouse IgG1 Fab and F(ab′)2 micropreparation kit (Thermo Fischer Scientific) after the manufacturer’s instructions. All other chemicals and materials were purchased from Sigma-Aldrich.
Mice and treatments.
8-wk-old C57BL/6 mice were purchased from The Jackson Laboratory. Sterile injury was induced by injection of 15 µM CTX in the tibialis anterior muscle. After the indicated times, muscles were isolated from mice and incubated for 2 h at 37°C in PBS containing protease inhibitors and Collagenase D (Roche); samples were centrifuged and supernatants were preserved at −80°C before Western blotting. Air pouches were established in 8-wk-old male C57BL/6 mice by dorsal subcutaneous injection of 5 and 3 ml of air at day 0 and day 3, respectively. At day 6, the air pouches were injected with 200 µl of PBS containing 10 pmol CXCL12, 300 pmol HMGB1 (WT or 3S) or CXCL12 10 pmol+HMGB1 300 pmol (WT or 3S), or with 200 µl of PBS containing 0, 1, or 10 nmol HMGB1 (WT or 3S). After 6 h, cells were collected from the air pouches, stained with anti-Ly6C (BD) and anti-CD11b (BioLegend) antibodies and analyzed by flow cytometry.
Animal experimentation was performed in Istituto Scientifico San Raffaele as approved by “Comitato Istituzionale per la Buona Sperimetazione Animale della Fondazione San Raffaele del Monte Tabor” on September 19, 2011.
Cells.
The mouse 3T3 fibroblast cell line and the THP-1 human acute monocytic leukemia cell line were purchased from American Type Culture Collection and cultured in DME supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. PBMCs were isolated from buffy coats of donor blood (Hospital of Magenta, Milan, Italy) by Ficoll gradient centrifugation (Lymphoprep; AXIS-SHIELD). CD14+ monocytes were isolated by positive immunoselection (CD14 Microbeads; Miltenyi Biotec) according to the manufacturer’s instructions, and differentiated into macrophages using X-Vivo medium supplemented with 1% heat inactivated human serum, GM-CSF, and M-CSF.
Production of recombinant WT and mutant HMGB1 proteins.
The WT HMGB1 used in the experiments is the rat protein, with no tags or additional amino acids, expressed in Escherichia coli and purified to homogeneity as previously described (Knapp et al., 2004). The plasmid encoding WT HMGB1 was freshly transformed into protease-deficient E. coli strain BL21 (Novagen) and incubated in 2-YT medium. Protein expression was induced at 23°C by addition of 1 mM IPTG overnight. Harvested cells were resuspended in 50 mM Tris-HCl, pH 8.0, 20 mM EDTA, and 0.5 mM PMSF (containing or not containing 1 mM DTT, as appropriate) and sonicated at 4°C. NaCl was added to a final concentration of 0.5 M, and the cell debris was discarded. Crystalline ammonium sulfate (0.39 g per ml) was added to the supernatant, and the protein precipitate was discarded. The supernatant was purified on a HiLoad 26/10 Phenyl Sepharose High Performance column (GE Healthcare) connected to a FPLC system (Akta Purifier; GE Healthcare). Proteins were eluted at room temperature by a continuous descending gradient of ammonium sulfate (39 to 0%) in 20 mM Hepes pH 7.9, 0.2 mM PMSF, 0.2 mM EDTA pH 8.0 (plus 0.5 mM DTT where appropriate). Fractions containing HMGB1 (identified by Coomassie staining after SDS-PAGE) were pooled, dialyzed overnight against 50 mM Hepes, pH 7.9, 0.2 mM PMSF, 20 mM NaCl (plus 0.5 mM DTT where appropriate), and loaded on a Hi-trap Q column (GE Healthcare). Proteins were eluted with an increasing ionic-strength buffer (from 20 mM to 1 M NaCl) in 50 mM Hepes, pH 7.9, 0.2 mM PMSF, 1 M NaCl (plus 0.5 mM DTT where appropriate). The purity and integrity of purified HMGB1 was verified by Coomassie blue staining after SDS-PAGE. We also tested HMGB1 before and after digestion with DNase I (100 U/ml, Invitrogen), and we observed no difference in activity. The protein was stored at –80°C. Buffers were always degassed before use. E106, 1S-, 2S-, and 3S-HMGB1 mutants were generated using the QuikChange XL Site-Directed Mutagenesis kit according to the manufacturer’s instructions (Stratagene), and were checked by sequencing. Mutant proteins were expressed, purified, and quality-controlled as WT HMGB1.
