Summary
Dscam1 potentially encodes 19,008 ectodomains of a cell recognition molecule of the immunoglobulin (Ig) superfamily through alternative splicing. Each ectodomain, comprising a unique combination of three variable (Ig) domains, exhibits isoform-specific homophilic binding in vitro. Although we have proposed that the ability of Dscam1 isoforms to distinguish between one another is crucial for neural circuit assembly, via a process called self-avoidance, whether recognition specificity is essential in vivo has not been addressed. Here we tackle this issue by assessing the function of Dscam1 isoforms with altered binding specificities. We generated pairs of chimeric isoforms that bind to each other (heterophilic) but not to themselves (homophilic). These isoforms failed to support self-avoidance or did so poorly. By contrast, co-expression of complementary isoforms within the same neuron restored self-avoidance. These data establish that recognition between Dscam1 isoforms on neurites of the same cell provides the molecular basis for self-avoidance.
Introduction
Brains comprise diverse neuronal cell types interconnected through precise patterns of synaptic connections to form functional neural networks. How different neurons distinguish between one another during circuit assembly is poorly understood. Several large families of homologous cell recognition proteins arising through alternative splicing or gene duplication have been shown to play important roles in neural circuit formation and function (Shapiro et al., 2007; Sudhof, 2008; Zipursky and Sanes, 2010). While different isoforms of several of these protein families, clustered protocadherins and neurexins in mammals and Dscam1 proteins in Drosophila, exhibit isoform-specific binding properties in vitro (Boucard et al., 2005; Schreiner and Weiner, 2010; Wojtowicz et al., 2007), whether this specificity is required in vivo remains unknown. Here we address whether the exquisite binding specificity of Dscam1 proteins is essential for their function in neural circuit assembly.
The Drosophila Dscam1 gene encodes many protein isoforms of the Ig superfamily through alternative splicing (Schmucker et al., 2000). This includes 19,008 potential ectodomains tethered to the membrane by two alternative transmembrane segments (Schmucker et al., 2000). Each isoform is defined by a unique combination of three variable Ig domains, numbered from the N-terminus as Ig2, Ig3, and Ig7 (Figure 1A). Biochemical studies showed that isoforms bind in trans to an identical isoform, but only weakly or not at all to different isoforms (Wojtowicz et al., 2004; Wojtowicz et al., 2007). These data together with structural studies led us to propose that modular matching at all three variable domains (Ig2:Ig2, Ig3:Ig3 and Ig7:Ig7) gives rise to exquisite homophilic binding specificity (Meijers et al., 2007; Sawaya et al., 2008; Wojtowicz et al., 2004; Wojtowicz et al., 2007).
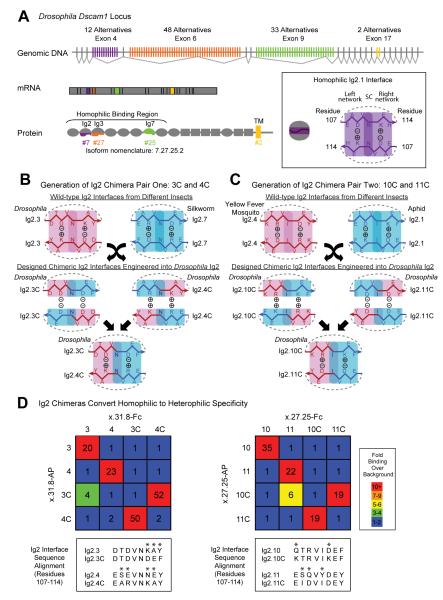
Figure 1. Design of chimeric Ig2 domains with altered binding specificity.
(A) Dscam1 encodes a large family of isoform-specific homophilic binding proteins. The Dscam1 gene contains 4 blocks of alternative exons, which encode the first halves of Ig2 (purple) and Ig3 (orange), the entire Ig7 (green), and the transmembrane domain (yellow). Only one alternative from each block is included in a mature mRNA. Ovals, Ig domains; grey rectangles, fibronectin type III domains (FNIII); yellow rectangle, transmembrane domain (TM). The inset shows schematic representations of the homophilic Ig2 interface using Ig2.1 as an example: The Ig2-Ig2 interface includes residues 107-114 (Meijers et al., 2007; Sawaya et al., 2008). All the amino acids within this segment for Ig2.1 are shown (i.e. both surface and inward-directed residues) on both sides of the symmetry center (SC). (B) Generation of chimeric Ig2.3C and Ig2.4C interfaces used in this study. Wild-type Drosophila (Drosophila melanogaster) Ig2.3 and silkworm (Bombyx mori) Ig2.7 interface sequences (residues 107-114) were chosen as “donor” for generating chimeras. Swapping the two halves of the interface segments gives rise to chimeric interface sequences that we predicted would not bind homophilically due to the lack of self-complementarity. Instead, chimeric Ig2 domains were predicted to bind heterophilically. The + and − signs refer to the charges at residues 109 and 112. Construction of these interfaces was done by modifying sequences within Drosophila Dscam1 Ig2.3 and Ig2.4 domains (to generate Ig2.3C and Ig2.4C, respectively, where “C” denotes chimera). (C) Design and interface sequences of Ig2.10C and Ig2.11C were analogous to Figure 1B. Yellow fever mosquito, Aedes aegypti; aphid, Acyrthosiphon pisum. (D) Binding properties of Dscam1 ectodomains containing wild-type and chimeric Ig2 domains represented as fold binding over background using an ELISA-based assay. Wild-type and chimeric Ig2.3 and Ig2.4 were tested in the context of Ig3.31 and Ig7.8. Wild-type and chimeric Ig2.10 and Ig2.11 were tested in the context of Ig3.27 and Ig7.25.
