Two unrelated children with HSE carry distinct heterozygous mutations in the gene encoding TANK-binding kinase 1.
Abstract
Childhood herpes simplex virus-1 (HSV-1) encephalitis (HSE) may result from single-gene inborn errors of TLR3 immunity. TLR3-dependent induction of IFN-α/β or IFN-λ is crucial for protective immunity against primary HSV-1 infection in the central nervous system (CNS). We describe here two unrelated children with HSE carrying different heterozygous mutations (D50A and G159A) in TBK1, the gene encoding TANK-binding kinase 1, a kinase at the crossroads of multiple IFN-inducing signaling pathways. Both mutant TBK1 alleles are loss-of-function but through different mechanisms: protein instability (D50A) or a loss of kinase activity (G159A). Both are also associated with an autosomal-dominant (AD) trait but by different mechanisms: haplotype insufficiency (D50A) or negative dominance (G159A). A defect in polyinosinic-polycytidylic acid–induced TLR3 responses can be detected in fibroblasts heterozygous for G159A but not for D50A TBK1. Nevertheless, viral replication and cell death rates caused by two TLR3-dependent viruses (HSV-1 and vesicular stomatitis virus) were high in fibroblasts from both patients, and particularly so in G159A TBK1 fibroblasts. These phenotypes were rescued equally well by IFN-α2b. Moreover, the IFN responses to the TLR3-independent agonists and viruses tested were maintained in both patients’ peripheral blood mononuclear cells and fibroblasts. The narrow, partial cellular phenotype thus accounts for the clinical phenotype of these patients being limited to HSE. These data identify AD partial TBK1 deficiency as a new genetic etiology of childhood HSE, indicating that TBK1 is essential for the TLR3- and IFN-dependent control of HSV-1 in the CNS.
HSV-1 encephalitis (HSE) remained a lethal disease for four decades after the establishment of its viral etiology in 1941 (Whitley, 2006). The advent of the antiviral drug acyclovir in 1981 greatly decreased mortality rates in children with HSE, but severe neurological sequelae are still commonly observed in surviving patients (Whitley et al., 1986). Childhood HSE occurs during primary infection with HSV-1, a double-stranded DNA (dsDNA) virus of the Herpesviridae family. HSV-1 follows a neurotropic infection route, as the virus reaches the central nervous system (CNS) from the nasal or oral epithelium via the cranial nerves, by retrograde axonal transport in the olfactory or trigeminal nerve (De Tiège et al., 2008; Abel et al., 2010). HSE is restricted to the CNS, with no detectable viremia or lesions of other tissues. About 85% of young adults worldwide are seropositive for HSV-1, and the vast majority are asymptomatic or present only benign herpes labialis (Stanberry et al., 1997). The incidence of HSE is ∼1–2 cases per 500,000 individuals per year (Whitley and Kimberlin, 2005). The occurrence of HSE, which is not epidemic, has never been shown to be associated with any particular HSV-1 isolate. HSE is nonetheless the most common sporadic viral encephalitis in the Western world. Childhood HSE thus remained a rare, unexplained, and devastating complication of primary infection with HSV-1, until our discoveries of single-gene inborn errors of TLR3 immunity in some children with HSE (Casrouge et al., 2006; Zhang et al., 2007, 2008; Pérez de Diego et al., 2010; Guo et al., 2011; Sancho-Shimizu et al., 2011b).
We have reported children with isolated HSE caused by autosomal-recessive (AR) UNC-93B deficiency (Casrouge et al., 2006), autosomal-dominant (AD) TLR3 deficiency (Zhang et al., 2007), AR TLR3 deficiency (Guo et al., 2011), AD TRAF3 deficiency (Pérez de Diego et al., 2010), AD TRIF deficiency, and AR TRIF deficiency (Sancho-Shimizu et al., 2011a,b). In addition, one patient with AR STAT-1 deficiency and impaired cellular responses to IFNs (Dupuis et al., 2003) and another patient with X-linked recessive NEMO deficiency and impaired TLR3-dependent production of IFNs (Audry et al., 2011) died of HSE, albeit after many other infectious illnesses. Conversely, patients with MyD88 and IRAK4 deficiencies but with an intact TLR3 pathway are not prone to HSE (Picard et al., 2003, 2010; Yang et al., 2005; Ku et al., 2007; von Bernuth et al., 2008). Collectively, these genetic data indicate that the TLR3-dependent induction of IFN-α/β and/or IFN-λ is essential for protective immunity to HSV-1 primary infection of the CNS during childhood. In dermal fibroblasts, genetic defects of the TLR3 pathway impair the production of IFN-β and IFN-λ after stimulation with the nonspecific, synthetic, TLR3 agonist polyinosinic-polycytidylic acid (poly(I:C)). These fibroblasts are also highly susceptible to infection with HSV-1 or vesicular stomatitis virus (VSV). This fibroblast phenotype has recently been shown to recapitulate the impairment of TLR3-dependent HSV-1 control in UNC-93B–deficient cells of the CNS (unpublished data). As only a fraction of children with HSE bear known defects in TLR3, UNC93B1, TRIF, or TRAF3, we searched for other genetic defects by screening HSE patients for mutations in other genes involved in the TLR3–IFN pathway.
RESULTS
Heterozygous TBK1 mutations in two patients with HSE
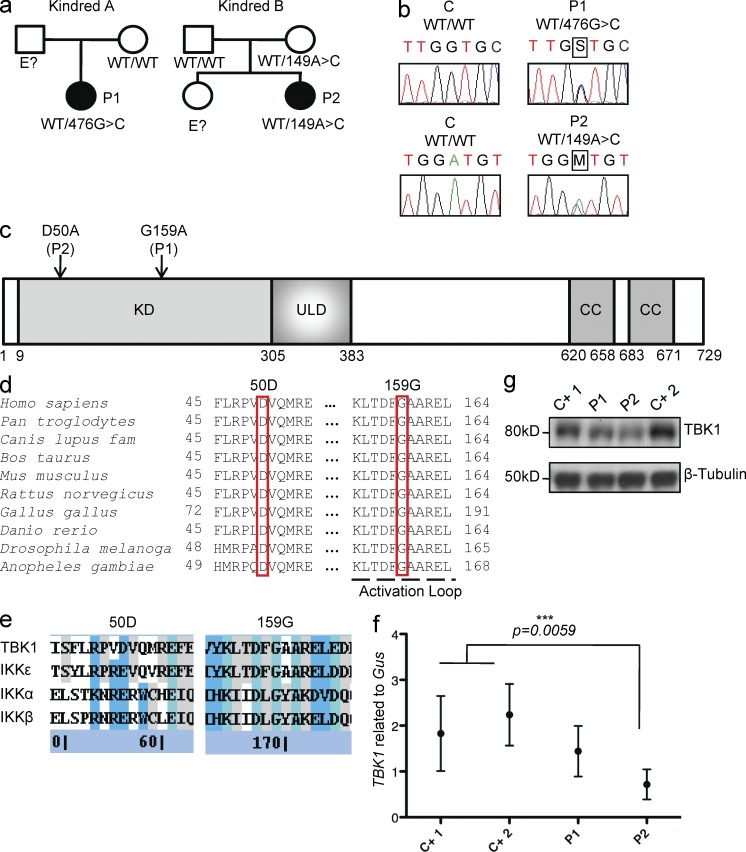
We investigated two unrelated patients with HSE from Poland (P1) and from France (P2; Fig. 1 a). These patients suffered from HSE at the ages of 7 yr (P1) and 11 mo (P2). They are now 17 and 26 yr old, respectively, and have suffered no other unusual infectious disease, viral in particular, in the absence of prophylaxis. We sequenced candidate TLR3 pathway genes from genomic DNA extracted from granulocytes and from simian virus 40 (SV40)–transformed fibroblasts (SV40-fibroblasts) from the two patients and cDNAs synthesized from messenger RNAs (mRNAs) extracted from SV40-fibroblasts. We found two different mutations in the coding region of the gene encoding TANK-binding kinase 1 (TBK1, also known as T2K/NAK [Tojima et al., 2000; Pomerantz and Baltimore, 1999; Bonnard et al., 2000]). P1 carries a heterozygous mutation in the fifth exon of TBK1, at position 476 of the cDNA: c.476G>G/C (Fig. 1 b). Her father was not tested and her mother does not carry this variant (Fig. 1 a). The mutation results in the nonconservative replacement of a glycine residue with an alanine residue at position 159 (G159A) in the immediate vicinity of the predicted activation loop of the kinase (Fig. 1 c; Xu et al., 2011). P2 carries a heterozygous mutation in the third exon of TBK1, c.149A>A/C (Fig. 1 b). Her mother is healthy and carries the same heterozygous mutation, whereas her father is WT for TBK1 (Fig. 1 a). This substitution results in a nonconservative change at position 50 in the kinase domain, replacing an aspartic acid residue with an alanine residue (D50A; Fig. 1 c). No mutations were found in the coding exons of TLR3, UNC93B1, TRAF3, TRIF (encoding Toll/IL-1 receptor domain–containing adaptor protein inducing IFN-β, TRIF) and TANK (encoding TRAF-interacting protein I, TANK; see Whole-exome sequencing in P1 and P2: TLR3–IFN pathway genes sequenced and found to be WT in the Materials and methods). No mutations were found in other genes known to be involved in the TLR3–IFN pathway, as detected by whole-exome sequencing (see Whole-exome sequencing in P1 and P2: TLR3–IFN pathway genes sequenced and found to be WT). Neither of the two TBK1 variants was found in single nucleotide polymorphism (SNP) databases (NCBI dbSNP 135 and UCSC) or on sequencing of a panel of 1,050 control human DNAs provided by the CEPH-HGD, ruling out the possibility of irrelevant polymorphisms. No other mutations were found in the remaining exons or flanking intronic regions of TBK1. Moreover, the D50 and G159 residues are strictly conserved across species (Fig. 1 d). Both substitutions are also predicted to be “probably damaging” by Polyphen-2 version 2.2.2 (Adzhubei et al., 2010) and “damaging” by SIFT (Kumar et al., 2009). Finally, the other human IKK-related kinases, IKK-ε (64% similar to human TBK1), IKK-α (46% similar to TBK1), and IKK-β (44% similar to TBK1), have a glutamic acid residue, which is similar to aspartic acid, at position 50 and a glycine residue at position 159 (Fig. 1, d and e). Collectively, these genetic data suggest that these two patients are heterozygous for rare, HSE-causing TBK1 missense alleles.
Figure 1.
Heterozygous TBK1 mutations in two children with HSE. (a) Family pedigrees and segregation. (b) Heterozygous TBK1 mutations 476G>C in P1 and 149A>C in P2. The PCR products sequenced were amplified from genomic DNA from the granulocytes of a control (C) and both patients. (c) Schematic diagram of the protein structure of TBK1, featuring its kinase domain (KD), ubiquitin-like domain (ULD), and coiled-coil (CC) regions. Both heterozygous substitutions, 159G>G/A (P1) and 50D>D/A (P2), affect the kinase domain of TBK1 (amino acids 9–305). (d) Multiple alignments of relevant amino acid sequences of the kinase domain of human TBK1 with its homologues from nine other species, with the residues mutated in P1 (G159) and P2 (D50) highlighted. (e) Multiple alignments of relevant amino acid sequences of the kinase domain of human TBK1 with the other IKK and IKK-related kinases, IKK-α (46% similar to TBK1), IKK-β (44% similar), and IKK-ε (64% similar). The residues mutated in P1 and P2 are conserved (G159) or similar (D50) across IKK or IKK-related kinases. (Blue signifies sequence similarity, teal signifies sequence identity, and gray signifies partial identity/similarity.) (f) TBK1 expression, as assessed by RT-qPCR on mRNA from the SV40-fibroblasts of patients (P1 and P2) and control lines (C+1 and C+2). Values represent mean values ± SD calculated from three independent experiments. (g) TBK1 levels, as assessed by Western blotting, in SV40-fibroblasts from patients (P1 and P2) and two control lines (C+ 1 and C+ 2). This Western blot result is representative of three experiments.
