In 1994 Leonard Adleman used DNA strands to encode cities and flights to show that itineraries satisfying special conditions could be constructed and isolated in the laboratory (1). The abstract of this seminal paper concludes, “This experiment demonstrates the feasibility of computation at the molecular level.” Within months Richard Lipton argued (2) that fundamental problems concerning the truth of logic statements also could be addressed in this new way. In this issue of PNAS, Faulhammer et al. (3) use laboratory techniques with relatively low error rates in an experiment realizing Lipton's earlier design.
However, within a year after Adleman's article, it already was known such approaches can require unrealistic quantities of materials, leading Stemmer (4) to suggest using existing laboratory procedures to evolve realistically sized molecular populations by inducing variation and selection. Such evolutionary computation is an established paradigm for conventional computers. Interest is beginning to focus on DNA-based implementations (5–7) of evolutionary computation because of its alleged robustness in the presence of errors and its ability to exploit the massive parallelism and memory inherent at the molecular level.
By its success in the laboratory, Adleman's work (1) sparked intense excitement and marked the birth of a new field, DNA computation. Adleman's insight was that the ability of single-stranded DNA to seek and bind to a complementary strand allows DNA to carry out massively parallel computation. Although it seems doubtful that biologically based computers will be suitable for general-purpose applications, there are some especially difficult problems where conventional computers lack the massive parallelism and huge memory capacity inherent in molecular computation. For example, the so-called “NP-complete” problems that apparently require exponentially increasing computing time with a linear increase in problem size are notoriously difficult for silicon computers to solve. Thus, are there special applications where biomolecular computation can outperform silicon-based computers? During the past few years there have been many proposals seeking new methods for using DNA to solve NP-complete problems or to construct programmable computers (8–10). Examples include: implementing blocked cellular automata by self-assembling DNA tiles (11), carrying out DNA computation on solid support surface rather than in solution (12), breaking the Data Encryption Standard (13), and boolean circuits (14). Guarnieri et al. (15) have developed a procedure to implement the addition of two non-negative binary numbers through a chain reaction, and Klein et al. (16) have demonstrated the implementation of logically reversible computation. Unfortunately, there are still few experimental results.
Before Adleman's article, molecular computation had been extensively studied (see ref. 17 for a survey). Also, DNA and associated laboratory techniques had been specifically proposed to implement computations of formal language theory (18). However, Adleman's computations for the Hamiltonian path problem (traveling salesman problem) seem to have caught most people's imagination. The Hamilton path problem is a famous intractable problem in computer science (19). This problem can be paraphrased as “find a flight itinerary for visiting each of the cities shown on an airline map exactly once” (Fig. 1).
Figure 1.
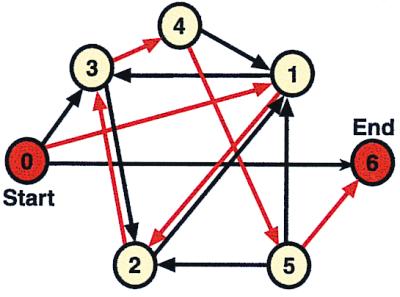
The Hamiltonian path problem. In this case, a salesman must find his way from city 0 to city 6, passing through each of the remaining cities only once. The map shown here has seven nodes and 14 connecting routes as used in Adleman's experiment (1). The red lines represent the only correct path, that is 0 → 1, 1 → 2, 2 → 3, 3 → 4, 4 → 5, 5 → 6.
Briefly, Adleman's experiments followed these six steps:
1. Encode cities and flights into DNA.
2. Assemble itineraries at random.
3. Select itineraries from initial city to final city.
4. The correct number of cities must be visited.
5. No city can be left out.
6. Is anything left?
These experiments were performed in two phases. First, randomly generate all potential solutions to the problem at hand. Second, isolate the correct solution through repeated removal or dilution of incorrect solutions. At the end of this series of separation steps it is only necessary to detect whether there are any DNA strands left. These necessarily represent correct solutions.