Genetic studies support the notion that Dscam1-mediated homophilic recognition plays a key role in neural circuit assembly by providing the molecular basis for self-avoidance (Hattori et al., 2009; Hattori et al., 2007; Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007; Wang et al., 2002; Zhan et al., 2004; Zhu et al., 2006). Self-avoidance refers to the tendency of neurites of the same cell to avoid each other (Kramer and Kuwada, 1983). Analysis of mutants encoding reduced numbers of isoforms established that thousands of isoforms are required for self-avoidance (Hattori et al., 2009; Hattori et al., 2007). Expression data from several neuronal cell types are consistent with each cell expressing a unique combination of Dscam1 isoforms, thereby endowing each neuron with a distinct cell-surface identity (Neves et al., 2004; Zhan et al., 2004). Based on these studies, we proposed that self-neurites express the same isoforms, bind to each other and are subsequently repelled. By contrast, as neurites of different neurons express different isoforms, Dscam1 does not mediate interactions between them (Hattori et al., 2008).
While isoform-specific homophilic recognition is the linchpin of models for Dscam1 function, whether this biochemical property is required in vivo is unknown. In this paper, we use a combined biochemical and genetic approach to address this issue.
Results and Discussion
Structure-based design of pairs of chimeric isoforms exhibiting interallelic complementation
To directly address the importance of binding specificity in vivo, we sought to generate pairs of Dscam1 isoforms that exhibit interallelic complementation; each protein would not bind to itself but would bind heterophilically to another isoform. If isoform-specific recognition were critical for self-avoidance, then expression of each homophilic binding deficient isoform would not rescue the mutant phenotype while expression of the two complementary isoforms in the same cell would. This is analogous to forward genetic approaches in bacteria to identify proteins interacting in vivo through the isolation of allele-specific extragenic suppressor mutations (Hartman and Roth, 1973).
To generate pairs of isoforms with altered binding specificities, we focused on the Ig2 interface, as it is the most extensively characterized of the three variable domain interfaces (Meijers et al., 2007; Sawaya et al., 2008; Wojtowicz et al., 2007). Each specificity interface of the Ig2 domains comprises a different 8 amino acid β-strand segment (positions 107–114). These unique sequences align in a two-fold symmetric fashion with a symmetry center and two identical complementary networks that fit together by shape and charge complementarity (Figure 1A).
As a first step towards generating pairs of novel isoforms in which specificity was converted from homophilic to heterophilic, we compared the Ig2 interface segments from Dscam1, and Dscam paralogs 2-4 in various insects and vertebrate paralogs DSCAM and DSCAML1 (i.e. 89 interface sequences from 39 species) to identify pairs of interface segments with the following properties: 1. They share the same symmetry center (position 111); 2. Each contains amino acids of opposite charge at interface residues flanking the symmetry center (i.e. positions 109 and 112); and 3. The charges at positions 109 and 112 in one interface are the opposite of those found at the other interface (Figures 1B and 1C). By swapping parts of interfaces with these properties, we reasoned that we could create chimeric interface segments that would disrupt self-pairing, while simultaneously directing pairing to a complementary yet different interface chimera.
One example of such an interface chimera is shown in Figure 1B. A Drosophila Ig2 and silkworm Ig2 interface share an asparagine at position 111, the Drosophila sequence has an aspartic acid at position 109 and a lysine at 112, and the silkworm sequence has an arginine at position 109 and an aspartic acid at 112. Two unique half-interface segments were then created by flanking the shared symmetry center with amino acids 108–110 and 112-114 from the Drosophila and silkworm sequences, respectively. We predicted that the resulting chimeras would not support self-binding due to charge incompatibility (Figure 1B) but that the two chimeras would bind to each other, as the contacts on each half interface were seen in a wild-type interface. Two pairs of complementary chimeric interface segments (indicated Ig2.3C/Ig2.4C and Ig2.10C/Ig2.11C) were introduced through mutagenesis of Drosophila Ig2 domains with the most similar interfaces (Figures 1B and 1C; also see sequence alignment in Figure 1D).