Differences in the expression of the mutant TBK1 alleles
We first assessed TBK1 mRNA levels by quantitative RT-PCR (RT-qPCR) in SV40-fibroblasts from the patients and in control cells (Fig. 1 f). TBK1 mRNA levels were similar in control cells and in fibroblasts from P1, whereas they were lower in fibroblasts from P2 (Fig. 1 f). Consistent with these findings, TBK1 protein levels were normal (similar to the control) in fibroblasts from P1, but low in fibroblasts from P2, as shown by Western blotting (Fig. 1 g). The heterozygous G159A mutation therefore seems to have no effect on TBK1 expression in the patients’ fibroblasts, whereas the D50A mutation seems to affect the amounts of both mRNA and protein for TBK1. As the level of expression of mutant alleles may be confounded by the expression of the WT allele in the patients’ heterozygous cells, we then compared the expression of the individual TBK1 alleles in HEK293T cells or TBK1 knockout (TBK1−/−) mouse embryonic fibroblasts (MEFs), transfected with either a mutant allele or the WT human TBK1 allele. Upon transient transfection with the G159A mutant allele, HEK293T cells and TBK1−/− MEFs produced G159A TBK1 in amounts similar to those obtained for the WT TBK1. In contrast, transfection with the D50A mutant allele resulted in the production of much smaller amounts of protein (Fig. 2, a and c). The D50A mutation in TBK1 may thus destabilize the protein, resulting in its premature degradation and the presence of smaller total amounts of TBK1 protein in the patient’s fibroblasts. These data strongly suggest that the TBK1 G159A allele is expressed normally, whereas the D50A allele is poorly expressed.
Figure 2.
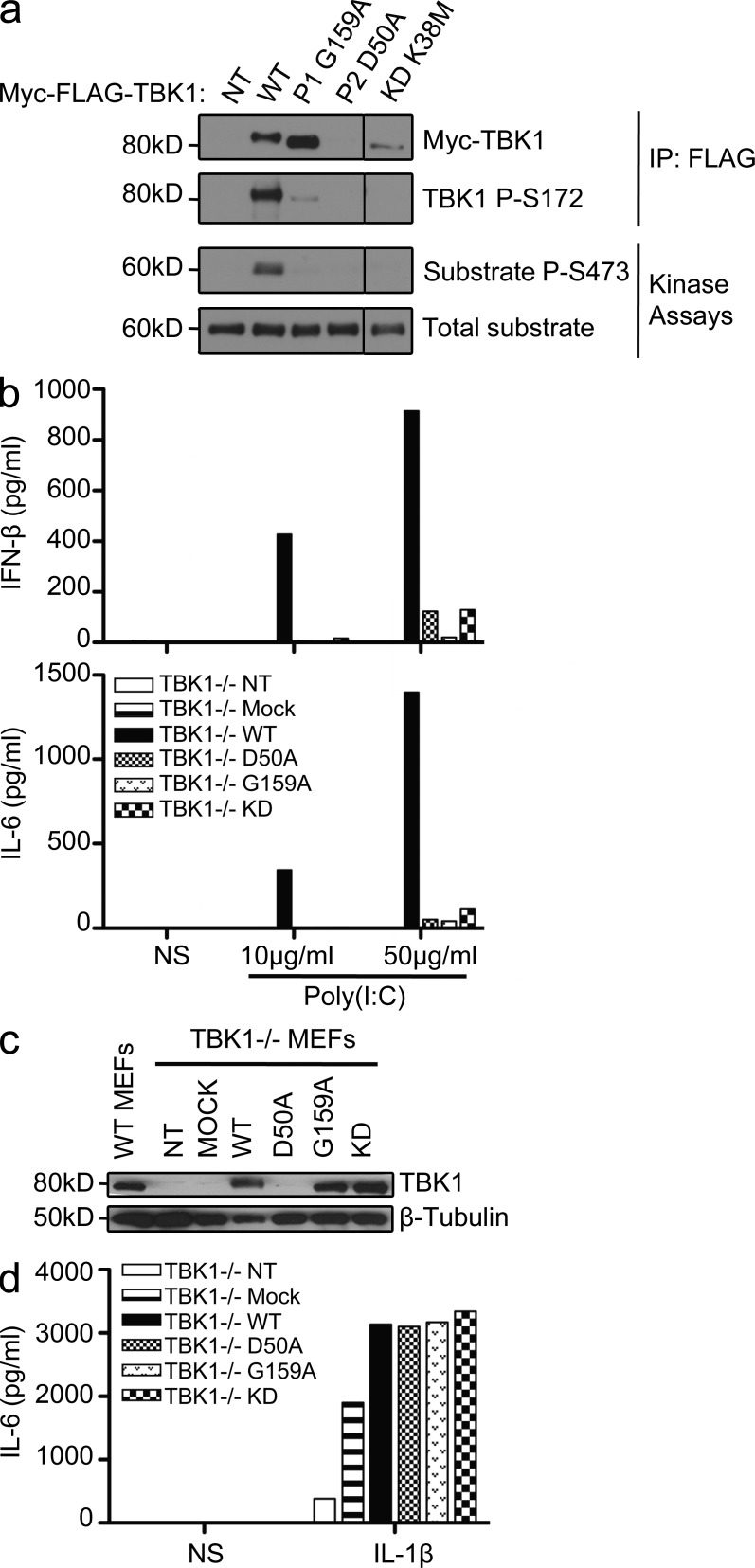
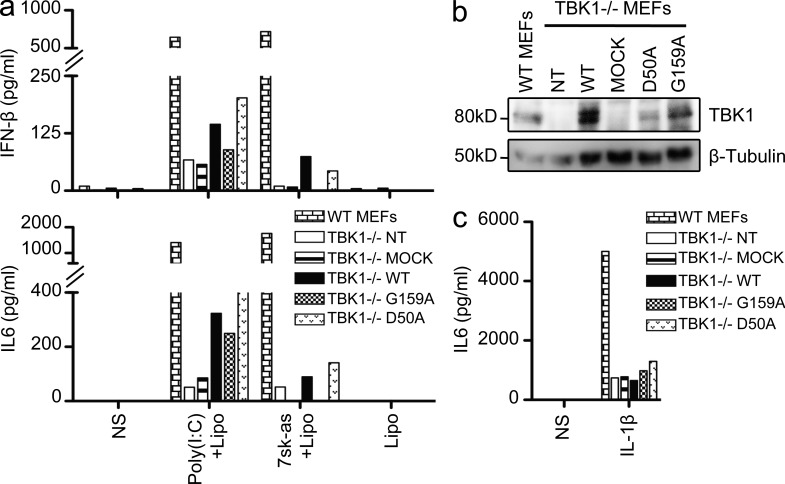
Both mutant TBK1 alleles are loss-of-function but through different mechanisms. (a) In vitro kinase assays: substrate (Akt) phosphorylation by both mutant kinases, G159A and D50A, compared with the phosphorylation levels for WT TBK1 and the kinase-dead (KD) K38M TBK1. A single experiment representative of three independent experiments performed is shown. HEK293T cells were transfected with WT and mutant constructs or left untransfected (NT). Black lines indicate that intervening lanes have been spliced out. IP, immunoprecipitation. (b) TBK1−/− MEFs were either left untransfected (NT) or were transfected with a mock vector, WT TBK1, or mutant constructs: D50A (mutation present in P2), G159A (the mutation in P1), and S172A (kinase-dead, KD). After 24 h, the cells were stimulated with 10 or 50 µg/ml poly(I:C). ELISA was performed to assess IFN-β and IL-6 production 24 h after stimulation. The data shown are representative of three independent experiments. (c) TBK1 expression from the transfected constructs was assessed by Western blotting with antibodies against TBK1, with β-tubulin used as a loading control. (d) TBK1−/− MEFs were either not transfected (NT) or transfected with a mock vector or a vector encoding WT TBK1 or the mutants, G159A, D50A, or S172A. 24 h later, cells were stimulated with 10 ng/ml IL-1β. IL-6 production was measured by ELISA. The data shown are representative of three independent experiments. NS, nonstimulated.
Both TBK1 mutant alleles are kinase-dead and loss-of-function
TBK1 is a Ser/Thr kinase of the IKK-related family, which, upon TLR3 activation, is recruited to the TRIF–TRAF3 complex (Sato et al., 2003; Oganesyan et al., 2006). After their activation by phosphorylation, TBK1 and IKK-ε (also known as IKKi [Shimada et al., 1999; Peters et al., 2000]) normally phosphorylate their target transcription factor, IRF3 (Fitzgerald et al., 2003; Sharma et al., 2003). In vitro kinase assays showed that the mutant TBK1 protein carrying the G159A substitution had no enzyme activity (Fig. 2 a). The levels of in vitro phosphorylation of the recombinant protein Akt, used as a substrate (Ou et al., 2011), were much lower with the G159A TBK1 protein than with WT TBK1 (Fig. 2 a). In addition, phosphorylation of the S172 residue of TBK1, which is required for its activation (Kishore et al., 2002), was severely impaired in the G159A mutant protein (Fig. 2 a). The kinase-dead K38M mutant was used as negative control in these in vitro assays (Fig. 2 a). As previously discussed (Fig. 1 g), D50A TBK1 was produced in much smaller amounts than the WT, G159A, or K38M TBK1 variants (Fig. 2 a). We then assessed the functionality of the two mutant proteins, in terms of TBK1 involvement in the TLR3 pathway, in TBK1−/− MEFs (McWhirter et al., 2004) transiently transfected with the D50A, G159A, or WT human TBK1 allele. The transfection of TBK1−/− MEFs with either the D50A or the G159A TBK1 allele failed to restore the poly(I:C)-induced IFN-β induction, whereas this induction was restored by transfection with the WT human TBK1 (Fig. 2 b). Indeed, G159A TBK1 behaved like the kinase-dead S172A mutant (Lei et al., 2010). The D50A allele was a loss-of-expression allele, as shown by Western blotting (Fig. 2 c), whereas the G159A and S172A TBK1 alleles resulted in the production of amounts of TBK1 protein similar to those produced from the WT allele (Fig. 2 c). All transfected cells were healthy and produced IL-6 in response to IL-1β (Fig. 2 d). Thus, the G159A TBK1 mutant allele is normally expressed but generates a kinase-dead protein, and the D50A allele is a loss-of-expression allele resulting in a loss of protein activity. Both mutants are therefore loss-of-function for TLR3-mediated cellular responses.