LPS in HMGB1 preparations.
Contaminating LPS was removed from protein preparations by Triton X-114 extraction (Knapp et al., 2004). We measured LPS in HMGB1 samples from all preparations using the Cambrex Limulus Amoebocyte Assay QCL-1000 (Lonza) before and after terminal digestion with trypsin (therefore excluding any interference of HMGB1 with the assay). LPS content was always below 0.4 ng/mg protein; this amount of LPS is totally ineffective in our assays when administered alone (unpublished data). Experiments were also performed in presence of Polymyxin B (10 µg/ml) to neutralize the biological activity of LPS, even though it was not found in HMGB1 preparations.
Preparation of reduced or terminally oxidized HMGB1.
WT or mutants HMGB1 were exposed to either 100 mM H2O2 or 5 mM DTT for 1 h and dialyzed before the addition to cells or other analyses.
Mass-spectrometric characterization of the cysteine oxidation status of recombinant or circulating HMGB1.
Reduced cysteine residues within HMGB1, either recombinant or obtained from THP-1 cells, were characterized by thiol-specific alkylation with 50 mM iodoacetamide for 30 min. Alkylation with iodoacetamide yields a mass-shift of 57 amu (atomic mass unit). After the incubation, recombinant HMGB1 preparations were precipitated with ice-cold methanol (MeOH). The pellet was recovered after centrifuging at 14,000 g at 4°C for 10 min. The presence of SOH residues in cysteines was investigated with a 20-min incubation with 0.1 mM dimedone (5,5-dimethyl-1,3-cyclohexanedione). SO2H and SO3H oxidative modifications were sought by looking for mass increases of 32 and 48 amu, respectively, on each particular cysteine residue. After the first alkylation step, remaining cysteine residues engaged in a disulfide bond were reduced with 1 mM DTT at 4°C for 15 min. Newly reduced cysteines were then alkylated with 50 mM N-ethylmalemide (NEM) for 5 min on ice, which yields a mass shift of 125 amu. HMGB1 was isolated by immunoprecipitation from THP-1 samples, as previously described (Antoine et al., 2009). Proteins were then separated by nonreducing SDS-PAGE, and protein bands corresponding to the molecular weight of HMGB1 were excised and subjected to tryptic digestion. The resulting peptides were characterized by liquid chromatography and tandem mass spectrometry (LC-MS/MS) as described previously (Antoine et al., 2009). Individual peptide fragmentation to produce b and y ions was used to determine the amino acid sequence and confirm the presence of specific modifications.
Western blotting.
Total protein content in muscle samples from control or CTX-treated mice was determined using the Protein Assay Dye Reagent (Bio-Rad Laboratories). Equivalent amounts of proteins were loaded on gels. For Western blotting, protein samples were separated on 12% SDS-PA gels and transferred on nitrocellulose membranes, which were blocked with 5% skim milk in Tris-buffered saline, pH 7.0, containing 0.1% Tween 20 (TBS-T). Blocked membranes were probed with rabbit anti-HMGB1 (1:1,000) in TBS-T plus 5% milk overnight at 4°C, washed several times with TBS-T, and incubated for 1 h with anti–rabbit peroxidase-conjugated antibody (1:10,000). Western blots were visualized using an enhanced chemiluminescence kit according to the manufacturer’s instructions (GE Healthcare).
Living cell microscopy.
Immortalized fibroblasts were derived from knock-in mice where the endogenous p65 gene was replaced by GFP-p65 (De Lorenzi et al., 2009). Cells were cultured on special chambered glass slides for confocal microscopy (Lab-Tek) in DME supplemented with 10% FCS. Cells were serum-starved for 16 h, and then stimulated with 80 nM HMGB1 produced with or without DTT, as indicated. Subsequently, living cells were imaged using a TCS SP5 AOBS confocal microscope (Leica) equipped with a ArgonPlus Ar-ion laser (220 mW nominal power; power measured at the objective: 11.4 mW for the 488 nm), a HCX PL Apo CS 63× oil immersion objective/1.4 NA, and a humidified thermostatic chamber (37°C and 5% CO2) as previously described (Sung et al., 2009); time-lapse images were taken and 30-min time-point images are shown.
Chemotaxis assays.
For fibroblast chemotaxis, modified Boyden chambers were used with filters (8 µm pores; Neuro Probe) coated with 50 µg/ml fibronectin (Roche). 50,000 cells in 200 µl were added to the upper chamber. HMGB1 or mutants were added to the lower chamber, and then cells were left to migrate for 3 h at 37°C. Nonmigrating cells were removed with a cotton swab, and migrated cells were fixed with ethanol and stained with Giemsa. All assays were done in triplicate, and migrated cells were counted in 10 random fields/filter.
