Abstract
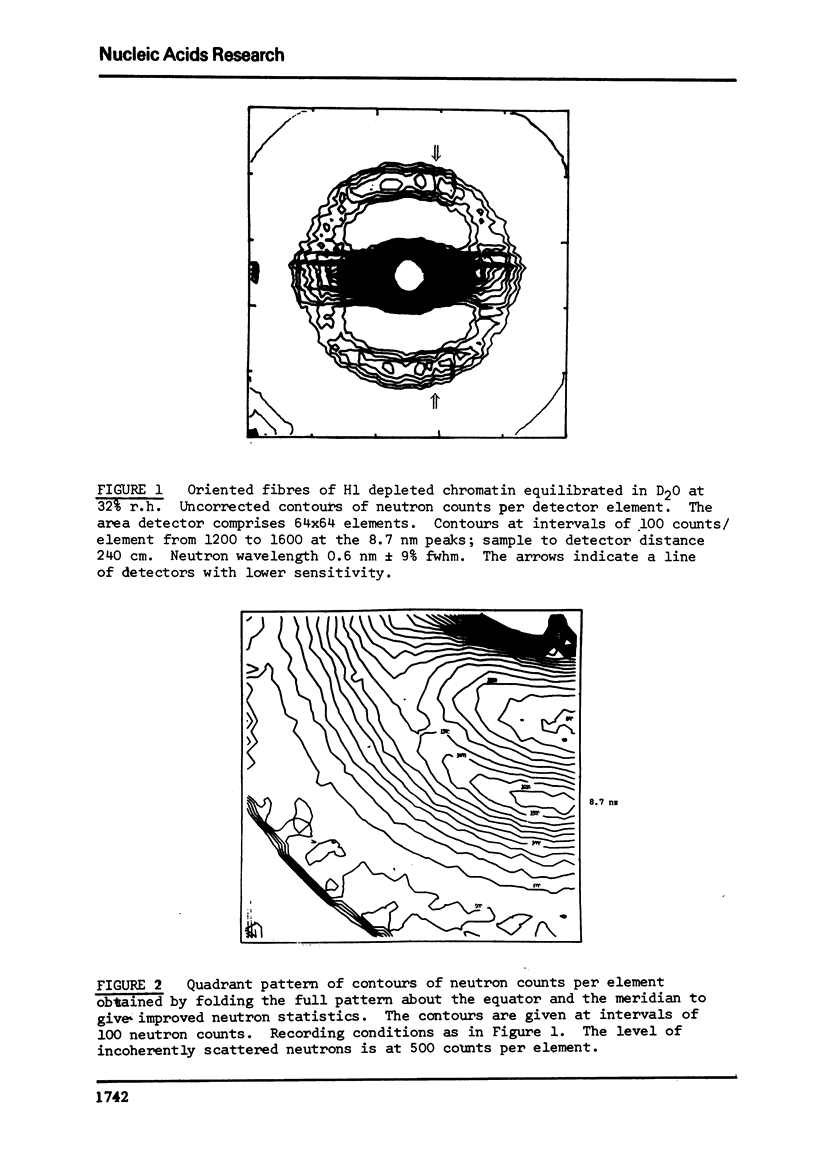
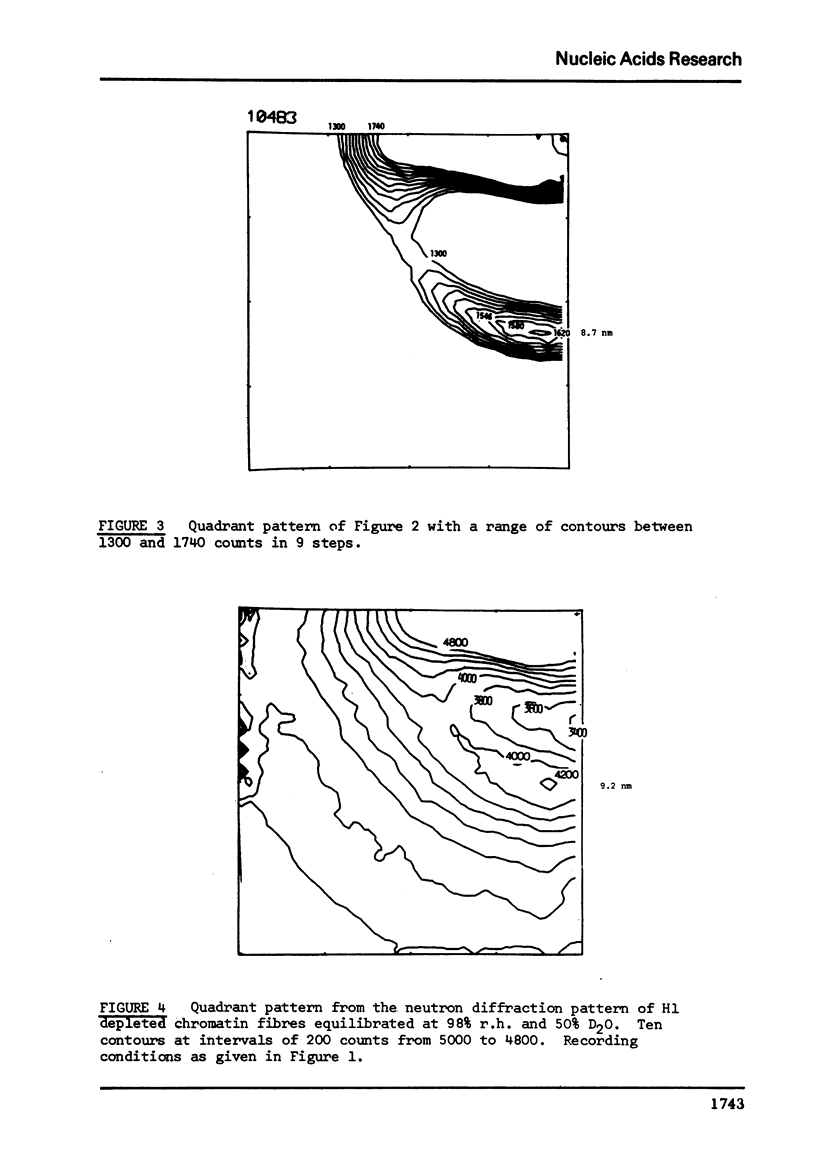
There is considerable current interest in the organisation of nucleosomes in chromatin. A strong X-ray and neutron semi-meridional diffraction peak at approximately 10 nm had previously been attributed to the interparticle specing of a linear array of nucleosomes. This diffraction peak could also result from a close packed helical array of nucleosomes. A direct test of these proposals is whether the 10 nm peak is truly meridional as would be expected for a linear array of nucleosomes or is slightly off the meridian as expected for a helical array. Neutron diffraction studies of H1-depleted chromatin support the latter alternative. The 10 nm peak has maxima which form a cross-pattern with semi-meridional angle of 8 to 9 degrees. This is consistent with a coil of nucleosomes of pitch 10 nm and outer diameter of approximately 30 nm. These dimensions correspond to about six nucleosomes per turn of the coli.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Bolund L. A., Johns E. W. The selective extraction of histone fractions from deoxyribonucleoprotein. Eur J Biochem. 1973 Jun 15;35(3):546–553. doi: 10.1111/j.1432-1033.1973.tb02871.x. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Cary P. D., Crane-Robinson C., Rattle H. W., Boublik M., Sautière P. Conformations and interactions of histone H2A (F2A2, ALK). Biochemistry. 1975 May 6;14(9):1876–1885. doi: 10.1021/bi00680a012. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Molgaard H. V., Stephens R. M., Bolund L. A., Johns E. W. X-ray studies of nucleoproteins depleted of lysine-rich histone. Eur J Biochem. 1972 Dec 18;31(3):474–482. doi: 10.1111/j.1432-1033.1972.tb02555.x. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Rattle H. W. Simple computer-aided approach for the analyses of the nuclear-magnetic-resonance spectra of histones. Fractions F1, Fsa1, F2B, cleaved halves of F2B and F2B-DNA. Eur J Biochem. 1972 May 23;27(2):270–281. doi: 10.1111/j.1432-1033.1972.tb01836.x. [DOI] [PubMed] [Google Scholar]
- Carlson R. D., Olins D. E. Chromatin model calculations: Arrays of spherical nu bodies. Nucleic Acids Res. 1976 Jan;3(1):89–100. doi: 10.1093/nar/3.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Panyim S., Bilek D., Chalkley R. An electrophoretic comparison of vertebrate histones. J Biol Chem. 1971 Jul 10;246(13):4206–4215. [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H. A super-coil model for nucleohistone. J Mol Biol. 1972 Jul 14;68(1):115–124. doi: 10.1016/0022-2836(72)90267-7. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Tatchell K., Van Holde K. E., Richards B. M. Low-angle neutron scattering from chromatin subunit particles. Nucleic Acids Res. 1975 Nov;2(11):2163–2176. doi: 10.1093/nar/2.11.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- Van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. A model for particulate structure in chromatin. Nucleic Acids Res. 1974 Nov;1(11):1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]