TLR3 responses to poly(I:C) are impaired in fibroblasts from P1 but not in those from P2
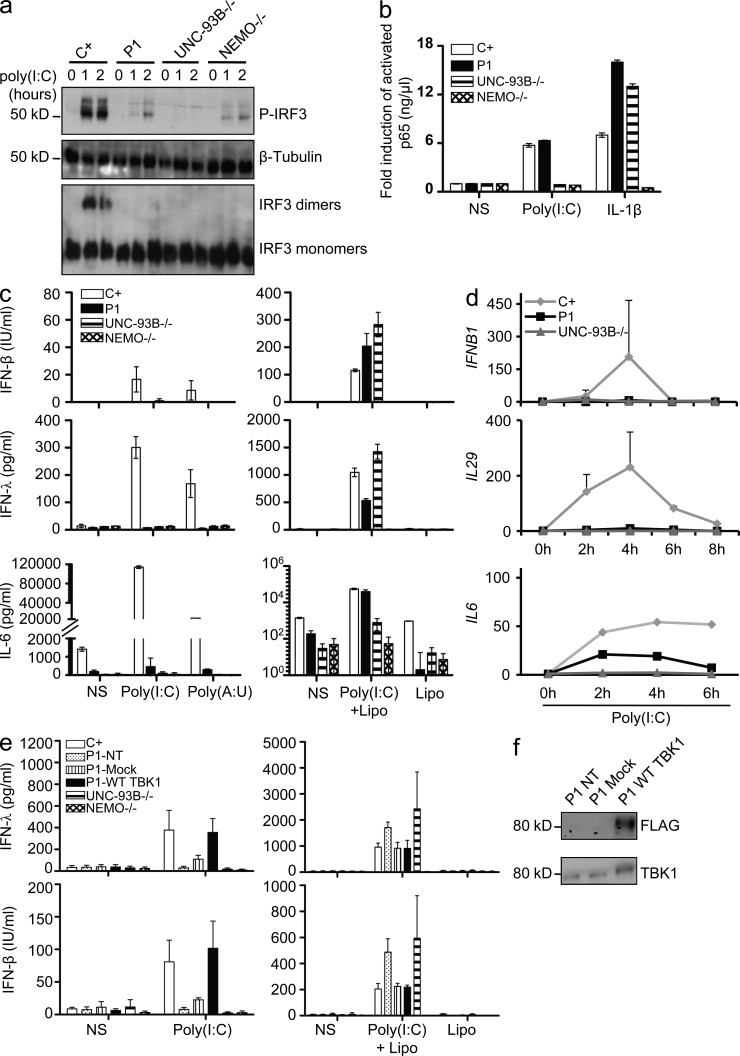
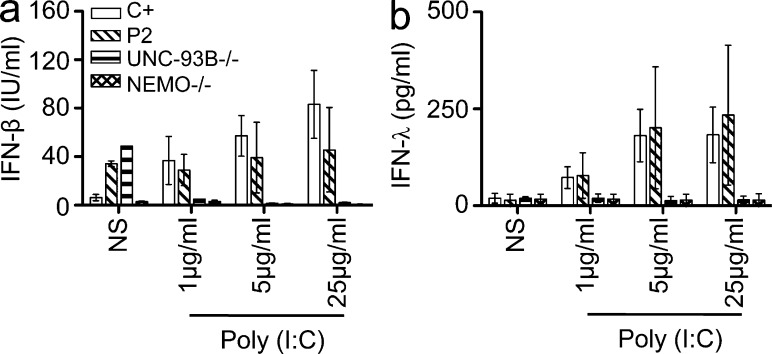
We tested the hypothesis that the loss-of-function G159A and D50A TBK1 alleles are associated with an AD form of HSE by first studying the response of the patients’ heterozygous fibroblasts to poly(I:C). Human dermal fibroblasts express TLR3 and respond to poly(I:C) in a TLR3-dependent manner (Matsumoto et al., 2002; Zhang et al., 2007; Guo et al., 2011). In control cells, the activation of IRF3 and NF-κB after the recruitment of TRIF to TLR3 (Sato et al., 2003) leads to the production of IFN-β, IFN-λ, and proinflammatory cytokines such as IL-6. We assessed IRF3 phosphorylation and subsequent dimerization by native PAGE and Western blotting after poly(I:C) stimulation. We found that cells from P1 (G159A/WT) did not support IRF3 phosphorylation or dimerization to levels observed in control cells, similar to UNC-93B– and NEMO-deficient cells, in which TLR3 signaling is abolished (Fig. 3 a). However, the fibroblasts of P1 displayed normal nuclear activation of NF-κB in response to poly(I:C) stimulation (Fig. 3 b), like healthy control cells but unlike UNC-93B– or NEMO-deficient fibroblasts. Consistent with the lack of IRF3 activation, SV40-transformed and primary fibroblasts from P1 did not produce IFN-β and IFN-λ mRNA or protein upon stimulation with extracellular poly(I:C) (Fig. 3, c and d; and not depicted). Consistent with the NF-κB activation observed, poly(I:C)-induced IL-6 production was low but detectable in P1’s fibroblasts (Fig. 3, c and d). Moreover, the lack of response of P1’s cells to poly(I:C) was rescued by WT TBK1 overexpression (Fig. 3, e and f). Similarly, the patient’s cells were able to produce IFN-β and IFN-λ upon stimulation with poly(I:C) in the presence of Lipofectamine (Fig. 3, c and e), which mediates the transfection of poly(I:C) into the cytosol, where it can be detected by cytosolic dsRNA sensors other than endosomal TLR3 (Kato et al., 2006, 2008). Cells from patients with AR UNC-93B and AR TLR3 deficiencies also respond to poly(I:C) in the presence of Lipofectamine (Fig. 3, c and e; Casrouge et al., 2006; Guo et al., 2011). Thus, the TBK1 defect in P1 impairs TBK1-dependent TLR3 signaling but does not seem to affect signaling by cytosolic dsRNA sensors. In contrast, in fibroblasts from P2 (D50A/WT), IRF3 dimerization was only marginally impaired upon stimulation with a low dose of poly(I:C) (1 µg/ml; not depicted), and no such impairment was observed at high doses (25 µg/ml; not depicted). Consequently, the production of IFN-β and IFN-λ in response to extracellular poly(I:C) was normal in fibroblasts from P2 (Fig. 4, a and b). Thus, TLR3 signaling was only marginally affected by the heterozygous D50A TBK1 allele in the fibroblasts of P2. Consistently, the TLR3-dependent poly(I:C) response was impaired in fibroblasts from P1 but not P2, as assessed by genome-wide transcription experiments (Fig. 5). Overall, heterozygosity for the G159A TBK1 allele severely impairs IRF3-dependent IFN-β and IFN-λ production in response to TLR3 activation, whereas the D50A TBK1 allele leads to no detectable impairment of TLR3-dependent poly(I:C) responses in human fibroblasts.
Figure 3.
The G159A TBK1 allele of P1 abolishes TLR3-mediated IFN induction in the patient’s fibroblasts. (a) IRF-3 phosphorylation in fibroblasts from P1, as assessed by Western blotting after stimulation with 25 µg/ml poly(I:C) for 0, 1, or 2 h, with β-tubulin as a loading control. Comparison with control cells (C+) and UNC-93B−/− and NEMO−/− fibroblasts. IRF3 dimerization was assessed by native Western blotting. Blots are representative of three independent experiments. Two different healthy control cell lines were tested and gave the same result. (b) Activated p65 levels, as determined by EMSA-ELISA after 30 min of stimulation with IL-1β or 1 h of poly(I:C) in control cells (C+; averaged from two control cell lines), in fibroblasts from P1 and from UNC-93B−/− and NEMO−/− patients. (c) IFN-β, IFN-λ, and IL-6 production, as assessed by ELISA, after 24 h of stimulation with poly(I:C) or poly(A:U) (polyadenylic-polyuridylic acid) or transfection of poly(I:C) mediated by Lipofectamine (Lipo) in control (C+) cells (averaged from two distinct control cell lines) and in fibroblasts from P1 and from UNC-93B−/− and NEMO−/− patients. (d) Induction of mRNA for IFNB1, IL29, and IL6 after stimulation with 25 µg/ml poly(I:C), as assessed by RT-qPCR in fibroblasts from P1, an UNC-93B−/− patient, and controls (C+; averaged from two distinct control cell lines). Graphs present the mean values ± SD of three independent experiments for IFNB1 and IL29. The graph presented for IL6 is representative of two independent experiments. (e) Fibroblasts from P1 were untransfected (NT), mock-transfected, or transfected with a vector encoding FLAG-tagged WT TBK1 and stimulated for 24 h with the indicated reagents (poly(I:C) or poly(I:C) transfection mediated by Lipofectamine). IFN-β and IFN-λ production was assessed by ELISA. (b, c, and e) Values represent mean values ± SD calculated from three independent experiments. (f) Western blot of transfected P1 cells with anti-TBK1 and anti-FLAG antibodies. NS, nonstimulated.
Figure 4.
TLR3-mediated IFN production is normal in P2’s fibroblasts. (a and b) WT (C+), P2, UNC-93B−/−, and NEMO−/− fibroblasts were stimulated with various doses of poly(I:C) for 24 h. The production of IFN-β (a) and IFN-λ (b) was assessed by ELISA. The graphs show the mean values ± SD for three independent experiments. NS, nonstimulated.
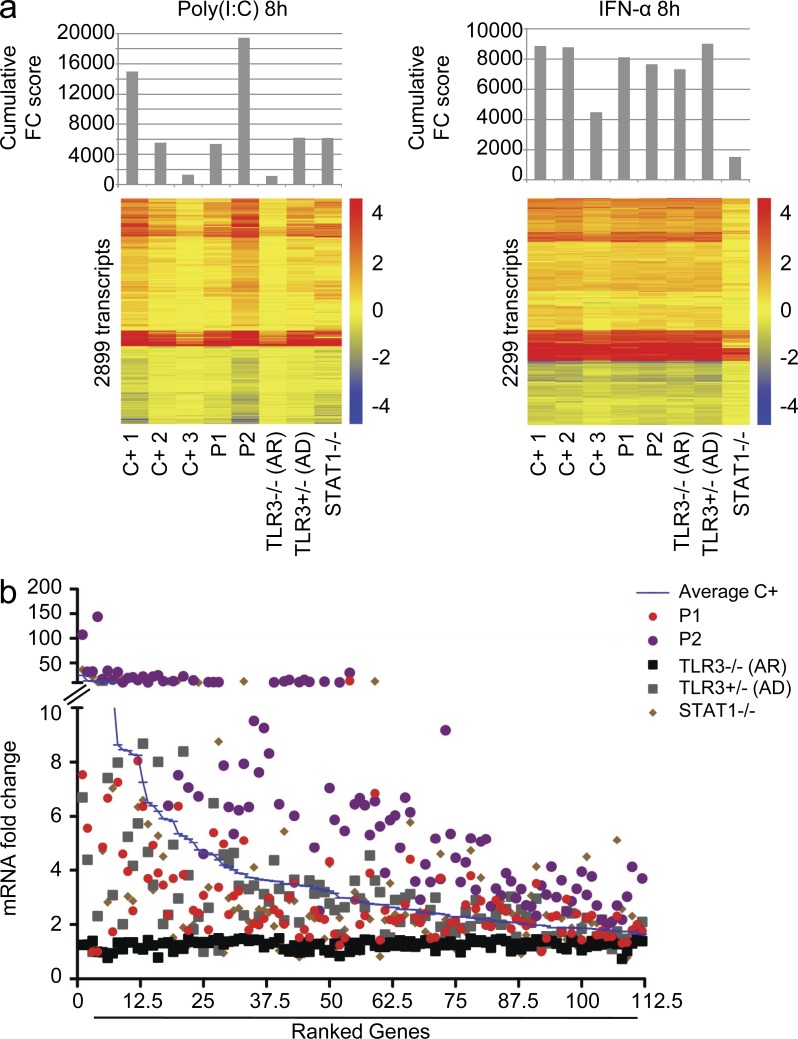
Figure 5.
Genome-wide transcriptional evaluation of the TLR3 pathway in primary fibroblasts. (a) Cumulative fold change (FC) score (top) and heat maps (bottom) of the transcripts regulated by 8 h of stimulation with poly(I:C) (left) or IFN-α2b (right) in primary fibroblasts from three healthy controls (C+), both TBK1 patients (P1 and P2), a patient with TLR3 AR deficiency (TLR3−/−), a patient with AD TLR3 deficiency (AD TLR3), and a patient with STAT-1 complete deficiency (STAT1−/−). The cumulative score is the sum of all the fold change values of >1.5 (up- or down-regulation). Heat maps show a hierarchical clustering of transcripts differentially expressed upon stimulation (based on 100 differences in intensity and 1.5-fold changes compared with nonstimulated condition in healthy controls). Changes with respect to the unstimulated condition are shown by a color scale: red, up-regulated; blue, down-regulated; yellow, no change. The probes displaying differences of >100 in intensity were used to calculate the cumulative score. (b) Ranking of the 112 transcripts up-regulated after 8 h of poly(I:C) stimulation, with a fold change of at least 1.5 in all three controls tested, in primary fibroblasts from three healthy controls averaged together (Average C+), both TBK1 patients (P1 and P2), a patient with TLR3 AR deficiency (TLR3−/−), a patient with AD TLR3 deficiency (AD TLR3), and a patient with STAT-1 complete deficiency (STAT1−/−).
