Abstract
The magnetosomes of many magnetotactic bacteria consist of membrane-enveloped magnetite crystals, whose synthesis is favored by a low redox potential. However, the cellular redox processes governing the biomineralization of the mixed-valence iron oxide have remained unknown. Here, we show that in the alphaproteobacterium Magnetospirillum gryphiswaldense, magnetite biomineralization is linked to dissimilatory nitrate reduction. A complete denitrification pathway, including gene functions for nitrate (nap), nitrite (nir), nitric oxide (nor), and nitrous oxide reduction (nos), was identified. Transcriptional gusA fusions as reporters revealed that except for nap, the highest expression of the denitrification genes coincided with conditions permitting maximum magnetite synthesis. Whereas microaerobic denitrification overlapped with oxygen respiration, nitrate was the only electron acceptor supporting growth in the entire absence of oxygen, and only the deletion of nap genes, encoding a periplasmic nitrate reductase, and not deletion of nor or nos genes, abolished anaerobic growth and also delayed aerobic growth in both nitrate and ammonium media. While loss of nosZ or norCB had no or relatively weak effects on magnetosome synthesis, deletion of nap severely impaired magnetite biomineralization and resulted in fewer, smaller, and irregular crystals during denitrification and also microaerobic respiration, probably by disturbing the proper redox balance required for magnetite synthesis. In contrast to the case for the wild type, biomineralization in Δnap cells was independent of the oxidation state of carbon substrates. Altogether, our data demonstrate that in addition to its essential role in anaerobic respiration, the periplasmic nitrate reductase Nap has a further key function by participating in redox reactions required for magnetite biomineralization.
INTRODUCTION
Magnetosomes are bacterial organelles synthesized by magnetotactic bacteria (MTB) for orientation in the Earth's magnetic field to facilitate the search for growth-favoring suboxic zones of stratified aquatic habitats (22). In the alphaproteobacterium Magnetospirillum gryphiswaldense MSR-1 (in the following referred to as MSR-1) and many other MTB, magnetosomes are membrane-enveloped magnetic crystals of magnetite (Fe3O4) which are aligned in chains along cytoskeletal structures (23, 24, 48). The intracellular biomineralization of magnetite is of substantial interdisciplinary interest not only for microbiology and cell biology but also for geobiology, biotechnology, and even astrobiology (22, 28, 48, 63).
Recent studies have shown that the biomineralization of magnetite crystals is under the control of a number of essential and accessory genes which have been speculated to all be encoded within a single genomic magnetosome island (31, 37, 45, 59). The synthesis of magnetosome crystals proceeds in several steps, which include the invagination of magnetosome membrane vesicles (24, 27) and the uptake of iron and its crystallization as magnetite within these vesicles (12, 44). Although the mechanism of biomineralization has not been fully elucidated, it has been suggested that the synthesis of the mixed-valence iron oxide magnetite (Fe3O4) occurs by coprecipitation from ferrous and ferric iron in supersaturating concentrations, which is favored by a low redox potential (11, 12, 33). It was observed early that Magnetospirillum (“Aquaspirillum”) magnetotacticum MS-1 (MS-1) is capable of microaerobic dissimilatory nitrate reduction and produces N2O or N2 as the final products (2), and in Magnetospirillum magneticum strain AMB-1 (AMB-1) nitrate also supported magnetosome formation at low oxygen concentrations (35, 36, 66). Oxystat experiments further demonstrated that magnetite synthesis was induced only when the oxygen concentration was below a threshold value of 2,000 Pa in MSR-1 and other magnetospirilla (19). Although molecular oxygen was initially assumed to be required for Fe3O4 biomineralization (6), it was later shown by isotope experiments that the oxygen bound in bacterially synthesized Fe3O4 is derived from water (32). In fact, in the marine vibrio strain MV-1 (“Magnetovibrio blakemorii”) magnetosomes can be biomineralized in the entire absence of oxygen during anaerobic respiration with N2O as an electron acceptor (3), and in Desulfovibrio magneticus RS-1 this can occur using either sulfate or fumarate as an electron acceptor (42). Although previous studies failed to demonstrate oxygen-independent growth and magnetosome synthesis in microaerophilic magnetospirilla MS-1 and MSR-1, earlier observations that magnetite synthesis is stimulated by nitrate suggested a potential link to denitrification also in these organisms (6, 19).