Chimeric interface segments convert homophilic to heterophilic binding specificity
To test the binding specificity of each altered variable domain complementary pairs of Ig2 interfaces was inserted into ectodomains comprising the same Ig3 and Ig7 domains to generate pairs of closely related chimeric isoforms. We first assessed interactions using the ELISA-based binding assay where Dscam1 protein ectodomains were clustered in cis in a limited fashion (presumably tetramers) (Wojtowicz et al., 2007). The binding of two ectodomains each comprising the N-terminal ten domains was tested as previously described (Wojtowicz et al., 2007). Wild-type isoforms exhibited strong homophilic interaction, but homophilic binding of each chimera was reduced to background levels (Figure 1D). Importantly, heterophilic binding of each chimera pair was observed at a similar level to that observed with homophilic binding of the control wild-type isoforms.
To gain a more quantitative measure of binding specificity we performed analytical ultracentrifugation (AUC). Binding of chimeric isoforms was assessed using fragments of the homophilic binding region of Dscam1 containing the N-terminal 8 Ig domains. From previous studies using different assays, these fragments, which encompass the 3 variable domains, exhibited binding properties indistinguishable from full-length ectodomains (Wojtowicz et al., 2004). Sedimentation equilibrium AUC experiments showed that the wild-type isoforms homodimerized strongly with KD values of 1 μM for Dscam110.27.25 (shorthand nomenclature for the isoform comprising Ig2.10, Ig3.27 and Ig7.25) and 2.1 μM for Dscam13.31.8 (Table 1). By contrast homodimers were not observed with isoforms containing chimeric Ig2 domains, as indicated by the results of both sedimentation equilibrium (Table 1) and sedimentation velocity (Figure S1) AUC experiments. We then measured heterophilic binding between complementary pairs of chimeras in both velocity and equilibrium AUC experiments. As predicted, each pair of complementary chimeras bound to each other with affinities similar to wild-type homodimers; the Ig2.3C containing isoform bound to Ig2.4C containing isoform with a KD of 1 μM and the Ig2.10C containing isoform bound to Ig2.11C containing isoform with a KD of 4.6 μM. These data argue that the loss of homophilic binding is not due to more global changes in protein conformation, but rather to an alteration in binding specificity. Our results indicate that matching Ig3 and Ig7 is not sufficient to form dimers within the detection limit for equilibrium AUC (i.e. <500 μM). These quantitative analytical studies support the view, based on our previous ELISA-based assays, that the vast majority of Dscam1 isoforms show little binding to isoforms with a markedly different interface at only a single variable domain.
Table 1.
Summary of AUC sedimentation equilibrium experiments
| Wild-type Dscam1 Isoforms | |
| Homodimeric Binding | KD (μM) |
| 3.31.8 | 2.12 ± 0.87 |
| 10.27.25 | 1.00 ± 0.42 |
| Chimeric Dscam1 Isoforms | |
| Homodimeric Binding | |
| 3C.31.8 | monomer |
| 4C.31.8 | monomer |
| 10C.27.25 | monomer |
| 11C.27.25 | monomer |
| Heterodimeric Binding | |
| 3C.31.8 mixed with 4C.31.8 | 0.99 ± 0.08 |
| 10C.31.8 mixed with 11C.31.8 | 4.60 ± 0.11 |
In summary, isoforms containing chimeric Ig2 domains exhibit altered recognition specificities with a profound loss of homophilic binding accompanied by a gain of heterophilic specificity between isoforms containing complementary chimeric Ig2 domains. Some degree of homophilic binding of chimeric isoforms was observed when isoforms were over-expressed in S2 cells (Figure S2). Whether this reflects a limited ability for interactions when proteins are presented on the cell surface, is a result of overexpression, or both is unknown.
Homophilic binding of Dscam1 isoforms is necessary for self-avoidance
The chimeric isoforms with altered binding specificities provided us with a unique opportunity to definitively test the notion that specific recognition between Dscam1 isoforms on sister neurites is necessary to promote self-avoidance. As a first step towards addressing this issue we sought to knock-in the chimeric isoforms into the endogenous locus; expression from the endogenous locus would ensure that the gene is expressed at the same level and spatiotemporal pattern as the wild-type gene. To do this, we combined knock-in technology (Figure S3A and Experimental Procedures) and intragenic mitotic recombination (iMARCM) (Figure S3B) (Hattori et al., 2007) to drive expression of a chimeric isoform from the endogenous locus in single cells.