IFN-dependent control of HSV-1 and VSV is impaired in the fibroblasts of both patients
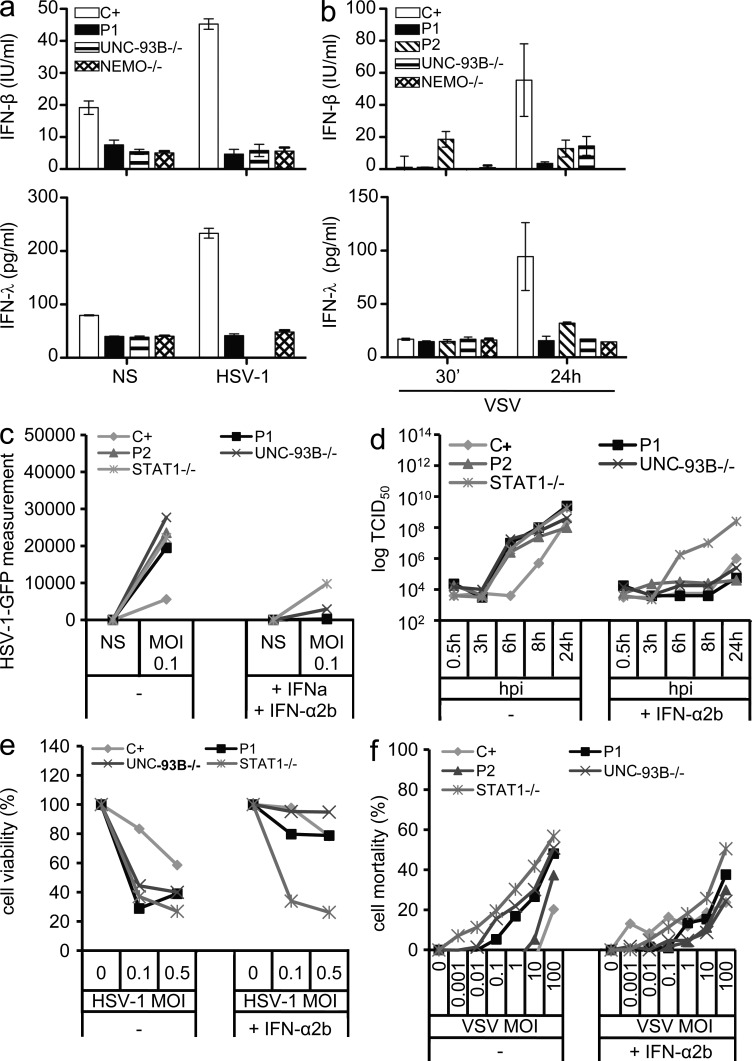
The IFN response to HSV-1 and VSV in human dermal fibroblasts has been shown to be partially dependent on TLR3. TLR3-, UNC-93B–, TRIF-, and TRAF3-deficient fibroblasts fail to contain the replication of HSV-1 and/or VSV as the result of a lack of IFN-β and IFN-λ production (Casrouge et al., 2006; Zhang et al., 2007; Pérez de Diego et al., 2010; Guo et al., 2011; Sancho-Shimizu et al., 2011b). We tested the hypothesis that the loss-of-function G159A and D50A TBK1 alleles were responsible for an AD form of HSE by investigating the IFN-mediated antiviral response in heterozygous fibroblasts from P1 and P2. The fibroblasts of P1 produced no IFN-β or IFN-λ 24 h after infection with HSV-1 (Fig. 6 a). The cells of P1 therefore displayed high levels of viral replication and cell death, like TLR3- and UNC-93B–deficient cells but unlike healthy control cells (Fig. 6, c and e). The addition of IFN-α2b to these cells restored virus containment to levels similar to those in control cells (Fig. 6, c and e). The IFN dependence of this phenotype was confirmed by the impaired response of STAT-1–deficient fibroblasts to prior treatment with IFN-α2b, in terms of viral control (Fig. 6 c). Moreover, fibroblasts from P1 and P2 also displayed very low levels of IFN-β and IFN-λ production after infection with VSV at a multiplicity of infection (MOI) of 10 (Fig. 6 b). VSV replication was not suppressed by cells from either of the patients but was suppressed by fibroblasts from a healthy control (Fig. 6 d). VSV-induced cell death levels were also higher in cells from both patients than in the control, as in TLR3- and UNC-93B–deficient cells (Fig. 6 f). The treatment of these cells with IFN-α2b restored viral containment and cell death to levels similar to those in control cells (Fig. 6, d and f). The IFN-dependent control of HSV-1 and VSV, which is at least partly dependent on TLR3 in human fibroblasts (Casrouge et al., 2006; Zhang et al., 2007; Pérez de Diego et al., 2010), was therefore impaired in the patients’ cells. In cells heterozygous for the G159A and D50A TBK1 alleles, the two loss-of-function TBK1 mutant alleles thus confer a dominant phenotype, in terms of the TLR3- and IFN-dependent control of viruses. Fibroblasts heterozygous for D50A TBK1 displayed an impaired control of viruses via TLR3, despite responding normally to poly(I:C) via TLR3. This suggests that the entire, intact, TBK1-dependent TLR3 pathway is required for HSV-1 and VSV control in human fibroblasts, although we cannot exclude a possible role for TBK1 in the TLR3-dependent or -independent inhibition of HSV-1 replication (Verpooten et al., 2009; Ma et al., 2012) or cell death (Ou et al., 2011). Both the G159A and D50A TBK1 alleles are thus loss-of-function and dominant for TLR3- and IFN-dependent antiviral immune responses. The D50A allele, which is weakly expressed, is dominant for viral control via TLR3. The G159A allele, which is normally expressed and encodes a kinase-dead protein, is dominant not only for viral control but also for poly(I:C) responses via TLR3.
Figure 6.
Impaired IFN-dependent control of HSV-1 and VSV infection in the patients’ fibroblasts. (a and b) IFN-β and IFN-λ production, as measured by ELISA, in the patients’ fibroblasts, control cells (C+; averaged from two distinct control cell lines), and UNC-93B−/− and NEMO−/− fibroblasts after 24 h of stimulation with HSV-1 (a) and 30 min or 24 h of stimulation with VSV (b). The graphs display means ± SD determined from three independent experiments. (c and d) Replication of the HSV-1–GFP virus at an MOI of 1 (c) and of VSV at an MOI of 10 (d) in the patients’ fibroblasts (P1 and P2) and in control cells (C+; averaged from two distinct control cell lines), and UNC-93B−/− and STAT-1−/− fibroblasts, as determined at the indicated hours after infection, with or without 18-h IFN-α2b pretreatment. One experiment representative of two independent experiments performed is shown. (e) Viability of control cells (C+; averaged from two different control cell lines) and in fibroblasts from P1, an UNC-93B−/− patient, and a STAT-1−/− patient after infection with HSV-1–GFP at MOIs of 0.1 and 0.5, with or without IFN-α2b treatment 18 h before infection. (f) Cell mortality after 24 h of VSV infection in control cells (C+; averaged from two distinct control cell lines) and in fibroblasts from P1, P2, a UNC-93B−/− patient, and a STAT-1−/− patient, with or without 18-h IFN-α2b pretreatment. NS, nonstimulated.
The two TBK1 mutant alleles are dominant through different mechanisms
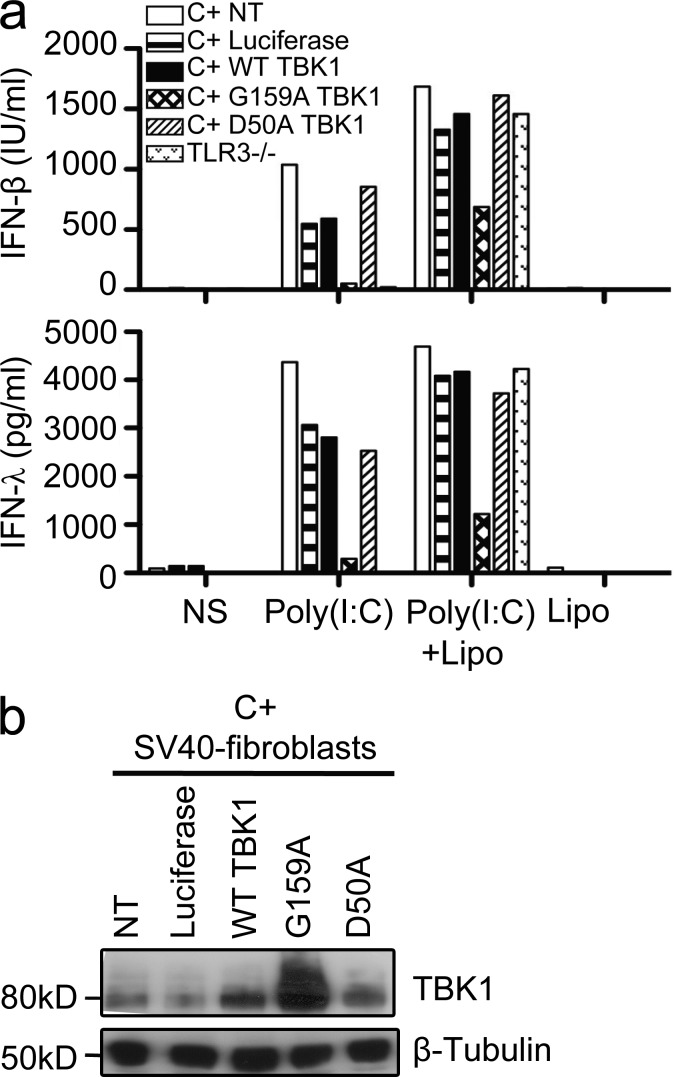
Our data suggest that the two TBK1 alleles may be dominant through different mechanisms. We investigated whether the mutant TBK1 alleles were dominant negative by transducing control human fibroblasts, which express endogenous WT TBK1, with lentiviral pseudoparticles encoding luciferase only (used as a negative control), WT TBK1, or one of the two mutant TBK1 proteins. After TLR3 stimulation by poly(I:C), we measured the IFN-β and IFN-λ production from those cells. Transient overexpression of the G159A TBK1 allele in healthy control fibroblasts decreased the IFN-λ and IFN-β production upon poly(I:C) stimulation (Fig. 7 a), but no such effect was observed after overexpression of the D50A or WT allele. Western blotting of the whole-cell lysates confirmed the overexpression of the G159A and WT TBK1 proteins (Fig. 7 b). The G159A TBK1 allele thus exerts a dominant-negative effect on WT TBK1 activity, which is normally expressed in control cells. In contrast, the amounts of IFN-λ and IFN-β produced in response to poly(I:C) were similar in control fibroblasts transduced with lentiviral pseudoparticles encoding luciferase or the WT or D50A TBK1 proteins. The D50A TBK1 allele is therefore most likely not dominant negative and exerts its dominant effect by haploinsufficiency. This is consistent with the previous observations of TBK1 instability and lower total amounts of TBK1 in the cells of P2 (Fig. 1, f and g and Fig. 2, a and c). Both the G159A and D50A TBK1 alleles are thus loss-of-function, but one is dominant over the WT allele by negative dominance and the other by haploinsufficiency. We then investigated the possible dominance of the two alleles in the context of other TLR3-independent, TBK1-mediated, IFN-inducing pathways.
