Abstract
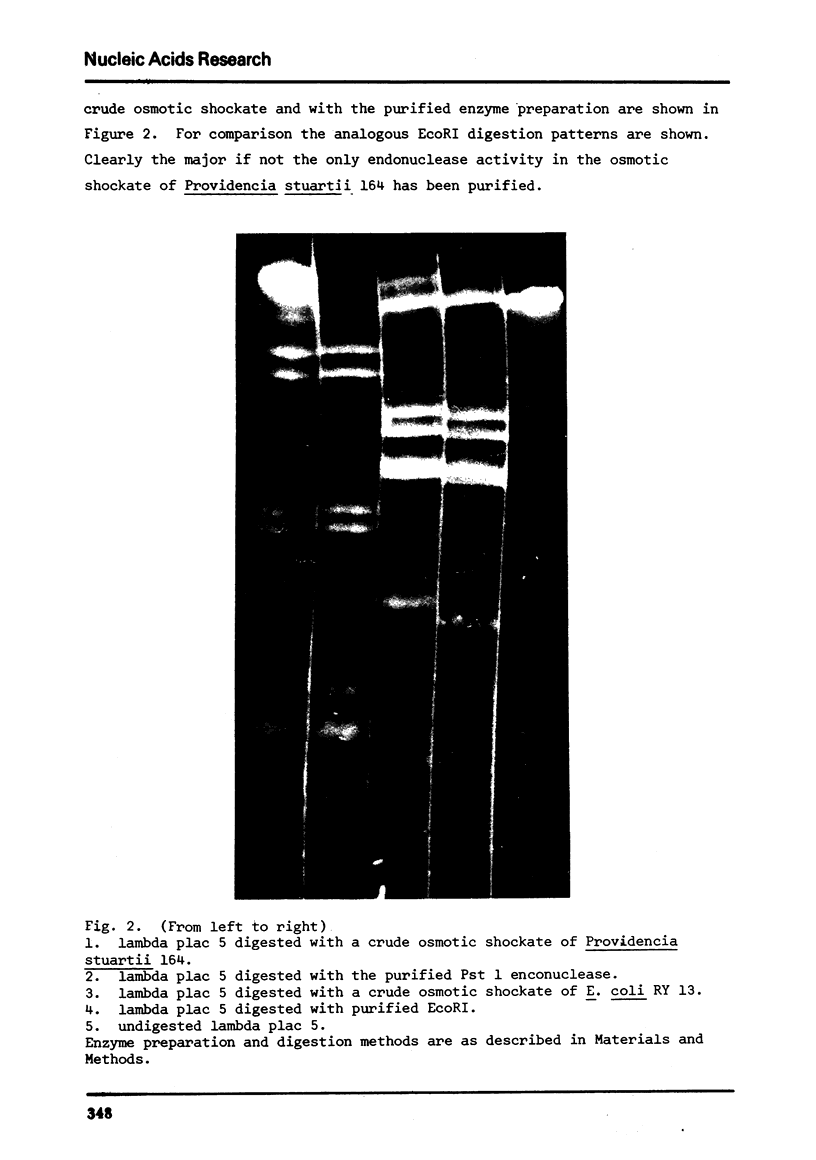
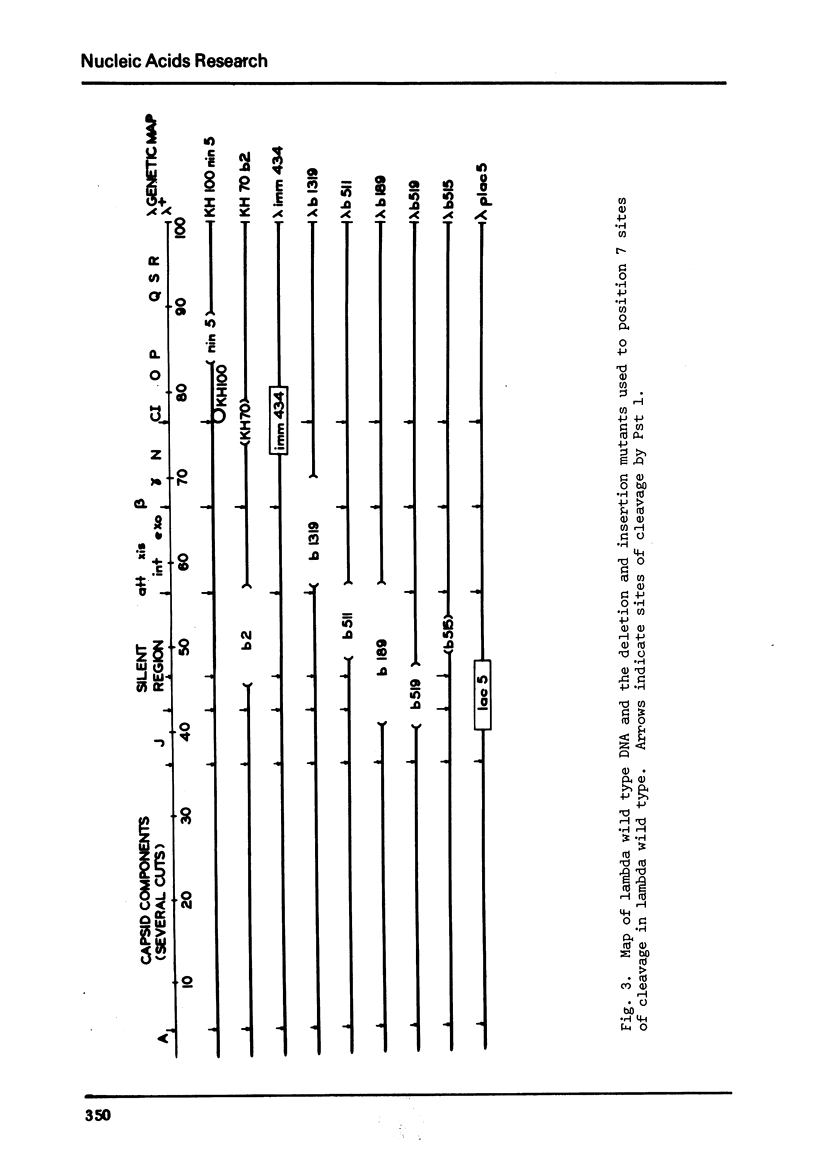
We describe the isolation of a new class 2 restriction endonuclease from Providencia stuartii 164. Using the procedure of osmotic shock treatment, we have partially purified this enzyme (Pst 1) and have begun preliminary work to characterize its specificity and requirements. Pst 1 requires Mg++ as the only cofactor and produces more than 18 cleavages in wild type lambda. We have determined the location of 7 of these cleavages by the use of deletion and insertion mutants of lambda.
Full text
PDF





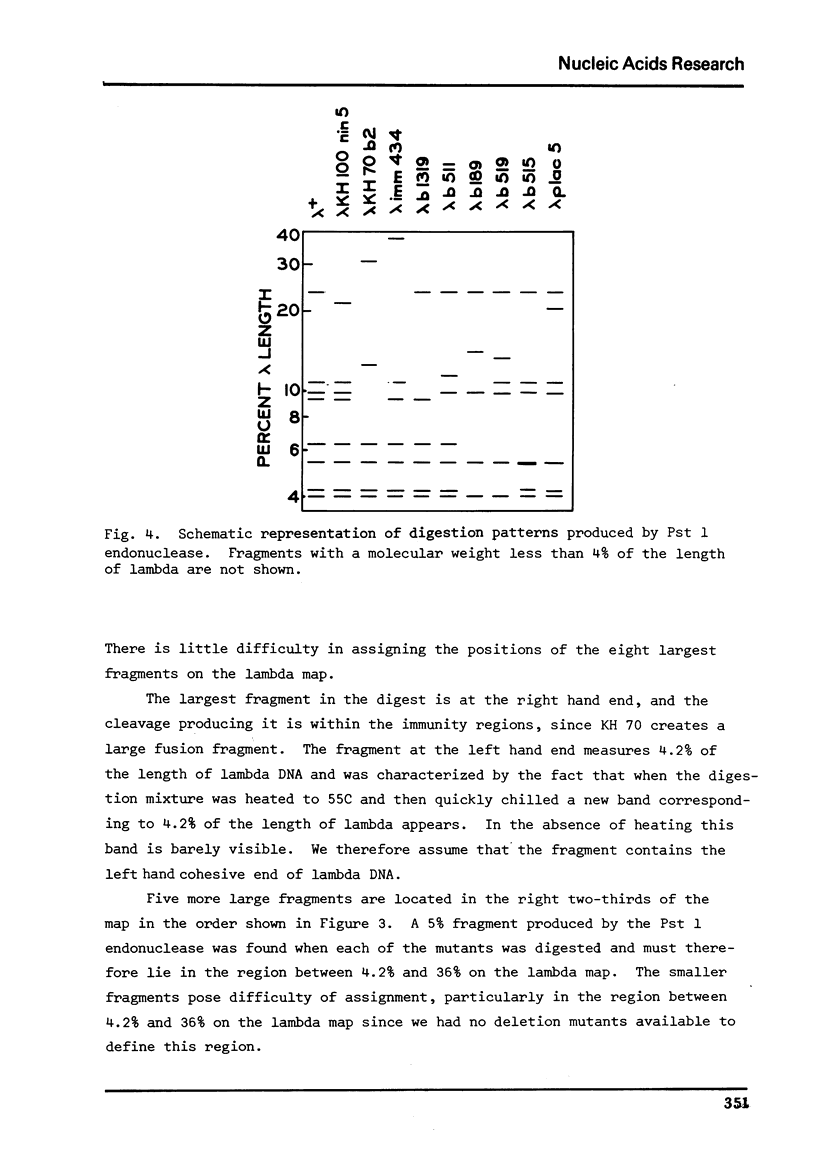


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blattner F. R., Fiandt M., Hass K. K., Twose P. A., Szybalski W. Deletions and insertions in the immunity region of coliphage lambda: revised measurement of the promoter-startpoint distance. Virology. 1974 Dec;62(2):458–471. doi: 10.1016/0042-6822(74)90407-3. [DOI] [PubMed] [Google Scholar]