Two chimeric isoforms, Dscam110C.27.25 and Dscam13C.31.8, were knocked into the endogenous locus. These alleles exhibited similar properties and, therefore, we refer to them collectively as Dscam1single chimera. For each chimera, the function of a control knock-in allele encoding the corresponding wild-type Ig2 domain and the same Ig3 and Ig7 domains was assessed. We refer to these alleles likewise as Dscam1single. Knock-in alleles were confirmed by genomic sequencing (Experimental Procedures). These alleles were generated in a two-step process. In the first step, a single cDNA fragment encoding one ectodomain was introduced into the locus replacing all of Dscam1 ectodomain diversity. This gene segment was maintained in the germline as an incomplete allele (Figure S3A). In a second step, termed iMARCM, intragenic recombination was induced in somatic cells to generate a fully resolved single isoform-encoding genomic allele in a single cell, in which this allele provided the only source of Dscam1 expression (Figure S3B). These single cells, selectively labeled with GFP, were surrounded by unlabeled neighboring cells containing the wild-type allele expressing the full complement of Dscam1 diversity. Fully resolved germline versions of both chimeric alleles were difficult to obtain. We generated a full-length germ-line version of one chimeric allele, however, encoding the Ig2.10C-containing isoform to assess protein expression (i.e. Dscam110C.27.25) (We were unable to generate a fully resolved germline allele for the other chimera, Dscam13C.31.8, for unknown reasons). Dscam110C.27.25 protein was expressed at the same level (Figure S4A) and in a similar distribution in the embryonic nervous system (Figure S4B) to both the corresponding control knock in with a single wild-type isoform and the wild-type endogenous locus expressing full Dscam1 diversity. Both chimeric alleles were analyzed using iMARCM to assess their ability to rescue self-avoidance in axons and dendrites.
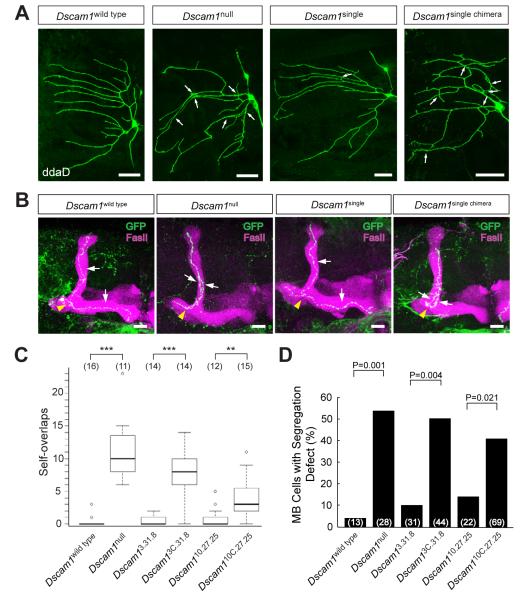
Dscam1 mediates self-avoidance between dendrites of dendritic arborization (da) neurons (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). There are four classes of da neurons, each identifiable by its cell body position and dendritic morphology (Grueber et al., 2002). To assess the role of homophilic binding in dendrite self-avoidance, we used iMARCM to generate single da sensory neurons expressing only one Dscam1 isoform surrounded by wild-type cells. As previously described, sister dendrites (i.e. dendrites from the same cell) from a Dscam1null da neuron overlapped extensively (Matthews et al., 2007) (Figures 2A and 2C). Dscam1single rescued the self-avoidance defects in class I neurons. By contrast, the ability of Dscam1single chimera to rescue the phenotype was compromised (Figures 2A and 2C). Similar results were obtained using iMARCM to assess the function of chimeric isoforms in class III neurons (Figure S5). In class I neurons, one of the Dscam1single chimera alleles, Dscam13C.31.8, did not rescue the phenotype significantly, while the other, Dscam110C.27.25, showed considerable rescue (see below).
Figure 2. Dscam1 homophilic recognition specificity is required for self-avoidance.
(A) The requirement for Dscam1 homophilic binding in dendrite self-avoidance. Labeled ddaD class I da sensory neurons for Dscam1wild type, Dscam1single, and Dscam1single chimera were generated through intragenic MARCM (iMARCM), and Dscam1null labeled neurons were produced through conventional MARCM. Clones were labeled with mCD8GFP. Arrows indicate crossing and fasciculation between self-dendrites. Cells were visualized in third instar larvae. Scale bar = 20μm. (B) The requirement for Dscam1 homophilic binding in axon self-avoidance. MB lobes and single labeled MB neurons, generated by iMARCM or MARCM, for Dscam1wild type, Dscam1null, Dscam1single, and Dscam1single chimera alleles were visualized by staining with anti-FasII (magenta) and anti-GFP (green) antibodies. Yellow arrowheads indicate the branch point. White arrows indicate sister branches. Scale bar = 20μm. (C) Quantification of overlaps between self-dendrites in (A). Data in boxplot is represented as median (dark line), 25%-75% quantiles (white box), and data within 1.5× quantile range (dashed bar). Small open circles indicate data points outside of this range. Numbers in parenthesis denote the number of class I (ddaD) neurons analyzed for each genotype. Statistical analysis was performed in R (R Development Team, 2006). **=p<0.01, and ***=p<0.001. See Figure S5 for dendrite self-avoidance in class III da neurons. (D) Quantification of MB neuron axon segregation defects in (B). Numbers in parenthesis represent the number of single labeled MB neurons examined for each genotype. Statistics were performed using a two-tailed Fisher’s exact test.