Figure 7.
The G159A allele exerts a dominant-negative effect, whereas the D50A allele does not. (a) IFN-β and IFN-λ production, as assessed by ELISA, in response to 50 µg/ml poly(I:C) stimulation or 25 µg/ml poly(I:C) transfection mediated by Lipofectamine (Lipo) in TLR3-deficient fibroblasts and in control fibroblasts (C+) transduced with lentivirus pseudoparticles encoding luciferase, WT TBK1, or one of the TBK1 mutants (G159A or D50A) or left untransduced (NT). Cells were transduced 6 d before stimulation. This experiment is representative of three independent experiments. NS, nonstimulated. (b) Western blot of transduced control fibroblasts (C+) with anti-TBK1 and antitubulin antibodies.
Impact of TBK1 mutations on other IFN-inducing pathways
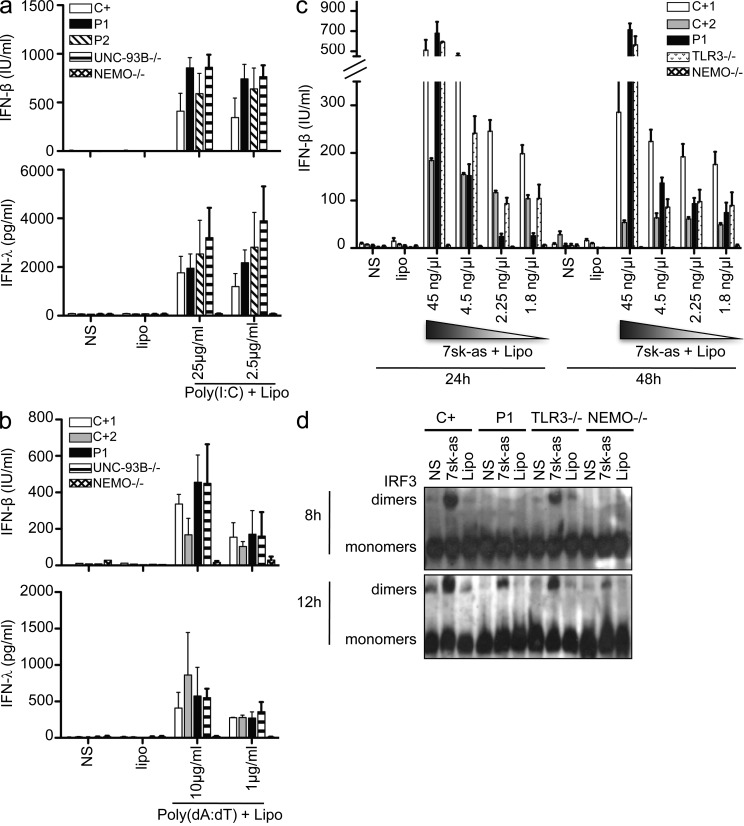
TBK1 is involved in the activation of IRF3 and IRF7 in multiple IFN-inducing signal transduction pathways (Chau et al., 2008), including cascades triggered by the activation of cytosolic RNA sensors (RIG-I and MDA-5), cytosolic DNA sensors (DAI and IFI16), and TLR4. We first investigated the response to TLR4 activation in PBMCs by assessing IFN-α production after 24 h of stimulation with LPS, which is recognized by TLR4 (Poltorak et al., 1998). The PBMCs of P1 responded normally to TLR4 activation (not depicted), consistent with the normal response induced by LPS stimulation in TBK1−/− and TBK1+/− MEFs (Hemmi et al., 2004). We further investigated the capacity of fibroblasts from the two patients to mediate signaling for other pathways known to be TBK1 dependent in mice by assessing the production of IFN-β and IFN-λ after transfection with the RIG-I–specific agonist 7sk-as (Pichlmair et al., 2006), with poly(I:C), which can activate both RIG-I and MDA-5 (Yoneyama et al., 2004), or with the synthetic dsDNA poly(dA:dT) (poly(deoxyadenylic-deoxythymidylic) acid), which can activate DAI or IFI16 (Takaoka et al., 2007; Unterholzner et al., 2010). The fibroblasts of both patients produced normal levels of IFN-β and IFN-λ after transfection with various amounts of dsDNA or poly(I:C) (Fig. 8, a and b; and not depicted) and large amounts of 7sk-as (Fig. 8 c and not depicted). However, the levels of IFN-β or IFN-λ production induced by lower doses of 7sk-as were much lower in cells from P1 than in control cells at 24 h but reached normal levels by 48 h (Fig. 8 c). Consistently, IRF3 activation was delayed after RIG-I stimulation after transfection with a low dose of 7sk-as: IRF3 dimerization was barely detectable at 8 h but normal after 12 h of RIG-I stimulation in the fibroblasts of P1 (Fig. 8 d). These results point to a mild impairment of RIG-I signaling in fibroblasts heterozygous for G159A TBK1. No major TLR3-independent phenotypes were observed in the cells of P1 and P2. We assessed the potential of the G159A and D50A TBK1 alleles to rescue the lack of IFN-β production observed in TBK1−/− MEFs upon transfection with poly(I:C) and 7sk-as (Fig. 9 a). Unlike the WT TBK1 allele, the G159A mutant allele could not rescue these responses, confirming that this allele was a loss-of-function allele. The D50A allele allowed residual responses, albeit probably caused by transient overexpression of the D50A TBK1 protein. On Western blot, cells transfected with the D50A allele were found to contain smaller total amounts of TBK1 (Fig. 9 b) than cells transfected with the WT or G159A alleles, consistent with our previous findings (Figs. 1 g and 2, a and c). All cells were healthy and produced IL-6 in response to IL-1β (Fig. 9 c). In TBK1−/− MEFs, both mutants are therefore loss-of-function for IFN-inducing pathways triggered by dsRNA and single-stranded RNA (ssRNA). It remains unclear whether the normal IFN response after the activation of TLR4 and cytosolic sensors of nucleic acids in our patients’ cells is caused by the residual TBK1 activity present in human cells with a partial TBK1 deficiency or caused by the contribution of TBK1 to these pathways not being essential in human PBMCs and fibroblasts.
Figure 8.
Normal responses to dsRNA, dsDNA, and ssRNA introduced by transfection in the patients’ fibroblasts. (a) Production of IFN-β and IFN-λ in response to various doses of poly(I:C), introduced by transfection into control (C+; averaged from two different control cell lines), P1, P2, and UNC-93B– and NEMO-deficient fibroblasts. Values represent mean values ± SD calculated from three independent experiments. (b) Production of IFN-β and IFN-λ in response to various doses of poly(dA:dT) introduced by transfection into control (C+), P1, P2, UNC-93B−/−, and NEMO−/− fibroblasts. Values from three different experiments were averaged and are presented here as mean values ± SD. (c) IFN-β production 24 or 48 h after transfection with the ssRNA 7sk-as in control (C+1 and C+2), P1, TLR3−/−, and NEMO−/− fibroblasts. Values represent mean values ± SD from three independent experiments. (d) IRF3 dimerization, assessed by native Western blotting upon stimulation with 2.25 ng/µl 7sk-as in fibroblasts from P1, at 8 and 12 h; comparison with control WT cells (C+), TLR3−/−, and NEMO−/− fibroblasts. The blots shown are representative of three independent experiments. NS, nonstimulated.
Figure 9.
Assessing the functionality of both mutant TBK1 alleles in the response to transfected poly(I:C) and ssRNA (7sk-as). (a) TBK1−/− MEFs were either left untransfected (NT) or were transfected with a mock vector, WT TBK1, or mutant constructs: D50A (mutation present in P2) or G159A (the mutation in P1). After 24 h, the cells were stimulated by transfection with Lipofectamine of 25 µg/ml poly(I:C) or 22.5 ng/µl ssRNA (7sk-as). ELISA was performed to assess IFN-β and IL-6 production 24 h after stimulation. The experiment shown is representative of three independent experiments. (b) TBK1 expression from the transfected constructs was assessed by Western blotting with an antibody against TBK1. β-Tubulin was used as a loading control. (c) IL-6 production as assessed by ELISA after stimulation with 10 ng/ml IL-1β for 24 h after transfection in TBK1−/− MEFs either not transfected (NT) or transfected with a mock vector or a vector encoding WT TBK1 or the mutants G159A and D50A TBK1. The experiment shown is representative of three independent experiments. NS, nonstimulated.
Normal IFN responses to viruses other than VSV and HSV-1 in the cells of both patients
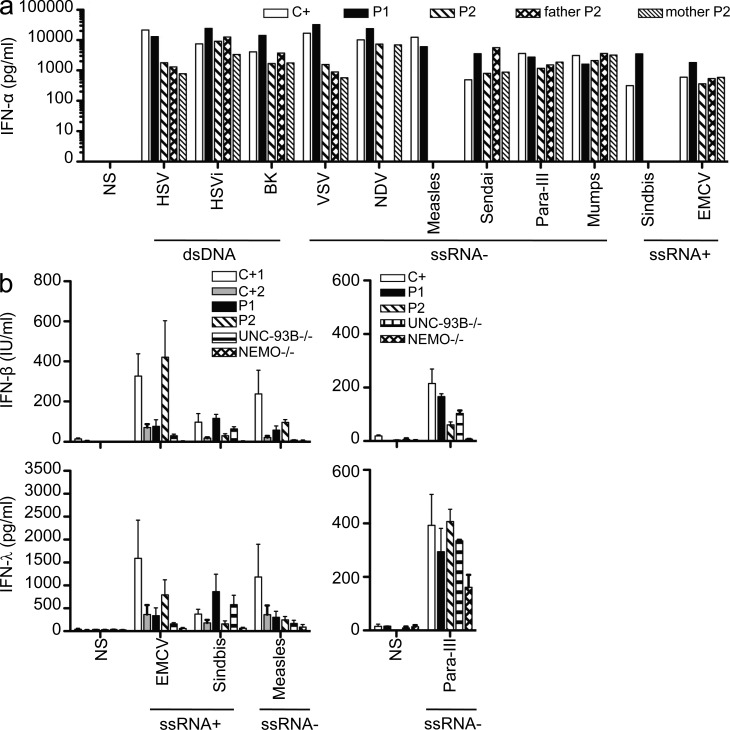
Both TBK1-deficient patients suffered from isolated HSE, with no other unusual infectious diseases, including viral diseases, during their 17 and 26 yr of life. We investigated the response of cells from P1, carrying the dominant-negative G159A TBK1 allele, to several viruses. We first challenged PBMCs from P1 and P2 with various types of viruses, including positive ssRNA viruses (encephalomyocarditis virus [EMCV] and Sindbis virus), negative ssRNA viruses (Sendai, human parainfluenza virus III, NDV [Newcastle disease virus], mumps virus, and measles virus), and dsDNA viruses (BK virus and HSV-1), all of which induced normal levels of IFN-α and IL-6 in these cells (Fig. 10 a and not depicted). This may reflect the partial nature of the genetic defects carried by the patients, the redundancy of TBK1 and IKKi in the activation of IRF3 and IRF7 in leukocytes (Fitzgerald et al., 2003; Sharma et al., 2003; Hemmi et al., 2004; McWhirter et al., 2004; Matsui et al., 2006), or both. Unlike leukocytes, fibroblasts do not produce IKKi (Shimada et al., 1999; Hemmi et al., 2004; Perry et al., 2004). When infected with human parainfluenza virus III, measles, EMCV, or Sindbis virus, the fibroblasts of both patients behaved like control cells in terms of their capacity to produce IFN-β and IFN-λ (Fig. 10 b). This result contrasts with the impaired response of these cells to infection with HSV-1 or VSV, both of which activate TLR3 to initiate IFN induction, as demonstrated by findings for the cells of patients with TLR3 (Guo et al., 2011), UNC-93B (Casrouge et al., 2006), TRIF (Sancho-Shimizu et al., 2011b), and TRAF3 (Pérez de Diego et al., 2010) deficiencies. The lack of viral control in the fibroblasts of patients heterozygous for TBK1 is thus limited to the neurotropic viruses HSV-1 and VSV. These results for PBMCs and fibroblasts are consistent with the narrow clinical phenotype observed in both TBK1-deficient patients. They suggest that the residual TBK1-dependent IFN induction by TLR3 and/or by other receptors in cells with a partial TBK1 deficiency may contribute to the protective immunity against most common viruses.