- Chevereau M., Daniels P. J., Davies J., LeGoffic F. Aminoglycoside resistance in bacteria medicated by gentamicin acetyltransferase II, an enzyme modifying the 2'-amino group of aminoglycoside antibiotics. Biochemistry. 1974 Jan 29;13(3):598–603. doi: 10.1021/bi00700a030. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick J. L., Smith A. J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968 Jul;126(1):155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D., Weil J. Recombination-deficient deletions in bacteriophage lambda and their interaction with chi mutations. Genetics. 1975 Feb;79(2):143–174. doi: 10.1093/genetics/79.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen K., Shapiro J. A., Beckwith J. R. Transposition of the lac region to the gal region of the Escherichia coli chromosome: isolation of lambda-lac transducing bacteriophages. J Bacteriol. 1971 Oct;108(1):5–9. doi: 10.1128/jb.108.1.5-9.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow J. F., Cohen S. N., Chang A. C., Boyer H. W., Goodman H. M., Helling R. B. Replication and transcription of eukaryotic DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1974 May;71(5):1743–1747. doi: 10.1073/pnas.71.5.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965 Sep;240(9):3685–3692. [PubMed] [Google Scholar]
- Parkinson J. S., Huskey R. J. Deletion mutants of bacteriophage lambda. I. Isolation and initial characterization. J Mol Biol. 1971 Mar 14;56(2):369–384. doi: 10.1016/0022-2836(71)90471-2. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R., Hughes S. G., Broda P. Plasmid identification using specific endonucleases. Mol Gen Genet. 1974;133(2):141–149. doi: 10.1007/BF00264835. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H., Carroll D., Brown D. D., Deutch A., Higashinakagawa T., Dawid I. B. Amplified ribosomal DNA from Xenopus laevis has heterogeneous spacer lengths. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2823–2827. doi: 10.1073/pnas.71.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]