The ability of the chimeric isoforms to rescue self-avoidance in axons was assessed in mushroom body (MB) neurons (Wang et al., 2002; Zhan et al., 2004). The MB is a central brain structure containing some 2,500 neurons. The axons of most MB neurons bifurcate with one axon branch extending dorsally and the other medially. In Dscam1null single cells in an otherwise wild-type background, the two branches frequently failed to segregate from each other and projected into the same lobe (Figures 2B and 2D). While a single arbitrarily chosen isoform rescued the mutant phenotype in iMARCM (Hattori et al., 2007), both Dscam1single chimera alleles showed little rescue activity (Figures 2B and 2D).
In summary, the ability of one chimeric isoform, Dscam13C.31.8, to rescue self-avoidance in either dendrites or axons was markedly disrupted, consistent with the biochemical properties of this isoform in vitro. While the ability of the second isoform, Dscam110C.27.25, to rescue MB axon self-avoidance and dendrite self-avoidance in class III da neurons (Figure S5) was markedly compromised, this isoform exhibited considerable rescue activity in dendrites of class I da neurons (Figures 2A and 2C). Whereas homophilic binding was not detected for this isoform in either AUC or the ELISA-based assay, substantial binding was observed in the cell aggregation assay (Figure S2). This finding raises the possibility that within the context of a cell membrane Dscam1 isoforms with the same Ig3 and Ig7 domains, but differing at Ig2, may in some cell types be sufficient to mediate recognition between sister neurites and, as a consequence, repulsion between them.
Heterophilic binding between complementary chimeras in the same cell restores self-avoidance
Presumably the chimeric Dscam1 isoforms fail to rescue the Dscam1null phenotypes as these isoforms were unable to bind to each other and thus to elicit a repulsive response. Alternatively, the chimeras may fail to rescue for other reasons unrelated to altered binding specificity. To definitively test whether binding between isoforms on opposing neurites of the same cell is essential for self-avoidance, we sought to assess whether cells expressing complementary chimeras reverse the effects of the branching defects seen in Dscam1null mutants.
To test for complementation, we used conventional MARCM analysis to generate single Dscam1 mutant labeled cells co-expressing cDNAs encoding complementary chimeric isoforms. These experiments were restricted to analyzing axon self-avoidance in MB neurons, as da neuron dendrite self-avoidance is only partially rescued by targeted expression of cDNAs encoding wild-type isoforms using MARCM (W.B. Grueber, personal communication). Presumably, this reflects the perdurance of GAL80, which prevents adequate levels or appropriate timing of expression of Dscam1 from cDNA transgenes in these cells to support self-avoidance. Note, however, that we were able to assess the requirement for homophilic binding in da neurons from the knock-in alleles using iMARCM reported in the previous section, as expression from the endogenous locus does not rely on GAL4. Unfortunately, iMARCM does not facilitate expression of two chimeric isoforms encoded at the endogenous locus in the same cell. Thus, to test for cell autonomous rescue of self-avoidance by complementary isoforms, we used MARCM analysis in MB neurons.
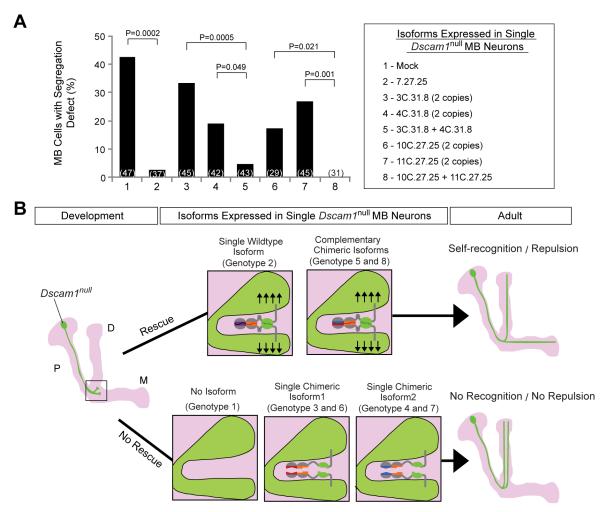
cDNAs encoding Dscam1 isoforms, both wild type and chimeras, were placed under the control of the UAS enhancer and were inserted into a defined genomic position through phiC31 site-specific recombination (Groth et al., 2004). Different isoforms were expressed at similar levels as assessed on western blots of extracts prepared from embryos in which UAS expression was driven by a pan-neuronal GAL4 transgene (data not shown). Consistent with iMARCM experiments (Figures 2 and S5), expression of two copies of any of the four chimeras only provided weak self-avoidance activity in Dscam1null MB neurons (Figure 3A). Expression of either pair of complementary isoforms (i.e. a single copy of each UAS transgene inserted into the same site on two homologous chromosomes), however, rescued the branch segregation defect to a similar extent to the wild-type transgenes (Figure 3A). Thus, Dscam1 acts in a cell autonomous fashion through direct binding of complementary protein domains on sister neurites of the same cell (Figure 3B). These data establish that binding between matching isoforms is essential for Dscam1 function in vivo.