Bacterial denitrification is a respiratory process to reduce nitrate stepwise to nitrogen gas (NO3− → NO2− → NO → N2O → N2) (67). In many Gram-negative bacteria reduction of nitrate is catalyzed by a membrane-bound nitrate reductase (Nar), whereas in several other bacteria this reaction is instead performed by a periplasmic nitrate reductase (Nap) (30). Two isofunctional periplasmic enzymes may catalyze the subsequent reduction of nitrite to nitric oxide: a homodimeric cytochrome cd1 nitrite reductase, NirS, and a monotrimeric copper-containing enzyme, NirK (30). The further reduction of nitrite to nitric oxide is then catalyzed by an integral membrane protein complex (67). Its catalytic subunit NorB is structurally homologous to oxygen-reducing heme-copper oxidases, whereas NorC is a membrane-anchored protein with a heme domain in the periplasmic face (62). The last step of the denitrification pathway is the reduction of nitrous oxide to dinitrogen gas, which is catalyzed by the periplasmic multicopper enzyme nitrous oxide reductase (Nos) (30).
Despite their potential importance for magnetite biomineralization, the genetics and biochemistry of denitrification processes have not been well studied in MTB. A cytochrome cd1-type nitrite reductase (NirS) was purified from MS-1 and shown to accelerate the oxidization of ferrous iron in the presence of nitrite under anaerobic conditions in vitro (65). Later, a soluble periplasmic nitrate reductase implicated in magnetite synthesis was purified from MS-1 (56). Wang et al. recently interrupted a gene (norB) for nitric oxide reductase in AMB-1 by transposon insertion and found that shorter magnetosome chains were produced under anaerobic conditions (61). However, except for this single study, no genetic evidence has been available for these possible functions in vivo so far, and the exact interrelation of these two pathways as well as the redox process governing magnetite biomineralization has largely remained unclear.
Here we started to explore the function of dissimilatory nitrate reduction in MSR-1 by expression analysis and mutagenesis of the periplasmic nitrate reductase Nap and comparison to the roles of downstream denitrification enzymes Nor and Nos. We found that Nap is important for biomineralization of fully functional magnetosomes in MSR-1 during both denitrification and microaerobic respiration. We demonstrate that in addition to its role in anaerobic respiration, Nap has a further key function by participating in redox reactions required for magnetite biomineralization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Tables S1 and S2 in the supplemental material. Escherichia coli strains were grown in lysogeny broth (LB) at 37°C. MSR-1 strains were grown at 30°C in modified flask standard medium (FSM) (19), which here was defined as nitrate medium if not specified otherwise. In ammonium medium, nitrate was replaced by 4 mM ammonium chloride. When necessary, antibiotics were added to the medium as follows: for E. coli, tetracycline (Tc) at 12 μg/ml, kanamycin (Km) at 25 μg/ml, ampicillin (Amp) at 50 μg/ml, and gentamicin (Gm) at 15 μg/ml, and for MSR-1, Tc at 5 μg/ml, Km at 5 μg/ml, and Gm at 30 μg/ml. When E. coli strain BW29427 was used as the donor in conjugation, 300 μM diaminopimelic acid (DAP) was added.
Growth experiments were carried out under microaerobic and anaerobic conditions in Hungate tubes containing 10 ml medium. For microaerobic conditions, Hungate tubes were sealed with butyl rubber stoppers under a microoxic gas mixture containing 2% O2 and 98% N2 before autoclaving. Anaerobic conditions were achieved by omitting O2 from the gas mixture, where some trace oxygen initially being potentially present did not support any detectable growth in the absence of nitrate. For aerobic conditions, cells were incubated in free gas exchange with air in 300-ml flasks containing 20 ml medium agitated at 200 rpm. Optical density (OD) and magnetic response (Cmag) were measured photometrically at 565 nm as previously described (47). For gas production assay, cells were inoculated and mixed with FSM medium with 0.3% agar in oxygen gradient tubes and incubated for 48 h exposed to the air. If not specified otherwise, inocula were prepared under aerobic conditions with a Cmag value of zero.
Genetic and molecular biology techniques.
Standard molecular biological techniques were performed for DNA isolation, digestion, ligation, and transformation (43). DNA products were sequenced using BigDye Terminator version 3.1 chemistry on an ABI 3700 capillary sequencer (Applied Biosystems, Darmstadt, Germany). Sequence data were analyzed with the software Vector NTI Advance 11.5.1 (Invitrogen, Darmstadt, Germany). All oligonucleotide sequences used in this work are available if required.