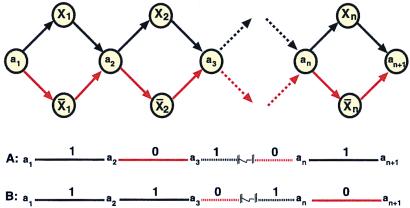
In 1995, Lipton (2) suggested a method for generating the set of all binary strings of DNA with length n. Lipton's procedure involves first creating a graph such that a path through the graph is a realization of a binary string of length n. The graph is constructed by the Adleman protocol and is constrained so that all paths start at a1, end at an+1, and encode an n bit binary string (Fig. 2). At each stage a path has exactly two choices: If it takes the vertex with an unbarred label, it will encode a 1; if it takes the vertex with a barred label, it will encode a 0. Therefore, the path [a1 X1 a2 X̄2 a3 … an Xn an+1] encodes the binary string [1, 0, … ,1] (as shown in Fig. 2A).
Figure 2.
Lipton's graph (2) for constructing binary numbers. The vertices and edges are constructed by using Adleman's algorithm so that the longest paths through the graph will represent an n-bit binary numbers. If a path goes through an X, then it has a 1 at that position (represented by black color); otherwise, it has a 0 at that position (represented by red color).
Lipton proposed that such binary encoding of DNA could be used to solve logic satisfaction problems. Such a problem has the form, “If a given logic statement contains n variables, is there a way to make the whole statement true by choosing the n variables to each be true or false?” Clearly such a choice corresponds to some binary string of length n. The only method known to verify logic satisfaction problems in all cases is simply to try all possibilities. This would quickly become intractable even for supercomputers when there are hundreds of variables. Lipton pointed out that in theory one could create strands of DNA (trillions or more molecules) representing all possible solutions of satisfiability problems having some tens of variables and then search through them all in parallel (2).
In the paper appearing in this issue of PNAS (3), implementing Lipton's earlier proposal, Faulhammer et al. show they can solve a 9-bit instance of a “knight problem,” which is a type of logic satisfaction problem, by constructing a 10-bit binary RNA library that encodes all of the 1,024 possible 10-bit binary strings, then systematically performing operations that destroy strings that fail to satisfy some parts of the problem (3). Faulhammer et al. successfully indicate that Adleman's approach to finding a solution to the Hamiltonian path problem could be generalized to solve the problem of finding a satisfying assignment for a general logic statement. It is notable that with a destructive algorithm (20), introducing complementary DNA oligonucleotides to selectively mark RNA strands for RNase H digestion (this enzyme requires a RNA–DNA duplex as substrate), Faulhammer et al. could destroy strands that did not fit the constraints of the desired solution. Thus, instead of a method that depends on an extraction procedure with potentially high error rates as used by Adleman (1), Faulhammer et al. show that their “mark and destroy” method can recover a set of “winning” molecules representing all of the solutions of the “knight problem” with low error rates. Forty-four distinct candidate solutions were isolated from the 10-bit RNA binary library, one of which was incorrect (3).
Both Adleman (1) and Faulhammer et al. (3) demonstrate the feasibility of carrying out computation at the molecular level. In this new field where the theoretical foundation has been strong, yet very few experimental results have been reported, the paper by Faulhammer et al. certainly has moved the molecular computational field to another milestone. However, it is important to notice that even though the experimental designs were elegant, both experiments have addressed only very small instances of two different problems. Their methods are not suitable for solving large instances of NP-complete problems. For example, Hartmanis (21) estimates that the same method Adleman used would, on a 200-node graph, require an amount of DNA weighing more than the Earth. Therefore, both demonstrations mentioned above are important not because they “solve” hard problems, but rather because they show a way to use the immense parallelism of molecular interactions.
Conventional computers often solve problems by using the paradigm of evolutionary computation, based on analogies with natural evolution. Evolution computation is particularly used for solving problems involving many interacting variables such as timetable construction, machine learning, and graph drawing layout (22). For example, running an airline involves a complex timetable construction problem. The most stable aspects involve scheduling of flights, arrival gates, crews, aircraft, maintenance, training, etc. Time-varying aspects are even more difficult to incorporate: weather delays, equipment breakdowns, strikes, etc.