Figure 10.
Both patients present normal responses to other viruses in PBMCs and fibroblasts. (a) PBMCs from a control (C+), P1, P2, and P2’s parents were challenged with several viruses with different types of genomes: dsDNA viruses (HSV-1, UV-inactivated HSV-1 or HSV-1i, and BK), negative-strand ssRNA viruses (VSV, NDV, measles, Sendai virus, human parainfluenza virus III [Para-III], and mumps), and positive-strand ssRNA viruses (Sindbis virus and EMCV). Levels of IFN-α were assessed by ELISA 24 h after infection. P2 and her parents’ cells were not tested for their response to infection with Sindbis and measles. This experiment was performed once. (b) Production of IFN-β and IFN-λ, as assessed by ELISA, in control (C+), P1, P2, UNC-93B−/−, and NEMO−/− fibroblasts 24 h after infection with positive-strand ssRNA viruses (Sindbis virus at an MOI of 10 and EMCV at an MOI of 1) and negative ssRNA viruses (measles at an MOI of 1 and parainfluenza virus III at an MOI of 1). Values represent mean values ± SD from three independent experiments. NS, nonstimulated.
DISCUSSION
We have identified AD TBK1 deficiency as a new genetic etiology of HSE in childhood. The patients’ fibroblasts displayed impaired TLR3 responses, confirming the essential role of human TBK1 as an IFN-inducing IRF3 kinase in the TLR3 pathway in these cells. Together with our previous discoveries of AR UNC-93B (Casrouge et al., 2006), AR and AD TLR3 (Zhang et al., 2007; Guo et al., 2011), AR and AD TRIF (Sancho-Shimizu et al., 2011b), and AD TRAF3 (Pérez de Diego et al., 2010) deficiencies, this finding highlights the nonredundant role of TLR3-dependent IFN induction in the CNS for the control of HSV-1 in the course of primary infection in childhood. The clinical penetrance of AD TBK1 deficiency is incomplete, like that of AD TLR3, AR UNC-93B, and AD TRIF deficiencies, consistent with the sporadic occurrence of HSE (Whitley and Kimberlin, 2005). The actual clinical penetrance cannot be assessed, as the frequencies of the TBK1 mutant alleles are not known. We know only that they were not found in 1,050 healthy controls (2,100 chromosomes) from 52 ethnic groups. Our discovery of rare morbid variants of the TLR3, UNC93B1, TRAF3, TRIF, and TBK1 genes, each found in only a small number of children with HSE, suggests that the determinism of HSE may depend on a collection of highly diverse and possibly immunologically related single-gene lesions (Casanova and Abel, 2007; Alcaïs et al., 2010). We were unable to investigate whether the two forms of TBK1 deficiency reported here displayed complete cellular penetrance, as documented in families with UNC-93B, TLR3, or TRIF deficiency (Casrouge et al., 2006; Zhang et al., 2007; Sancho-Shimizu et al., 2011b). Consistent with this possibility, no unreported variants other than that of TBK1 were found by whole-exome sequencing of genes involved in the TLR3–IFN axis in P2 and her asymptomatic mother (see Whole-exome sequencing in P1 and P2: TLR3–IFN pathway genes sequenced and found to be WT). Interestingly, the two unrelated patients with TBK1 deficiency had different cellular phenotypes, that of the patient with the G159A mutation being more severe than that of the patient with the D50A mutation because the G159A allele is dominant negative, whereas D50A is dominant by haploinsufficiency. Regardless of these differences, our discovery of two forms of AD partial TBK1 deficiency adds weight to the argument that HSE should not be considered only a viral disease. Instead, it should also be seen as a genetic disorder caused by an array of rare, single-gene inborn errors of TLR3 immunity, in at least a fraction of affected children (Casanova and Abel, 2005; Alcaïs et al., 2010).
The identification of TBK1 deficiency as a genetic etiology of HSE is surprising, as mouse TBK1 has been shown to be involved in many antiviral pathways other than the TLR3 cascade, including the RIG-I/MDA-5 (Fitzgerald et al., 2003; Sharma et al., 2003; Hemmi et al., 2004), DAI (DLM-1/ZBP1; Takaoka et al., 2007), and IFI16/STING (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Unterholzner et al., 2010) pathways. In vitro studies have shown that the IFN-α/β response is defective in TBK1−/− MEFs stimulated with poly(I:C) (Hemmi et al., 2004) or infected with Sendai virus (Hemmi et al., 2004; Perry et al., 2004) or VSV (Hemmi et al., 2004). IFN induction is also defective in mouse TBK1−/− macrophages challenged with HSV-1 and dsDNA (Miyahira et al., 2009), LPS, and poly(I:C) (Perry et al., 2004). TBK1−/− mice die in the perinatal period as the result of massive liver degeneration and apoptosis (Bonnard et al., 2000). The role of TBK1 in host defense in vivo has not, therefore, been studied in mice. Our data suggest that TBK1 haplotype insufficiency may also occur in mice. TBK1+/− mice are viable but have not been characterized immunologically or in terms of their response to infection (Marchlik et al., 2010). Nevertheless, our two patients with AD partial TBK1 deficiency were found to be resistant to most common viruses. Like TLR3, UNC-93B, TRIF, and TRAF3 deficiencies, but unlike STAT-1 (Yang et al., 2005) and NEMO (Audry et al., 2011) deficiencies, AD TBK1 deficiency is associated with isolated HSE. The residual TBK1 activity is not sufficient to sustain optimal TLR3 responses, against HSV-1 in particular, but is sufficient for the TLR3-independent pathways tested. AD TBK1 deficiency does not impair the response to transfected dsDNA, dsRNA, or ssRNA in human fibroblasts or the response to TLR4 in leukocytes. This may account for the effective control of the viruses tested in AD TBK1-deficient fibroblasts and for the patients’ lack of susceptibility to other viral diseases. However, we cannot exclude the possibility that TBK1 is redundant for certain pathways and/or in certain cells. In any case, this experiment of nature neatly illustrates how subtle mutations of human genes at the crossroads of multiple signaling pathways may result in partial biochemical defects, accounting for narrow cellular and clinical phenotypes.
MATERIALS AND METHODS
Case reports
P1 is a Polish child from a nonconsanguineous family with no family history of encephalitis or herpes labialis. Both parents and the child were healthy until P1 suffered from HSE at the age of 7 yr. The diagnosis was based on the positive detection by PCR of HSV-1 DNA in the child’s cerebrospinal fluid and on typical HSE-like brain pattern aberrations on MRI. P1 tested positive for IgG and IgM antibodies against HSV-1 at the time of diagnosis. She was treated with acyclovir, systemic steroids, intravenous gammaglobulins, phenobarbital, valproic acid, benzodiazepines, mannitol, and antibiotics for 4 mo. P1 subsequently presented cognitive disability and drug-resistant epilepsy. She has since required multidrug anti-epilepsy treatment, which has not influenced her immunological status, as immunological screenings gave normal results. Since her HSE episode, P1 has developed only mild upper respiratory tract infections despite attending kindergarten and primary school. P1 has been exposed to VZV (varicella zoster virus), CMV, EBV, B19, and influenza A, as demonstrated by positive serological results, in the absence of acute events. P1 was also immunized with hepatitis B, diphtheria/tetanus/polio type 1, 2, and 3, and mumps vaccines with no adverse effect.
P2 is a French patient from a nonconsanguineous family with no family history of encephalitis. P2 suffered from HSE at 11 mo of age, presenting initially with otitis and hyperthermia, followed by convulsions and fever the next day. 6 d later, a 15-d course of treatment with zovirax was initiated. At this time, P2’s condition had deteriorated and she presented right-sided hemiparesis and erythema on her arms and hands. A scan revealed typical encephalitis-like patterns for the left temporal area of the brain. At the end of zovirax treatment, P2 still had right-sided hemiparesis but no longer suffered from convulsions. In the next month, EEGs and scans confirmed the presence of lesions on the left side of the brain (temporal, frontoinsular, and parietal). P2 subsequently suffered from obesity and motor and cognitive disabilities, including epilepsy during treatment for her motor and cognitive disabilities. P2 has been exposed to VZV, CMV, B19, and influenza A, as demonstrated by positive serological results, with no acute events. P2 was also immunized with the live mumps/rubella/measles vaccines and with the diphtheria toxoid, inactivated poliovirus type 1, poliovirus type 2, poliovirus type 3, and tetanus toxoid vaccines with no adverse effect. P2’s mother and sister suffer from bouts of herpes labialis once or twice a year but have no other significant infectious phenotype. P2’s father suffered from measles and chicken pox as a child but has no other significant infectious phenotype.
Both children have been living in and followed up in their countries of origin (Poland for P1 and France for P2). Informed consent was obtained in the home country of each patient in accordance with local regulations and with institutional review board (IRB) approval. The experiments described here were conducted in the United States and in France in accordance with local regulations and with the approval of the IRB of The Rockefeller University and Institut National de la Santé et de la Recherche Médicale.
Human molecular genetics
Total RNA and genomic DNA were extracted from the PBMCs of P1, P2, and a control with TRIZOL (Gibco). RNA was reverse-transcribed with SuperScript II RT (Invitrogen) according to the manufacturer’s instructions. The cDNAs for all the genes sequenced and for the genomic coding regions of TBK1 were amplified with specific primers. PCR was performed with Taq polymerase (Invitrogen). PCR products were analyzed by electrophoresis in 1% agarose gels, purified by ultracentrifugation through Sephadex G-50 Superfine resin (GE Healthcare), sequenced with the Big Dye Terminator cycle sequencing kit (Applied Biosystems), and analyzed on an ABI Prism 3700 machine (Applied Biosystems).
Cell culture and transfections
Primary human fibroblasts were obtained from biopsies of patients or healthy controls and were cultured in DME (Invitrogen) supplemented with 10% FBS. They were transformed with an SV40 vector, as previously described (Chapgier et al., 2006), to create SV40-immortalized fibroblasts (SV40-fibroblasts). The SV40-fibroblast cell lines were activated in 24-well plates at a density of 105 cells/well for 24 h, unless otherwise specified. TBK1−/− and WT MEFs were supplied by M.A. White. MEFs were transformed with an SV40 vector, as for human fibroblasts, and cultured in DME (Invitrogen) supplemented with 10% FBS. Cells were transiently transfected with expression vectors with a calcium phosphate transfection kit from Sigma-Aldrich, used according to the manufacturer’s instructions. The cells were dispensed into 12-well plates at a density of 0.9 × 104 cells/well for transfection. The pcDNA3.1-FLAG-TBK1 construct was obtained by subcloning the FLAG-TBK1 sequence from the vector pCMV5-FLAG-TBK1 into pcDNA3.1+ (Invitrogen); pCMV5-FLAG-TBK1 was provided by P. Cohen (University of Dundee, Dundee, Scotland, UK). Point mutations encoding D50A, G159A, or S172A TBK1 were introduced in pcDNA3.1-FLAG-TBK1 by site-directed mutagenesis using the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies).