Construction of mutant strains.
All PCRs were performed using Phusion polymerase (NEB). Enzymes, including restriction enzymes and T4 DNA ligase, were purchased from Fermentas. For the interruption of napA, an internal fragment of napA was digested with ApaI and SacI and then ligated into pCM184 to yield pLYJ27. pLYJ27 was then inserted into MSR-1 by conjugation as described previously (49) to obtain a napA::kanR mutant strain. To generate the unmarked deletion mutant of the entire nap operon, a modified cre-lox method for large deletions was used (60). A 2-kb upstream PCR fragment of the nap operon was generated and cloned into EcoRI/NotI-digested pAL01 to obtain pLYJ85, the plasmid pLYJ85 was conjugationally integrated into the chromosome of MSR-1, and colonies screened positive by PCR for the presence of the kanamycin marker were designated Δnap-up. Subsequently, the plasmid pLYJ92 containing a 2-kb downstream fragment of the nap operon was integrated into the chromosome of Δnap-up by conjugation. After the presence of kanamycin and gentamicin markers was verified by screening PCR, the strain was designated Δnap-up-down. The lox-mediated excision of the nap operon was initiated by conjugational transformation of pCM157 (34). Precise excision was further confirmed by PCR amplification and sequencing. The plasmid pCM157 was lost by passaging cells several times in fresh nitrate medium. Finally, this strain was designated the Δnap mutant.
A two-step, cre-lox-based method (58) was used to generate unmarked deletions of norCB and nosZ. For norCB deletion, the 2-kb upstream PCR product was cloned into pCM184 between Acc65I and NotI sites, generating pLYJ31. A 2-kb downstream fragment of norCB was then ligated into MluI/SacI-digested pLYJ31 to obtain pLYJ34. For the deletion of nosZ, the upstream PCR product of the nosZ gene was cloned into NdeI/NotI-digested pCM184 to yield pLYJ32. The 2-kb downstream fragment of nosZ was digested with MluI and SacI and then ligated into pLYJ32 to obtain pLYJ35. Allelic exchange vectors pLYJ34 and pLYJ35 were then transformed into MSR-1 by conjugation. Deletion mutants were first screened on replica plates with kanamycin and tetracycline. Screening PCR was further performed for colonies which did not grow on tetracycline plates. To generate unmarked deletion mutants, pCM157 was conjugated into each mutant and subsequently cured from each mutant by several transfers in nitrate medium. Finally, the unmarked mutants were designated the ΔnorCB and ΔnosZ mutants, respectively.
Complementation experiments.
For genetic complementation of Δnap, ΔnorCB, and ΔnosZ, a series of pBBR1MCS-2-based plasmids were generated. Plasmid pLYJ80, which contains the nap cluster, including its own promoter region, was constructed in three steps as illustrated in Fig. S1 in the supplemental material. For complementation of norCB and nosZ, pLYJ75 and pLYJ76, respectively, were used, in which the norCB and nosZ gene sequences with their own promoter regions were ligated into ApaI/SacI-digested pBBR1MCS-2.
Construction and analysis of transcriptional gusA fusions.
To generate the transcriptional nap-gusA, nirS-gusA, nor-gusA, and nosZ-gusA fusion plasmids, the gusA gene from pK19mobGII was amplified and cloned between the HindIII and SmaI sites of pBBR1MCS-2 to obtain pLYJ97. Then nap, nirS, norCBQD, and nosZ promoter regions were cloned into Acc65I/HindIII-digested pLYJ97, designated pLYJ98, pLYJ94, pLYJ99, and pLYJ100, respectively. Plasmids were then introduced into wild-type (WT) MSR-1 by conjugation.
Cells in post-exponential phase were centrifuged, broken, and suspended in phosphate-buffered saline (PBS) for enzyme assay at 4°C. The protein concentration was determined by the method of Bradford (7). β-Glucuronidase activity was determined at 37°C as described by Wilson et al. (64). Units were expressed as nanomoles product formed per minute per milligram protein. Triplicate assays were performed, and the values reported were averaged by using at least two independent cultures.
Chemical analysis.