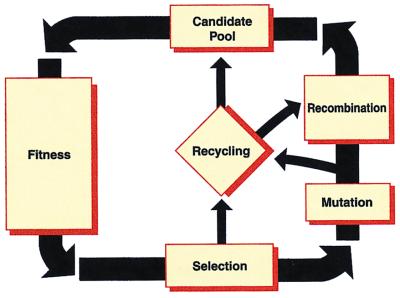
With conventional computers, evolution computation for a particular problem generally maintains a population of candidate solutions, often represented as a string of binary bits. In each generation, less promising candidates are likely to be eliminated. Their replacements are likely to be generated from relatively promising candidates by cloning and/or mutation and/or recombination. Less frequently, other genetic analogies may be used: inversion, gene hopping, multiple parents, etc. Evolutionary computation comes in many, many different varieties (see ref. 23 and http://alife.santafe.edu/∼joke/encore/www/). Most are variations and/or elaborations on the following very loose outline also illustrated in Fig. 3.
Figure 3.
Schematic of evolutionary computations. Fitness-based selection is universal. Mutation, recombination, and recycling may be used in various combinations.
Begin with an initial population of candidate solutions, perhaps chosen randomly.
Repeat the following steps until convergence:
1. Evaluate fitness of candidates.
2. Select among more fit candidates to breed and among less fit to be replaced.
3. (Optional) Induce variation by breeding.
DNA molecules seem particularly suited to certain versions of evolutionary computation, because they can encode populations of bit strings suitable for both crossover and pointwise mutation. Also, there are known molecular biological protocols (24–27) for manipulating DNA in ways likely to be useful for implementing evolutionary computation with DNA. Three other advantages of using DNA to implement evolutionary computation are: First, there is the massive information storage potential of DNA molecules. For example, a gram of DNA contains about 1021 bases. The information content is approximately 2 × 1021 bits, greatly exceeding the 200-petabyte storage of all digital magnetic tape produced in 1 year (R. Williams, http://www.ccsf.caltech.edu/∼roy/dataquan). Second, with DNA molecules one might process, in parallel, populations billions of times larger than is usual for conventional computers. One may process grams of DNA per day, with each gram encoding about 1021 bits of information. A state-of-the-art teraflop supercomputer performs about 1012 operations per sec, so it would need about 100 days to process 1021 bits in 100-bit blocks. Third, biolaboratory operations on DNA molecules inherently involve errors. Such errors are sometimes tolerable when executing evolutionary computation (28, 29), unlike when executing deterministic algorithms.
Laboratory work requires effort and money. At the current state of DNA computing, despite recent encouraging theoretical advances, there are still very few biochemical experiments reported. However, most would agree that the future of DNA computing certainly will depend on whether the experimental challenges can be met. Is DNA computing doomed to be useless? We believe this is unlikely. Even though it may be impossible to implement a programmable DNA computer capable of running most applications faster than a conventional computer, the massively parallel nature of DNA computing could be essential for some practical problems, such as evolutionary computations. Therefore, we believe that molecular computation could complement rather than replace silicon as a computational medium in the future.
Acknowledgments
Our research is partially supported by Defense Advanced Research Planning Agency/National Science Foundation Grant 9725021 and National Science Foundation Grant 9980092.
Footnotes
See companion article on page 1385.