Lentivirus pseudoparticles were generated by transfection of HEK293-T cells, mediated by Fugene 6 (Roche), in DME supplemented with 3% FBS and NEAA (nonessential amino acids from Life Technologies) with three plasmids: pTRIP, encoding TBK1 or luciferase, HIV gag-pol, and VSV envelope protein G (VSV-G) plasmids at a weight ratio of 1:0.8:0.2 (Schoggins et al., 2011). Mutations were introduced in TBK1 (D50A and G159A) by site-directed mutagenesis, as described above. Supernatants were collected at 48 and 72 h and supplemented with 4 µg/ml polybrene (Santa Cruz Biotechnology, Inc.) and 20 mM Hepes (Life Technologies).
TLR activation, signal transduction experiments, and cytokine measurement
We used the following TLR agonists: a synthetic analogue of dsRNA, poly(I:C) (GE Healthcare; used at a concentration of 25 µg/ml unless otherwise specified) and LPS (Re 595 from Salmonella minnesota, obtained from Sigma-Aldrich, and used at a concentration of 10 µg/ml); IL-1β was obtained from R&D Systems. All agonists and reagents were endotoxin free. For all TLR agonists except LPS, cells were incubated for 30 min with 10 µg/ml polymyxin B at 37°C before activation. TLR agonists were used to stimulate PBMCs at a density of 2 × 106 cells/ml in RPMI 1640/10% FCS, RPMI/1% FCS, or in AIMV medium, or in whole blood diluted 1:2 in RPMI 1640, as indicated. The SV40-fibroblast cell lines were activated in 24-well plates at a density of 105 cells/well for 20 h with HSV-1, VSV, 25 µg/ml poly(I:C) (unless otherwise specified), 20 ng/ml TNF, or 20 ng/ml IL-1β.
TransAM NF-κB p65 ELISA (Active Motif) was performed with an NF-κB p65-specific DNA probe with 10 µg nuclear extract. Native PAGE of total cell extract (25 µg of protein) was performed in a gel containing 1% sodium deoxycholate (DOC; Sigma-Aldrich) for the detection of IRF3 dimerization. After electrophoresis in a 7.5% polyacrylamide gel, the gel was blotted onto a membrane, which was then probed with the anti–IRF-3 antibody (FL-425; Santa Cruz Biotechnology, Inc.).
ELISA was performed for human IFN-α (AbCys SA), IFN-β (TFB; Fujirebio, Inc.), IL-6 (Pelipair kit), mouse IL-6 (R&D Systems), and mouse IFN-β (Verikine, PBL) according to the kit manufacturer’s instructions. The IFN-λ ELISA was developed in the laboratory. In brief, plates were coated by incubation overnight at 4°C with 1 µg/ml mAb against human IFN-λ (R&D Systems), and the levels of IFN-λ in the supernatant were determined by incubation with a biotinylated anti–human IFN-λ secondary mAb (R&D Systems) at a concentration of 400 µg/ml.
RT-qPCR and Western blotting
RNA was reverse-transcribed with the SuperScript II RT (Invitrogen) and oligo (dT) (Invitrogen) to determine TBK1 expression levels. In all RT-qPCR, β-glucuronidase (Gus) was used for normalization. For RT-qPCR analysis assessing the induction of IFNs upon poly(I:C) stimulation, RT was performed with random hexamer primers. RT-qPCR was performed with TaqMan reverse transcription reagents (Applied Biosystems) in an ABI Prism 7700 sequence detection system (Applied Biosystems). All the probes used were taken from the TaqMan Gene Expression Assays (Applied Biosystems). Results are expressed according to the ΔΔCt method, as described by the manufacturer.
Equal amounts of protein for each sample were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membranes (Bio-Rad Laboratories), which were then probed with primary antibodies followed by peroxidase-conjugated secondary antibodies and ECL Western blotting substrate (Thermo Fisher Scientific). The primary antibodies used included TBK1/NAK antibody (Cell Signaling), anti-DDK mAb (Origene), IRF3 phospho-pS386 antibody (Epitomics), rabbit polyclonal anti–IRF-3 FL-425 antibody (Santa Cruz Biotechnology, Inc.), and anti–β-tubulin antibody (Sigma-Aldrich).
Viral infection and quantification in fibroblasts
For VSV infection, 105 SV40-fibroblasts were dispensed into each individual well of 24-well plates and infected with VSV at an MOI of 10 in DME supplemented with 2% FBS. The plates were incubated for 30 min, and the cells were then washed and incubated in 500 µl of medium. Supernatants were obtained and frozen at the 0.5-, 3-, 6-, 8-, and 24-h time points. VSV titers were determined by calculating the 50% end point (TCID50), as described by Reed and Muench (1938), after the inoculation of Vero cell cultures in 96-well plates. For HSV-1–GFP infection, 104 SV40-fibroblasts were dispensed into each individual well of 96-well plates and infected with HSV-1–GFP (strain KOS; Desai and Person, 1998) at various MOIs in DME supplemented with 10% FBS. After 2 h, cells were washed and incubated in 100 µl of culture medium. The GFP fluorescence of the samples was quantified at the 2-, 16-, 24-, and 48-h time points. For assays of cell protection upon viral stimulation, cells were treated with 105 IU/ml IFN-α2b for 18 h before infection, as appropriate.
Cell viability assay
The viability of SV40-fibroblasts was assessed by resazurin oxidoreduction (TOX-8; Sigma-Aldrich). Cells were plated, in triplicate, in 96-well flat-bottomed plates (2 × 104 cells/well) in DME supplemented with 2% FBS. After 24 h, cells were infected by incubation for 24 h with VSV or for 72 h with HSV-1 at the indicated MOI. Resazurin dye solution was then added (5 µl per well) to the culture medium, and the samples were incubated for an additional 2 h at 37°C. Fluorescence was then measured at a wavelength of 590 nm, using an excitation wavelength of 531 nm. Background fluorescence, calculated for dye and complete medium alone (in the absence of cells), was then subtracted from the values for all the other samples; 100% viability corresponds to the fluorescence of uninfected cells.
In vitro protein kinase assays
TBK1 protein kinase assays were performed as previously described (Ou et al., 2011).
Whole-exome sequencing
3 µg DNA was extracted from leukocyte cells from the individuals and was sheared with an S2 Ultrasonicator (Covaris) for massively parallel sequencing. An adaptor-ligated library was prepared with the TruSeq DNA Sample Preparation kit (Illumina). Exome capture was performed with the SureSelect Human All Exon kit (Agilent Technologies; Byun et al., 2010). Paired-end sequencing was performed on a HiSeq 2000 (Illumina) that generated 100-bp reads. For sequence alignment, variant calling, and annotation, the sequences were aligned to the human genome reference sequence (GRCh37 build) using the Burrows-Wheeler Aligner (Li and Durbin, 2009). Downstream processing was performed with the Genome analysis toolkit (GATK; McKenna et al., 2010), SAMtools (Li et al., 2009), and Picard (http://picard.sourceforge.net). Variant calls were made with a GATK Unified Genotyper. All calls with a read coverage ≤2× and a Phred-scaled SNP quality of ≤20 were removed from consideration. All variants were annotated with a software system developed in-house.
Whole-exome sequencing in P1 and P2: TLR3–IFN pathway genes sequenced and found to be WT.
Genes sequenced by whole-exome sequencing were TAB3 (NAP1), IKBKE (IKKe), TBKBP1 (SINTBAD), TRAF6, IRF3, IRF7, HMGB1, HMGB2, CC2D1A (TAPE), TRAF2, TRAF4, TRAF1, ZMYND11 (BS69), RIPK1, IKBKG, IKBKB, CHUK, NFKBIA, NFKB1, RELA, NFKB2, CAMK2G, CAMK2A, PIAS4, TRADD, DDX3X, TNFAIP3, GAB1, TNIK*, and TRIM21. Genes sequenced by the Sanger method were TLR3, TICAM1 (TRIF), TRAF3, UNC93B1, and TANK. P1 carries an unreported heterozygous intronic variation in TNIK (asterisk) located 125 nucleotides 3′ to exon 4. A genomic DNA sample from P2’s mother was sequenced by whole-exome sequencing. All genes known to be involved in the TLR3–IFN axis covered by whole-exome sequencing and found to be WT in P2’s mother are indicated in bold typeface.
Genome-wide transcriptional profile experiments in primary fibroblasts
Data acquisition.
Fibroblasts from patients and controls were stimulated with IL-1β or poly(I:C) or IFN-α for 2 or 8 h or left unstimulated. Total RNA was isolated (RNeasy kit; QIAGEN) and RNA integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies). Biotinylated cRNA targets were prepared from 250 ng of total RNA, using the Illumina TotalPrep RNA Amplification kit (Ambion). The labeled cRNAs (750 ng) were then incubated for 16 h to HT-12 version 4 BeadArrays (48,323 probes). BeadChip arrays were then washed, stained, and scanned on a HiScanSQ (Illumina) according to the manufacturer’s instructions.
Data preprocessing.
After background subtraction, the raw signal values extracted with BeadStudio (version 2; Illumina) were scaled using quantile normalization. Minimum intensity was set to 10, and all the intensity values were log2 transformed. Only the probes called present in at least one sample (P < 0.01) were retained for downstream analysis (n = 28,553).
Data analysis.
Transcripts differentially regulated upon stimulation were defined based on a minimum twofold change (up- or down-regulation) and a minimum absolute raw intensity difference of 100 with respect to the respective unstimulated sample. Heat maps were generated using R (version 2.12.2). Cumulative fold change scores were calculated using the absolute fold change values from the transcripts passing the cutoffs defined above.
Data availability.
Raw data for the microarray analyses performed in this study are available from the public repository of GEO DataSets (accession no. GSE38652). Networks were resolved with MetaCore version 6.10 (GeneGo).
Statistical methods
Statistical significance was determined by Student’s t test. P-values of <0.05 were considered statistically significant.
Acknowledgments
We thank the members of both branches of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions, especially Vanessa Bryant for guidance and assistance with the construction of lentiviral pseudoparticles and Yuval Itan for discussions about microarrays. We thank the children and their families for their participation in this study.
J.-L. Casanova was an International Scholar of the Howard Hughes Medical Institute from 2005 to 2008. This work was conducted in the two branches of the Laboratory of Human Genetics of Infectious Diseases and was funded by the Rockefeller University Center for Clinical and Translational Science grant number 5UL1RR024143-04, the Rockefeller University, Institut National de la Santé et de la Recherche Médicale, Paris Descartes University, the St. Giles Foundation, the Eppley Foundation, the Jeffrey Modell Foundation and Talecris Biotherapeutics, the Agence Nationale de la Recherche, the Thrasher Research Fund, and the March of Dimes. Work conducted in M. White’s laboratory was funded by The National Cancer Institute (CA129451), the Robert Welch Foundation (I-1414), and the Cancer Prevention Research Institute of Texas.