For monocytes, chemotaxis was assayed in 48-well Boyden microchambers (Neuro Probe), as previously described (Uguccioni et al., 1995). In brief, freshly isolated monocytes (5 × 104) were diluted in RPMI 1640 supplemented with 20 mM Hepes, pH 7.4, 1% pasteurized plasma protein solution (5% PPL SRK), and chemoattractants were diluted in the same buffer supplemented with Polymyxin (10 µg/ml). Cells were then added to the upper wells. After 90 min of incubation, the membrane was removed, washed on the upper side with PBS, fixed, and stained. All assays were done in triplicate, and for each well the migrated cells were counted at 1,000-fold magnification in randomly selected fields.
Hybrid ELISA.
The heterocomplex between CXCL12 and HMGB1 (all-thiol or disulfide-HMGB1) was detected by a hybrid ELISA. The plates (Costar 3596) were coated overnight with an anti-CXCL12 capture antibody (human CXCL12/SDF-1 DuoSet ELISA; R&D Systems), and then blocked with diluent solution (1% BSA in PBS) for 3 h. CXCL12 and HMGB1 (2:1 stoichiometric ratio) were preincubated at 37°C for 15 min in agitation, to promote the formation of the heterocomplex. The samples were distributed in the wells in diluent solution and incubated for 2 h. After 2 h of incubation with an Enzyme-conjugated anti-HMGB1 antibody (HMGB1 ELISA; IBL International), the reaction was visualized by the addition of the substrate solution and stopped with stop solution (HMGB1 ELISA; IBL International). Optical density was measured at 450 nm. The procedure was performed at room temperature, and 4 washes were repeated before each step of the procedure using 0.1% Tween 20 in PBS.
Quantitative real-time polymerase chain reaction.
Total RNAs were isolated using the Illustra RNAspin Mini kit (GE Healthcare), and complementary DNA (cDNA) was obtained by retro-transcription with Oligo(dT) primers (Invitrogen) and SuperScript II Reverse transcription (Invitrogen) following the manufacturers’ instructions. Quantitative real-time PCR was then performed in duplicates using LightCycler480 (Roche), SYBR Green I master mix, and the following primers: β-actin, 5′-TGACGGGGTCACCCACACTGTGCCC-3′ and 5′-CTAGAAGCATTGCGGTGGACGATGG-3′; TNF, 5′-AGCCCATGTTGTAGCAAACC-3′ and 5′-AGGACCTGGGAGTAGATGAGG-3′; IL-6, 5′-TACCCCCAGGAGAAGATTCC-3′ and 5′-TTTTCACCAGGCAAGTCTCC-3′; IL-8, 5′-TGCCAAGGAGTGCTAAAG-3′ and 5′-CTCCACAACCCTCTGCAC-3′; MIP-2, 5′-TGCCAGTGCTTGCAGAC-3′ and 5′-TCTTAACCATGGGCGATGC-3′. The ΔCt method was used for quantification, and the β-actin gene was used for normalization.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism software. Differences between treatment groups were determined by Student’s t or Mann-Whitney U tests; P values <0.05 were considered statistically significant. For experiments with multiple doses and air pouch experiments, ANOVA was performed, followed by Dunnett’s posttest.
Online supplemental material.
Fig. S1 is a summary of HMGB1 redox status versus activity. Fig. S2 shows the redox status of intracellular and extracellular HMGB1 in THP-1 cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120189/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Giovanna Musco for discussions on the structure and redox state of HMGB1.
E. Venereau was supported by a fellowship from Fondazione Italiana Ricerca sul Cancro. This work was supported by grants from the Associazione Italiana Ricerca sul Cancro (to M.E. Bianchi) and EndoStem (to HMGBiotech), the European Union FP7 (ADITEC 280873 and TIMER 281608 to M. Uguccioni), the San Salvatore Foundation, and the Institute for Arthritis Research.
The authors have no conflicting financial interests. However, M.E. Bianchi is founder and part owner of HMGBiotech, a company that provides goods and services related to HMGB proteins, and M. Casalgrandi, A. Preti, and F. De Marchis were partially supported by HMGBiotech.