For nitrate and nitrite analysis, MSR-1 cells were grown under anaerobic conditions for 20 h. Nitrate was detected using Szechrome reagents (Polysciences, Inc.). Diluted 20-fold samples of cultures were prepared, and Szechrome reagents were then added. After half an hour, the absorbance at 570 nm was recorded. When nitrate was no longer detectable, cultures without dilution were used to confirm the absence of nitrate. A nitrate standard curve (0 to 350 μM) was obtained to convert absorbance values to concentrations.
Nitrite was measured with the modified Griess reagent (Sigma). One hundred microliters of 20-fold-diluted samples of cultures were mixed with equal modified Griess reagent and the absorbance recorded at 540 nm after 15 min. When no nitrite was detected, cultures without dilution were used to confirm the absence of nitrite. A nitrite standard curve (0 to 70 μM) was generated to calculate final nitrite concentration.
Transmission electron microscopy (TEM).
WT MSR-1 and mutants were grown at 25°C under anaerobic or microaerobic conditions up to an OD at 565 nm (OD565) of 0.1 and then were concentrated and adsorbed onto carbon-coated copper grids. Samples were viewed and recorded with a TECNAI FEI20 microscope (FEI, Eindhoven, Netherlands) at 200 kV or with a Morgagni 268 microscope (FEI, Eindhoven, Netherlands) at 80 kV as previously described (25). For magnetosome analysis, more than 200 crystals and 100 cells were detected for each strain.
Bioinformatic analysis.
Denitrification genes were identified by BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) homology searches in the genomes of MSR-1 (GenBank accession no. CU459003.1), AMB-1 (GenBank accession no. AP007255.1), MS-1 (NCBI reference sequence NZ_AAAP00000000.1), and Magnetococcus marinus MC-1 (MC-1) (GenBank accession no. CP000471.1) with an expectation value E of <1e−06 and an amino acid similarity of >50%.
RESULTS
MSR-1 is capable of anaerobic growth by a complete denitrification pathway.
Various nitrogen sources were tested for their ability to support anaerobic growth and magnetite synthesis in MSR-1. As expected, no growth was observed in the presence of only NH4+ (see Table S3 in the supplemental material). Only very poor growth (OD of 0.01 or less) was observed with the denitrification intermediate nitrite at concentrations of ≤1 mM (Table 1), as well as with N2O (see Table S3 in the supplemental material). However, significant anaerobic growth and magnetite formation was found in the presence of nitrate. Growth was concentration dependent up to 8 mM, and approximately 6 mM nitrate was utilized after 20 hours of incubation (Table 1). Excess nitrate (>10 mM) gradually decreased growth yields to zero at 20 mM. Growth at 4 mM nitrate led to transient accumulation of 0.1 mM nitrite, which was consumed as growth proceeded (see Fig. S2 in the supplemental material). Compared to anaerobic growth, MSR-1 incubated under microaerobic conditions and in the presence of 4 mM nitrate reached higher densities (see Table S3 in the supplemental material) while larger amounts of nitrite (about 1 mM) accumulated, likely due to repression of nitrite reduction by oxygen (described below). If nitrate was replaced by an equal amount of ammonium (4 mM), under microaerobic conditions the final cell yield was slightly reduced (see Table S3 in the supplemental material), whereas highest yields were reached under fully aerobic conditions in both nitrate and ammonium media. This suggested that denitrification and aerobic respiration cooccurred simultaneously under microaerobic conditions.
Table 1.