Figure 3. Complementary chimeras rescue the self-avoidance defects in Dscam1null MB neurons.
(A) Single Dscam1null MB axon branch segregation phenotypes with different cDNA rescue constructs. Quantification is shown as a bar graph (two-tailed Fisher’s exact test). The number of single labeled MB neurons examined for each genotype is indicated in parenthesis. All UAS-encoded isoforms contained a TM2 domain. (B) Interpretation of rescue phenotypes in (A). The left panel shows schematic of a Dscam1null (green) MB neuron during development. Light magenta outlines the entire MB comprising the peduncle (P) region, dorsal (D) and medial (M) lobes. The box highlights a proposed axon bifurcation occurring during development. Middle panels illustrate the isoform expression and the resulting repulsive signaling at the branch point. Right panels show schematics of the outcome of adult Dscam1null MB neurons after rescue.
Ectopic expression of complementary isoforms induces repulsion between processes of different cells
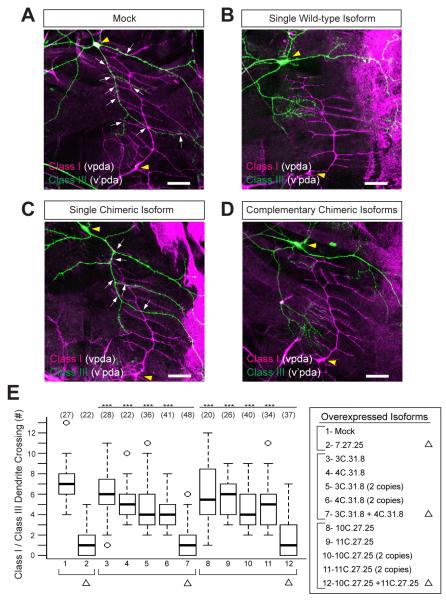
If Dscam1 isoform-specific recognition does, indeed, play an instructive role in self recognition, then expressing two different, yet complementary, isoforms on neurites of different cells should also elicit a repulsive response between them. To test this, we expressed chimeric isoforms alone or in combination with a complementary isoform in da neurons and explored the dendritic arbor patterns elaborated by class III (v’pda) neurons relative to the dendrites of class I (vpda) neurons (Figure 4). In wild-type animals, the class I dendritic arbor pattern is established first (Soba et al., 2007). Subsequently, the class III neurons elaborate dendrites, which overlap with the dendrites of class I neurons. (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007) (Figures 4A and 4E). Expression of a wild-type Dscam1 isoform in both cells induced repulsion and as a result, there were few overlaps between their dendrites (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007) (Figures 4B and 4E). Only weak ectopic repulsion was seen in response to expression of each Dscam1 chimera (Figures 4C and 4E). While we speculated previously that weaker binding between isoforms might promote an adhesive interaction (Wojtowicz et al., 2004; Zhan et al., 2004), the modest decrease in overlap seen in response to ectopic expression of each chimera on its own suggests that even weak binding between isoforms promotes repulsion, albeit at an attenuated level. By contrast, co-expression of complementary chimeras induced ectopic repulsion between the dendrites of different cells similar to wild-type isoforms (Figures 4D and 4E). Thus, selective recognition between isoforms is sufficient to induce ectopic repulsion between processes of different cells.
Figure 4. Heterophilic-specific binding induces ectopic repulsion between dendrites of class I and class III da neurons.
Representative images of class I (vpda, magenta) and class III (v’pda, green) da neurons with an empty UAS vector (mock) inserted into the same genomic site as constructs tested in panels B-D (A), a single wild-type isoform (B), a single chimeric isoform (C), or two complementary chimeric isoforms (D). Neurons were differentially labeled using a FLP-out cassette driven by a pan-da neuron GAL4 driver, and visualized by staining with anti-CD2 (magenta) and anti-GFP (green) antibodies. White arrows indicate the crossing between class I and class III da neuron dendrites. Yellow arrowheads indicate the cell body of vpda and v’pda. Scale bar = 20μm. (E) Quantification of crossing between vpda and v’pda neuron dendrites. Statistical analysis was performed in R (R Development Team, 2006). All UAS-encoded isoforms contained a TM2 domain. ***=p<0.001.