Author contributions: E. Venereau designed and performed most experiments; M. Casalgrandi and A. Preti produced all proteins. M. Schiraldi performed the air pouch experiments. A. Cattaneo and D.J. Antoine performed mass spectrometry, independently and in different locations. L. Raeli performed the hybrid ELISA experiments. F. De Marchis, J. Liu, S. Samadi Shams, L. Varani, and A. Antonelli participated in some experiments. U. Andersson, D.J. Antoine, H. Yang, K.J. Tracey, M. Uguccioni, and A. Bachi discussed results and participated in the writing of the manuscript. E. Venereau and M.E. Bianchi wrote most of the manuscript, and M.E. Bianchi was responsible for general organization.
Footnotes
Abbreviations used:
- CTX
- cardiotoxin
- DAMP
- damage-associated molecular pattern
- DTT
- dithiothreitol
- HMGB1
- high-mobility group box 1
- LC-MS/MS
- liquid chromatography tandem mass spectrometric analysis
- NEM
- N-ethylmaleimide
- RAGE
- receptor for advanced glycation end products
- ROS
- reactive oxygen species
- TLR
- Toll-like receptor
- TNF
- tumor necrosis factor
References
- Andersson U., Tracey K.J. 2011. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu. Rev. Immunol. 29:139–162 10.1146/annurev-immunol-030409-101323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson U., Wang H., Palmblad K., Aveberger A.C., Bloom O., Erlandsson-Harris H., Janson A., Kokkola R., Zhang M., Yang H., Tracey K.J. 2000. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J. Exp. Med. 192:565–570 10.1084/jem.192.4.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrassy M., Volz H.C., Igwe J.C., Funke B., Eichberger S.N., Kaya Z., Buss S., Autschbach F., Pleger S.T., Lukic I.K., et al. 2008. High-mobility group box-1 in ischemia-reperfusion injury of the heart. Circulation. 117:3216–3226 10.1161/CIRCULATIONAHA.108.769331 [DOI] [PubMed] [Google Scholar]
- Antoine D.J., Williams D.P., Kipar A., Jenkins R.E., Regan S.L., Sathish J.G., Kitteringham N.R., Park B.K. 2009. High-mobility group box-1 protein and keratin-18, circulating serum proteins informative of acetaminophen-induced necrosis and apoptosis in vivo. Toxicol. Sci. 112:521–531 10.1093/toxsci/kfp235 [DOI] [PubMed] [Google Scholar]
- Bianchi M.E. 2007. DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukoc. Biol. 81:1–5 10.1189/jlb.0306164 [DOI] [PubMed] [Google Scholar]
- Bianchi M.E. 2009. HMGB1 loves company. J. Leukoc. Biol. 86:573–576 10.1189/jlb.1008585 [DOI] [PubMed] [Google Scholar]
- Biscetti F., Ghirlanda G., Flex A. 2011. Therapeutic potential of high mobility group box-1 in ischemic injury and tissue regeneration. Curr. Vasc. Pharmacol. 9:677–681 10.2174/157016111797484125 [DOI] [PubMed] [Google Scholar]
- Carta S., Castellani P., Delfino L., Tassi S., Venè R., Rubartelli A. 2009. DAMPs and inflammatory processes: the role of redox in the different outcomes. J. Leukoc. Biol. 86:549–555 10.1189/jlb.1008598 [DOI] [PubMed] [Google Scholar]
- De Lorenzi R., Gareus R., Fengler S., Pasparakis M. 2009. GFP-p65 knock-in mice as a tool to study NF-kappaB dynamics in vivo. Genesis. 47:323–329 10.1002/dvg.20468 [DOI] [PubMed] [Google Scholar]
- Gardella S., Andrei C., Ferrera D., Lotti L.V., Torrisi M.R., Bianchi M.E., Rubartelli A. 2002. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 3:995–1001 10.1093/embo-reports/kvf198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jube S., Rivera Z.S., Bianchi M.E., Powers A., Wang E., Pagano I., Pass H.I., Gaudino G., Carbone M., Yang H. 2012. Cancer Cell Secretion of the DAMP protein HMGB1 supports progression in malignant mesothelioma. Cancer Res. 72:3290–3301 10.1158/0008-5472.CAN-11-3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R., Livesey K.M., Zeh H.J., III, Lotze M.T., Tang D. 2011. HMGB1 as an autophagy sensor in oxidative stress. Autophagy. 7:904–906 10.4161/auto.7.8.15704 [DOI] [PubMed] [Google Scholar]