Effects of different nitrate and nitrite concentrations on growth and magnetic response after a 20-hour anaerobic incubationa
| Added nitrogen source and concn (mM) | Growth (ΔOD565)b | Cmag | Nitrate left (mM) | Nitrite left (mM) |
|---|---|---|---|---|
| Nitrate | ||||
| 0 | 0.00 | 1.7 ± 0.1 | 0 | 0 |
| 1 | 0.06 ± 0.00 | 1.8 ± 0.1 | 0 | 0 |
| 2 | 0.12 ± 0.00 | 1.9 ± 0.0 | 0 | 0 |
| 4 | 0.20 ± 0.00 | 1.8 ± 0.1 | 0 | 0 |
| 6 | 0.25 ± 0.01 | 1.7 ± 0.1 | 0.36 ± 0.07 | 0 |
| 8 | 0.32 ± 0.02 | 1.8 ± 0.0 | 2.23 ± 0.25 | 0 |
| 10 | 0.29 ± 0.01 | 1.8 ± 0.1 | 3.89 ± 0.66 | 0 |
| 11 | 0.14 ± 0.02 | 1.9 ± 0.0 | 5.42 ± 0.44 | 0 |
| 12 | 0.09 ± 0.04 | 1.8 ± 0.1 | 6.53 ± 0.12 | 0 |
| 15 | 0.08 ± 0.00 | 1.8 ± 0.1 | 7.83 ± 0.62 | 0 |
| 20 | −0.03 ± 0.01 | 1.3 ± 0.1 | 19.88 ± 0.52 | 0 |
| Nitrite | ||||
| 0 | 0.00 | 1.7 ± 0.1 | 0 | 0 |
| 0.5 | 0.01 ± 0.00 | 1.7 ± 0.0 | 0 | 0 |
| 1.0 | 0.01 ± 0.00 | 1.7 ± 0.1 | 0 | 0 |
| 1.5 | −0.01 ± 0.00 | 0.4 ± 0.0 | 0 | 1.3 ± 0.1 |
| 2.0 | −0.02 ± 0.00 | 0 | 0 | 1.9 ± 0.0 |
| 2.5 | −0.02 ± 0.00 | 0 | 0 | 2.4 ± 0.0 |
Values are means and standard deviations for experiments with triplicate cultures and repeated three times.
Negative values represent a decrease in cell density compared to the initial value on inoculation of 0.04.
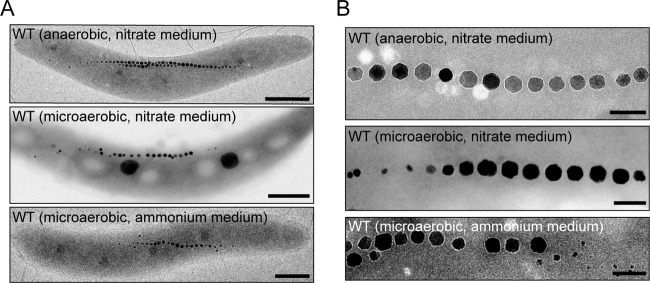
Anaerobic magnetosome formation was independent of nitrate concentrations up to 15 mM (Table 1). However, although the Cmag value was slightly lower in anaerobically grown cells than in microaerobically grown cells (see Table S3 in the supplemental material), probably due to subtle effects on cell length (anaerobic, 4.4 ± 0.2 μm; microaerobic, 4.2 ± 0.2 μm) that are known to affect Cmag readings (26), anaerobically grown cells had higher magnetosome numbers (29, versus 25 in microaerobically grown cells), larger crystals (49.9 ± 5.0 nm, versus 41.1 ± 2.5 nm for microaerobically grown cells), and more regular crystal morphologies and chain alignment (Fig. 1). In microaerobic ammonium medium, cells biomineralized similar amounts of magnetite (23 crystals, 41.9 ± 2.0 nm) (Fig. 1; see Table S3 in the supplemental material), confirming that nitrate reduction is not essential for magnetosome formation under microaerobic conditions. Under aerobic conditions, cells were nonmagnetic (Cmag = 0) in both nitrate and ammonium media, similar to earlier findings (19).
Fig 1.
Effect of oxygen and nitrogen sources on magnetosome formation. (A) TEM micrographs of whole cells of the WT under the indicated conditions. Scale bars, 500 nm. (B) Close-up views of the magnetosome crystals shown in panel A. Scale bars, 100 nm.
Identification of denitrification genes in MSR-1 and other MTB.
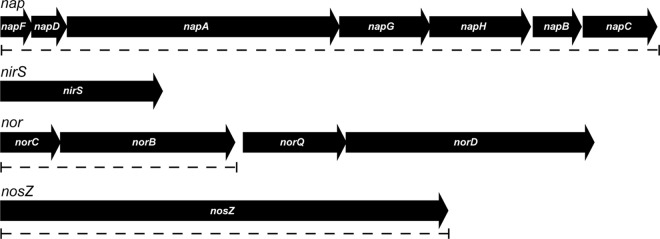
Using the respective protein sequences from Pseudomonas stutzeri as queries in BLASTP analysis, we reconstructed a complete pathway of denitrification from the genome of MSR-1. A putative nap operon containing napFDAGHBC genes for a periplasmic nitrate reductase was identified (Fig. 2; see Table S4 in the supplemental material), whereas we failed to detect nar genes encoding a membrane-bound nitrate reductase complex. nap operons but no nar genes are also present in other MTB, including the magnetospirilla AMB-1 and MS-1 as well as MC-1 (see Table S4 in the supplemental material). The organization of the napFDAGHBC operon resembles those of nap genes from E. coli (21), Haemophilus influenzae (17), and Rhodobacter sphaeroides 2.4.3 (18), in which this type of clusters has been designated nap-β (53). In many bacteria, this Nap cluster is used for anaerobic growth via nitrate respiration (17).