References
- 1.Adleman L M. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 2.Lipton R J. Science. 1995;268:542–545. doi: 10.1126/science.7725098. [DOI] [PubMed] [Google Scholar]
- 3.Faulhammer D, Cukras A R, Lipton R J, Landweber L F. Proc Natl Acad Sci USA. 2000;97:1385–1389. doi: 10.1073/pnas.97.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stemmer W P C. Science. 1995;270:1510–1510. doi: 10.1126/science.270.5241.1510. [DOI] [PubMed] [Google Scholar]
- 5.Deaton R, Murphy R C, Rose J A, Garzon M H, Franceschetti D R, Stevens S E., Jr . IEEE International Conference on Evolutionary Computation. Washington, DC: Institute of Electrical and Electronics Engineers; 1997. pp. 267–271. [Google Scholar]
- 6.Bäck T, Kok J N, Rozenberg G. In: Preliminary Proceedings of DIMACS Workshop on Evolution as Computation. Landweber L, Wintree E, Lipton R, Freeland S, editors. Piscataway, NJ: Center for Discrete Mathematics and Theoretical Computer Science; 1999. pp. 67–88. [Google Scholar]
- 7.Wood D H, Chen J, Antipov E, Lemieux B, Cedeño W. In: DNA-Based Computers V. Gifford D, Wintree E, editors. Providence, RI: Am. Math. Soc.; 2000. , in press. [Google Scholar]
- 8.Puaun G, Rozenberg G, Salomaa A. DNA Computing: New Computing Paradigms. New York: Springer; 1998. [Google Scholar]
- 9.Dove A. Nat Biotechnol. 1998;16:830–832. doi: 10.1038/nbt0998-830. [DOI] [PubMed] [Google Scholar]
- 10.Pool R. New Scientist. 1996;151:26–31. [Google Scholar]
- 11.Winfree E, Liu F, Wenzler L A, Seeman N C. Nature (London) 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q, Wang L, Frutos A G, Condon A E, Corn R M, Smith L M. Nature (London) 2000;403:175–178. doi: 10.1038/35003155. [DOI] [PubMed] [Google Scholar]
- 13.Boneh D, Dunworth C, Lipton R J. In: Proceedings of DIMACS Workshop on DNA Computing. Lipton R, Baum E, editors. Providence, RI: Am. Math. Soc.; 1995. pp. 37–65. [Google Scholar]
- 14.Boneh D, Dunworth C, Sgallj J. Discrete Appl Math. 1996;71:79–94. [Google Scholar]
- 15.Guarnieri E, Fliss M, Bancrok C. Science. 1996;273:220–223. doi: 10.1126/science.273.5272.220. [DOI] [PubMed] [Google Scholar]
- 16.Klein J P, Leete T H, Rubin H. BioSystems. 1999;52:123–128. doi: 10.1016/s0303-2647(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 17.Conrad M. Adv Computers. 1990;31:235–324. [Google Scholar]
- 18.Head T. Bull Math Biol. 1987;49:737–759. doi: 10.1007/BF02481771. [DOI] [PubMed] [Google Scholar]
- 19.Garey M R, Johnson S D. Computers and Intractability: A Guide to the Theory of NP-Completeness. New York: Freeman; 1979. [Google Scholar]
- 20.Amos M, Gibbons A, Hodgson D. In: DNA-Based Computers II. Baum E B, Lipton R J, editors. Providence, RI: Am. Math. Soc.; 1998. pp. 151–161. [Google Scholar]
- 21.Hartmanis J. Bull Eur Assoc Theor Comput Sci. 1995;55:136–138. [Google Scholar]
- 22.Zbigniew M. Genetic Algorithms + Data Structures = Evolution Programs. New York: Springer; 1992. [Google Scholar]
- 23.Bäck T, Fogel D B, Michalewicz Z. Handbook of Evolutionary Computation. Philadelphia: Institute of Physics Publishing; 1997. [Google Scholar]
- 24.Beaudry A A, Joyce G E. Science. 1992;257:635–641. doi: 10.1126/science.1496376. [DOI] [PubMed] [Google Scholar]
- 25.Sassanfar M, Szostak J W. Nature (London) 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 26.Lorsch J R, Szostak J W. Nature (London) 1994;371:31–36. doi: 10.1038/371031a0. [DOI] [PubMed] [Google Scholar]
- 27.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg D E, Miller B L. Evol Comput J. 1996;4:113–131. [Google Scholar]
- 29.Goldberg D E, Deb K, Clark J M. Complex Systems. 1992;6:333–362. [Google Scholar]