The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used:
- AD
- autosomal dominant
- AR
- autosomal recessive
- CNS
- central nervous system
- EMCV
- encephalomyocarditis virus
- HSE
- HSV-1 encephalitis
- MEF
- mouse embryonic fibroblast
- MOI
- multiplicity of infection
- mRNA
- messenger RNA
- poly(I:C)
- polyinosinic-polycytidylic acid
- RT-qPCR
- quantitative RT-PCR
- SNP
- single nucleotide polymorphism
- ssRNA
- single-stranded RNA
- SV40
- simian virus 40
- VSV
- vesicular stomatitis virus
References
- Abel L., Plancoulaine S., Jouanguy E., Zhang S.Y., Mahfoufi N., Nicolas N., Sancho-Shimizu V., Alcaïs A., Guo Y., Cardon A., et al. 2010. Age-dependent Mendelian predisposition to herpes simplex virus type 1 encephalitis in childhood. J. Pediatr. 157:623–629: 629: e1 10.1016/j.jpeds.2010.04.020 [DOI] [PubMed] [Google Scholar]
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaïs A., Quintana-Murci L., Thaler D.S., Schurr E., Abel L., Casanova J.L. 2010. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann. N. Y. Acad. Sci. 1214:18–33 10.1111/j.1749-6632.2010.05834.x [DOI] [PubMed] [Google Scholar]
- Audry M., Ciancanelli M., Yang K., Cobat A., Chang H.H., Sancho-Shimizu V., Lorenzo L., Niehues T., Reichenbach J., Li X.X., et al. 2011. NEMO is a key component of NF-κB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J. Allergy Clin. Immunol. 128:610–617: e1–e4 10.1016/j.jaci.2011.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnard M., Mirtsos C., Suzuki S., Graham K., Huang J., Ng M., Itié A., Wakeham A., Shahinian A., Henzel W.J., et al. 2000. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 19:4976–4985 10.1093/emboj/19.18.4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun M., Abhyankar A., Lelarge V., Plancoulaine S., Palanduz A., Telhan L., Boisson B., Picard C., Dewell S., Zhao C., et al. 2010. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J. Exp. Med. 207:2307–2312 10.1084/jem.20101597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., Abel L. 2005. Inborn errors of immunity to infection: the rule rather than the exception. J. Exp. Med. 202:197–201 10.1084/jem.20050854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., Abel L. 2007. Primary immunodeficiencies: a field in its infancy. Science. 317:617–619 10.1126/science.1142963 [DOI] [PubMed] [Google Scholar]
- Casrouge A., Zhang S.Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N., et al. 2006. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 314:308–312 10.1126/science.1128346 [DOI] [PubMed] [Google Scholar]
- Chapgier A., Wynn R.F., Jouanguy E., Filipe-Santos O., Zhang S., Feinberg J., Hawkins K., Casanova J.L., Arkwright P.D. 2006. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J. Immunol. 176:5078–5083 [DOI] [PubMed] [Google Scholar]
- Chau T.L., Gioia R., Gatot J.S., Patrascu F., Carpentier I., Chapelle J.P., O’Neill L., Beyaert R., Piette J., Chariot A. 2008. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 33:171–180 10.1016/j.tibs.2008.01.002 [DOI] [PubMed] [Google Scholar]
- De Tiège X., Rozenberg F., Héron B. 2008. The spectrum of herpes simplex encephalitis in children. Eur. J. Paediatr. Neurol. 12:72–81 10.1016/j.ejpn.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Desai P., Person S. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., et al. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388–391 10.1038/ng1097 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- Guo Y., Audry M., Ciancanelli M., Alsina L., Azevedo J., Herman M., Anguiano E., Sancho-Shimizu V., Lorenzo L., Pauwels E., et al. 2011. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J. Exp. Med. 208:2083–2098 10.1084/jem.20101568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H., Takeuchi O., Sato S., Yamamoto M., Kaisho T., Sanjo H., Kawai T., Hoshino K., Takeda K., Akira S. 2004. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199:1641–1650 10.1084/jem.20040520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Barber G.N. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 455:674–678 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ma Z., Barber G.N. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 461:788–792 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 10.1084/jem.20080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore N., Huynh Q.K., Mathialagan S., Hall T., Rouw S., Creely D., Lange G., Caroll J., Reitz B., Donnelly A., et al. 2002. IKK-i and TBK-1 are enzymatically distinct from the homologous enzyme IKK-2: comparative analysis of recombinant human IKK-i, TBK-1, and IKK-2. J. Biol. Chem. 277:13840–13847 10.1074/jbc.M110474200 [DOI] [PubMed] [Google Scholar]
- Ku C.L., von Bernuth H., Picard C., Zhang S.Y., Chang H.H., Yang K., Chrabieh M., Issekutz A.C., Cunningham C.K., Gallin J., et al. 2007. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J. Exp. Med. 204:2407–2422 10.1084/jem.20070628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P.C. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4:1073–1081 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Lei C.Q., Zhong B., Zhang Y., Zhang J., Wang S., Shu H.B. 2010. Glycogen synthase kinase 3β regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity. 33:878–889 10.1016/j.immuni.2010.11.021 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Jin H., Valyi-Nagy T., Cao Y., Yan Z., He B. 2012. Inhibition of TANK binding kinase 1 by herpes simplex virus 1 facilitates productive infection. J. Virol. 86:2188–2196 10.1128/JVI.05376-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchlik E., Thakker P., Carlson T., Jiang Z., Ryan M., Marusic S., Goutagny N., Kuang W., Askew G.R., Roberts V., et al. 2010. Mice lacking Tbk1 activity exhibit immune cell infiltrates in multiple tissues and increased susceptibility to LPS-induced lethality. J. Leukoc. Biol. 88:1171–1180 10.1189/jlb.0210071 [DOI] [PubMed] [Google Scholar]
- Matsui K., Kumagai Y., Kato H., Sato S., Kawagoe T., Uematsu S., Takeuchi O., Akira S. 2006. Cutting edge: Role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J. Immunol. 177:5785–5789 [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Kikkawa S., Kohase M., Miyake K., Seya T. 2002. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem. Biophys. Res. Commun. 293:1364–1369 10.1016/S0006-291X(02)00380-7 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter S.M., Fitzgerald K.A., Rosains J., Rowe D.C., Golenbock D.T., Maniatis T. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA. 101:233–238 10.1073/pnas.2237236100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahira A.K., Shahangian A., Hwang S., Sun R., Cheng G. 2009. TANK-binding kinase-1 plays an important role during in vitro and in vivo type I IFN responses to DNA virus infections. J. Immunol. 182:2248–2257 10.4049/jimmunol.0802466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan G., Saha S.K., Guo B., He J.Q., Shahangian A., Zarnegar B., Perry A., Cheng G. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 439:208–211 10.1038/nature04374 [DOI] [PubMed] [Google Scholar]
- Ou Y.H., Torres M., Ram R., Formstecher E., Roland C., Cheng T., Brekken R., Wurz R., Tasker A., Polverino T., et al. 2011. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol. Cell. 41:458–470 10.1016/j.molcel.2011.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellat-Deceunynck C., Jego G., Harousseau J.L., Vié H., Bataille R. 1999. Isolation of human lymphocyte antigens class I-restricted cytotoxic T lymphocytes against autologous myeloma cells. Clin. Cancer Res. 5:705–709 [PubMed] [Google Scholar]
- Pérez de Diego R., Sancho-Shimizu V., Lorenzo L., Puel A., Plancoulaine S., Picard C., Herman M., Cardon A., Durandy A., Bustamante J., et al. 2010. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 33:400–411 10.1016/j.immuni.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry A.K., Chow E.K., Goodnough J.B., Yeh W.C., Cheng G. 2004. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J. Exp. Med. 199:1651–1658 10.1084/jem.20040528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R.T., Liao S.M., Maniatis T. 2000. IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex. Mol. Cell. 5:513–522 10.1016/S1097-2765(00)80445-1 [DOI] [PubMed] [Google Scholar]
- Picard C., Puel A., Bonnet M., Ku C.L., Bustamante J., Yang K., Soudais C., Dupuis S., Feinberg J., Fieschi C., et al. 2003. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 299:2076–2079 10.1126/science.1081902 [DOI] [PubMed] [Google Scholar]
- Picard C., von Bernuth H., Ghandil P., Chrabieh M., Levy O., Arkwright P.D., et al. 2010. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine. 89:403–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 314:997–1001 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- Poltorak A., He X., Smirnova I., Liu M.Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., et al. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 282:2085–2088 10.1126/science.282.5396.2085 [DOI] [PubMed] [Google Scholar]
- Pomerantz J.L., Baltimore D. 1999. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 18:6694–6704 10.1093/emboj/18.23.6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- Sancho-Shimizu V., Perez de Diego R., Jouanguy E., Zhang S.Y., Casanova J.L. 2011a. Inborn errors of anti-viral interferon immunity in humans. Curr Opin Virol. 1:487–496 10.1016/j.coviro.2011.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Shimizu V., Pérez de Diego R., Lorenzo L., Halwani R., Alangari A., Israelsson E., Fabrega S., Cardon A., Maluenda J., Tatematsu M., et al. 2011b. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 121:4889–4902 10.1172/JCI59259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Sugiyama M., Yamamoto M., Watanabe Y., Kawai T., Takeda K., Akira S. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304–4310 [DOI] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 472:481–485 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., tenOever B.R., Grandvaux N., Zhou G.P., Lin R., Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science. 300:1148–1151 10.1126/science.1081315 [DOI] [PubMed] [Google Scholar]
- Shimada T., Kawai T., Takeda K., Matsumoto M., Inoue J., Tatsumi Y., Kanamaru A., Akira S. 1999. IKK-i, a novel lipopolysaccharide-inducible kinase that is related to IkappaB kinases. Int. Immunol. 11:1357–1362 10.1093/intimm/11.8.1357 [DOI] [PubMed] [Google Scholar]
- Stanberry L.R., Jorgensen D.M., Nahmias A.J. 1997. Herpes simplex virus 1 and 2. Viral Infections of Humans: Epidemiology and Control. Fourth Edition. Evans A.S., Kaslow R.A., Plenum Publishing Corporation, New York: 419–454 [Google Scholar]
- Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., et al. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 448:501–505 10.1038/nature06013 [DOI] [PubMed] [Google Scholar]
- Tojima Y., Fujimoto A., Delhase M., Chen Y., Hatakeyama S., Nakayama K., Kaneko Y., Nimura Y., Motoyama N., Ikeda K., Karin M., Nakanishi M. NAK is an IkappaB kinase-activating kinase. Nature. 2000. 404:778–82 http://www.ncbi.nlm.nih.gov/pubmed/?term=%2010783893 [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S., et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11:997–1004 10.1038/ni.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpooten D., Ma Y., Hou S., Yan Z., He B. 2009. Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J. Biol. Chem. 284:1097–1105 10.1074/jbc.M805905200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bernuth H., Picard C., Jin Z., Pankla R., Xiao H., Ku C.L., Chrabieh M., Mustapha I.B., Ghandil P., Camcioglu Y., et al. 2008. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 321:691–696 10.1126/science.1158298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R.J. 2006. Herpes simplex encephalitis: adolescents and adults. Antiviral Res. 71:141–148 10.1016/j.antiviral.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Whitley R.J., Kimberlin D.W. 2005. Herpes simplex encephalitis: children and adolescents. Semin. Pediatr. Infect. Dis. 16:17–23 10.1053/j.spid.2004.09.007 [DOI] [PubMed] [Google Scholar]
- Whitley R.J., Alford C.A., Hirsch M.S., Schooley R.T., Luby J.P., Aoki F.Y., Hanley D., Nahmias A.J., Soong S.J. 1986. Vidarabine versus acyclovir therapy in herpes simplex encephalitis. N. Engl. J. Med. 314:144–149 10.1056/NEJM198601163140303 [DOI] [PubMed] [Google Scholar]
- Xu G., Lo Y.C., Li Q., Napolitano G., Wu X., Jiang X., Dreano M., Karin M., Wu H. 2011. Crystal structure of inhibitor of κB kinase β. Nature. 472:325–330 10.1038/nature09853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Puel A., Zhang S., Eidenschenk C., Ku C.L., Casrouge A., Picard C., von Bernuth H., Senechal B., Plancoulaine S., et al. 2005. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and -lambda Is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity. 23:465–478 10.1016/j.immuni.2005.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., et al. 2007. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 317:1522–1527 10.1126/science.1139522 [DOI] [PubMed] [Google Scholar]
- Zhang S.Y., Boisson-Dupuis S., Chapgier A., Yang K., Bustamante J., Puel A., Picard C., Abel L., Jouanguy E., Casanova J.L. 2008. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol. Rev. 226:29–40 10.1111/j.1600-065X.2008.00698.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data for the microarray analyses performed in this study are available from the public repository of GEO DataSets (accession no. GSE38652). Networks were resolved with MetaCore version 6.10 (GeneGo).