- Kazama H., Ricci J.E., Herndon J.M., Hoppe G., Green D.R., Ferguson T.A. 2008. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 29:21–32 10.1016/j.immuni.2008.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S., Müller S., Digilio G., Bonaldi T., Bianchi M.E., Musco G. 2004. The long acidic tail of high mobility group box 1 (HMGB1) protein forms an extended and flexible structure that interacts with specific residues within and between the HMG boxes. Biochemistry. 43:11992–11997 10.1021/bi049364k [DOI] [PubMed] [Google Scholar]
- Muhammad S., Barakat W., Stoyanov S., Murikinati S., Yang H., Tracey K.J., Bendszus M., Rossetti G., Nawroth P.P., Bierhaus A., Schwaninger M. 2008. The HMGB1 receptor RAGE mediates ischemic brain damage. J. Neurosci. 28:12023–12031 10.1523/JNEUROSCI.2435-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova V.V., Choi E.Y., Xie C., Chavakis E., Bierhaus A., Ihanus E., Ballantyne C.M., Gahmberg C.G., Bianchi M.E., Nawroth P.P., Chavakis T. 2007. A novel pathway of HMGB1-mediated inflammatory cell recruitment that requires Mac-1-integrin. EMBO J. 26:1129–1139 10.1038/sj.emboj.7601552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ownby C.L., Fletcher J.E., Colberg T.R. 1993. Cardiotoxin 1 from cobra (Naja naja atra) venom causes necrosis of skeletal muscle in vivo. Toxicon. 31:697–709 10.1016/0041-0101(93)90376-T [DOI] [PubMed] [Google Scholar]
- Palumbo R., Galvez B.G., Pusterla T., De Marchis F., Cossu G., Marcu K.B., Bianchi M.E. 2007. Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-κB activation. J. Cell Biol. 179:33–40 10.1083/jcb.200704015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lippard S.J. 2011. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry. 50:2567–2574 10.1021/bi2000214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubartelli A., Sitia R. 2009. Stress as an intercellular signal: the emergence of stress-associated molecular patterns (SAMP). Antioxid. Redox Signal. 11:2621–2629 10.1089/ars.2009.2377 [DOI] [PubMed] [Google Scholar]
- Schiraldi M., Raucci A., Muñoz L.M., Livoti E., Celona B., Venereau E., Apuzzo T., De Marchis F., Pedotti M., Bachi A., et al. 2012. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J. Exp. Med. 209:551–563 10.1084/jem.20111739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia G., Iannacone M., Müller S., Bianchi M.E., Guidotti L.G. 2007. Treatment with HMGB1 inhibitors diminishes CTL-induced liver disease in HBV transgenic mice. J. Leukoc. Biol. 81:100–107 10.1189/jlb.0306173 [DOI] [PubMed] [Google Scholar]
- Sitia G., Iannacone M., Aiolfi R., Isogawa M., van Rooijen N., Scozzesi C., Bianchi M.E., von Andrian U.H., Chisari F.V., Guidotti L.G. 2011. Kupffer cells hasten resolution of liver immunopathology in mouse models of viral hepatitis. PLoS Pathog. 7:e1002061 10.1371/journal.ppat.1002061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M.H., Salvatore L., De Lorenzi R., Indrawan A., Pasparakis M., Hager G.L., Bianchi M.E., Agresti A. 2009. Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PLoS ONE. 4:e7163 10.1371/journal.pone.0007163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uguccioni M., D’Apuzzo M., Loetscher M., Dewald B., Baggiolini M. 1995. Actions of the chemotactic cytokines MCP-1, MCP-2, MCP-3, RANTES, MIP-1 alpha and MIP-1 beta on human monocytes. Eur. J. Immunol. 25:64–68 10.1002/eji.1830250113 [DOI] [PubMed] [Google Scholar]
- Urbonaviciute V., Meister S., Fürnrohr B.G., Frey B., Gückel E., Schett G., Herrmann M., Voll R.E. 2009. Oxidation of the alarmin high-mobility group box 1 protein (HMGB1) during apoptosis. Autoimmunity. 42:305–307 10.1080/08916930902831803 [DOI] [PubMed] [Google Scholar]
- Yang H., Hreggvidsdottir H.S., Palmblad K., Wang H., Ochani M., Li J., Lu B., Chavan S., Rosas-Ballina M., Al-Abed Y., et al. 2010. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc. Natl. Acad. Sci. USA. 107:11942–11947 10.1073/pnas.1003893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lundbäck P., Ottosson L., Erlandsson-Harris H., Venereau E., Bianchi M.E., Al-Abed Y., Andersson U., Tracey K.J., Antoine D.J. 2012. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol. Med. 18:250–259 10.2119/molmed.2011.00389 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.