Concluding Remarks
Dscam1 is among a small group of very large families of cell recognition molecules (e.g. neurexins and clustered protocadherins) with diverse binding specificities, which are important for the assembly and function of neural circuits. To critically assess whether it is the isoform specificity of these interactions that is crucial for their function in vivo, it will be necessary to selectively manipulate binding specificity between isoforms. As we describe here, the use of structural and biochemical data to generate pairs of complementary isoforms with altered specificities provides an effective way to directly address the biological relevance of this recognition.
Experimental Procedures
Fly stocks
Chimeric knock-in alleles were generated and maintained as previously described (Hattori et al., 2007). The stocks used in misexpression experiments in da sensory neurons are UAS-Dscam1 stocks and hsFLP; Gal4109(2)80; UAS>CD2>mCD8-GFP. The stocks used in MARCM were hsFLP, elav-Gal4, UAS-mCD8-GFP; FRT42D, tub-Gal80/CyO, and in iMARCM were hsFLP, elav-Gal4, UAS-mCD8-GFP; Dscam1FRT, tub-Gal80/CyO.
Biochemical characterization
Mutations were introduced into the corresponding wild-type isoforms with the QuickChange Site-Directed Mutagenesis Kit (Stratagene). The ELISA-based binding assay was performed as previously described (Wojtowicz et al., 2007). Cell aggregation assays were performed as previously described (Matthews et al., 2007). Immunoblots were performed using mAb anti-Dscam1 (11G4) at 1:2000 dilution.
Analytical ultracentrifugation (AUC)
Sedimentation equilibrium measurements
AUC equilibrium experiments were performed at 25°C, using a Beckman XL-A/I ultracentrifuge equipped with a Ti60An rotor. Data were collected using UV absorbance at 280 nm. Samples of each protein, at concentrations of 0.7, 0.46 and 0.24 mg/ml, were dialyzed in a PBS buffer, pH 7.4 for 16 hours at 4°C, and 120 μl aliquots of each concentration were loaded into six-channel equilibrium cells with parallel sides and sapphire windows. Samples were spun at 8000 rpm for 20 hours, after which four scans were collected at a rate of 1 per hour. The rotor speed was then increased to 10,000 rpm for 10 hours, after which four additional scans were collected at the same rate. The speed was further increased to 12,000 rpm for another 10 hours and four more scans were recorded under the same conditions. During the last step, the rotor speed was increased to 14,000 rpm for four more scans, resulting in a total of 16 scans for each concentration and 48 scans per protein. Each experiment was reproduced at least twice. The data were processed and analyzed using HeteroAnalysis 1.1.44 software (http://www.biotech.uconn.edu/auf) and buffer density and protein v-bar values were calculated using the SednTerp (Alliance Protein Laboratories) software. The data for all concentrations and speeds were globally fit using nonlinear regression to either a monomer-dimer equilibrium model (A+A for homodimeric and A+B for heterodimeric interactions) or an ideal monomer model.
Sedimentation velocity measurements
AUC velocity measurements were performed in a Beckman XL-A/I ultracentrifuge, using a Ti60An rotor. Interference at 660 nm was used for detection. Protein samples at 1mg/ml were loaded into 12 mm two-channel tapered cells with sapphire windows and the rotor containing the samples was subsequently spun at 40,000 rpm at 25°C for four hours. A minimum of 300 scans were recorded at two-minute intervals. The velocity data was processed using the SedFit ver. 12.1b software (https://sedfitsedphat.nibib.nih.gov).
Transgenes
A Dscam1 cDNA encoding the full-length isoform 7.27.25.2 with 2× flag tags inserted in frame into exon 18 was isolated as a 6kb XbaI restriction fragment blunt end ligated into the XbaI site of the Drosophila transgene vector pUASTB (Groth et al., 2004). Expression constructs encoding other Dscam1 cDNAs were subsequently created by replacing the 2kb Acc65I-SapI fragment containing the 7.27.25 sequence, with a 2kb Acc65I-SapI fragment encoding other wild-type or chimeric isoform ectodomain sequences. Transgenes were generated through a phiC31 recombinase-mediated system into the attP2 site on the 3rd chromosome (Groth et al., 2004).
Homologous Recombination
Dscam1 homologous recombinant alleles were generated through a gene targeting strategy essentially the same as previously described (Hattori et al., 2007). The intended knock-in gene structure of Dscam110C.27.25 was verified by sequencing 14kb from the Dscam1 locus. Flies carrying the complete resolved Dscam13C.31.8 allele did not survive to be established as stocks. Therefore, 5′ intermediate alleles of Dscam13C.31.8 over CyO were maintained as stocks. The genomic organization for Dscam13C.31.8 was verified in its 5′ intermediate allele.
Mosaic analysis
For Dscam1 misexpression experiments in da sensory neurons, UAS-Dscam1 stocks were crossed to hsFLP; Gal4109(2)80; UAS>CD2>mCD8-GFP. The progeny were heat shocked to achieve differential labeling in different neurons as described previously (Matthews et al., 2007). For iMARCM, clones were generated by using heat-shock-mediated expression of FLP recombinase to trigger mitotic recombination between FRT sites on the modified Dscam1 locus. iMARCM analysis in MB neurons was performed as previously described (Hattori et al., 2007). iMARCM analysis in da sensory neurons was performed using essentially the same developmental stage and heat-shock procedure as previously described for MARCM analysis (Matthews et al., 2007). Dscam1null clones in MB and da neurons were generated as previously described (Zhan et al., 2004; Matthews et al., 2007) .
Immunohistochemistry
The following antibodies were used for immunohistochemistry: mAb anti-rat CD2 (1:100, Serotec), mAb anti-FasII (1D4) (1:10), mAb anti-Dscam1 (11G4, 1:500), rabbit anti-GFP (1:1000, Molecular Probes), Cy5-conjugated goat anti-HRP (1:200, Jackson ImmunoResearch laboratories), Alexa488-conjugated goat anti-mouse (1:200, Molecular Probes), and Alexa568-conjugated goat anti-rabbit (1:200, Molecular Probes). For mushroom body imaging, late pupal or adult brains were dissected and immunostained as previously described (Zhan et al., 2004). For da sensory neuron imaging, third instar larvae were dissected and immunostained as previously described (Grueber et al., 2002). Stage 16 embryos were fixed and immunostained as previously described (Kidd et al., 1998).
Image acquisition and analysis
Images were acquired on a Zeiss 510 Meta confocal miscroscope. Statistical analysis of da neuron phenotypes was performed in R (R Development Core Team, 2006). Quantification of MB neuron phenotype was done using a two-tailed Fisher’s exact test.
Supplementary Material
Acknowledgements
We thank Angela Ho and Jost Vielmetter of the CalTech Protein Expression Facility for production of Dscam11-8 proteins used for AUC, Phini Katsamba and Barry Honig for helpful discussions about biophysical measurements, and Wes Grueber for helpful suggestions on da neuron analysis. We thank Thomas Rogerson for assistance at an early stage of the project, Howon Kim for providing control transgenes for the ectopic repulsion assay, and Dorian Gunning for providing the Dscam1 ectodomain antibody. We thank members of Zipursky lab for comments on the manuscript and helpful discussions. We particularly thank Daisuke Hattori, Josh Sanes, and Woj Wojtowicz, for critical reading of the manuscript. This work was supported by grants from the NIH (DC006485 to S.L.Z., and GM62270 to L.S.). S.L.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental data Supplementary material includes five figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Hartman PE, Roth JR. Mechanisms of suppression. Adv Genet. 1973;17:1–105. doi: 10.1016/s0065-2660(08)60170-4. [DOI] [PubMed] [Google Scholar]
- Hattori D, Chen Y, Matthews BJ, Salwinski L, Sabatti C, Grueber WB, Zipursky SL. Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature. 2009;461:644–648. doi: 10.1038/nature08431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Millard SS, Wojtowicz WM, Zipursky SL. Dscam-mediated cell recognition regulates neural circuit formation. Annu Rev Cell Dev Biol. 2008;24:597–620. doi: 10.1146/annurev.cellbio.24.110707.175250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd T, Russell C, Goodman CS, Tear G. Dosage-sensitive and complementary functions of roundabout and commissureless control axon crossing of the CNS midline. Neuron. 1998;20:25–33. doi: 10.1016/s0896-6273(00)80431-6. [DOI] [PubMed] [Google Scholar]
- Kramer AP, Kuwada JY. Formation of the receptive fields of leech mechanosensory neurons during embryonic development. J Neurosci. 1983;3:2474–2486. doi: 10.1523/JNEUROSCI.03-12-02474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, Schmucker D. Structural basis of Dscam isoform specificity. Nature. 2007;449:487–491. doi: 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- R Development Core Team R: A Language and Environment for Statistical Computing. 2006 http://www.R-project.org.
- Sawaya MR, Wojtowicz WM, Andre I, Qian B, Wu W, Baker D, Eisenberg D, Zipursky SL. A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms. Cell. 2008;134:1007–1018. doi: 10.1016/j.cell.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci U S A. 2010;107:14893–14898. doi: 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Love J, Colman DR. Adhesion molecules in the nervous system: structural insights into function and diversity. Annu Rev Neurosci. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Sanes JR. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 2010;143:343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.