Fig 2.
Molecular organization of identified denitrification genes in MSR-1. Dashed lines indicate the extent of deletions in mutant strains.
A nirS gene, encoding a homodimeric cytochrome cd1 nitrite reductase, was found in MSR-1 next to several other nir genes of partially unknown functions located on a short contig of the incomplete genome assembly of MSR-1 (data not shown). norC and norB, encoding nitric oxide reductase subunits, are part of an operon also comprising norQ and norD in MSR-1, AMB-1, and MC-1, whereas we failed to detect nor genes in strain MS-1, possibly due to the incompleteness of its genome assembly. A singular nosZ gene was identified in MSR-1 and AMB-1 but not in MC-1, whereas no other nos genes that are usually colocated with nosZ in the same operon (5, 8, 20) were found in magnetospirilla. In MS-1 two complementary nosZ fragments were detected on two different contigs of the incomplete genome assembly.
Expression of nir, nor, and nos but not nap is upregulated by nitrate and downregulated by oxygen.
Since maximum magnetite synthesis in MSR-1 occurs at low oxygen tensions and in the presence of nitrate (Table 1; see Table S3 in the supplemental material), we tested whether the expression of denitrification genes is correlated with magnetosome formation. To this end, transcriptional gene fusions of napFDAGHBC, nirS, norCBQD, and nosZ with gusA, encoding β-glucuronidase, were constructed and transferred into WT MSR-1 by conjugation. As shown in Table 2, cells containing the transcriptional nap-gusA reporter gene fusion exhibited an approximately 2-fold increase of β-glucuronidase activity under aerobic compared to microaerobic conditions, whereas nitrate had no obvious effect on nap-gusA expression. In contrast, about a 6-fold-higher level of β-glucuronidase activity was observed under microaerobic conditions with nitrate than without nitrate, whereas increased oxygen concentrations resulted in decreased β-glucuronidase activity. WT MSR-1 carrying nor-gusA showed the same pattern as for nirS-gusA, i.e., a higher level of norCBQD expression under microaerobic conditions in the presence of nitrate (762.7 ± 37.0 U) than in the absence of nitrate (221.5 ± 52.4 U), whereas β-glucuronidase activity was lowered by increasing oxygen concentrations. nosZ-gusA also exhibited an approximately 5-fold-higher β-glucuronidase activity under microaerobic conditions in the presence of nitrate than in its absence, and it was downregulated by oxygen.
Table 2.
Effects of oxygen and nitrate on transcriptional expression of denitrification genes nap, nirS, nor, and nos fused with gusA
| Promoter | β-Glucuronidase activity (U)a |
|||
|---|---|---|---|---|
| Microaerobic conditions |
Aerobic conditions |
|||
| With NO3− | Without NO3− | With NO3− | Without NO3− | |
| nap | 16.2 ± 1.4 | 15.9 ± 0.8 | 30.8 ± 2.6 | 28.6 ± 2.8 |
| nirS | 124.0 ± 5.5 | 21.2 ± 9.6 | 14.2 ± 7.9 | 18.3 ± 7.8 |
| norCBQD | 762.8 ± 37.0 | 221.5 ± 52.4 | 204.4 ± 41.1 | 151.1 ± 10.5 |
| nosZ | 519.0 ± 43.4 | 118.3 ± 33.3 | 146.6 ± 34.7 | 152.5 ± 21.9 |
Values are averages and standard deviations for at least replicate cultures.
Notably, the finding that the expression of nap was different from that of other denitrification genes suggested that Nap might have a distinct function. In addition, whereas nirS and nosZ are absent from the genome of nondenitrifying strain MC-1 (46), nap and nor genes are more widely conserved within alphaproteobacterial MTB. Therefore, further genetic analysis was focused mostly on these genes.
NosZ and NorCB are required for complete denitrification but have only minor roles in magnetite synthesis.
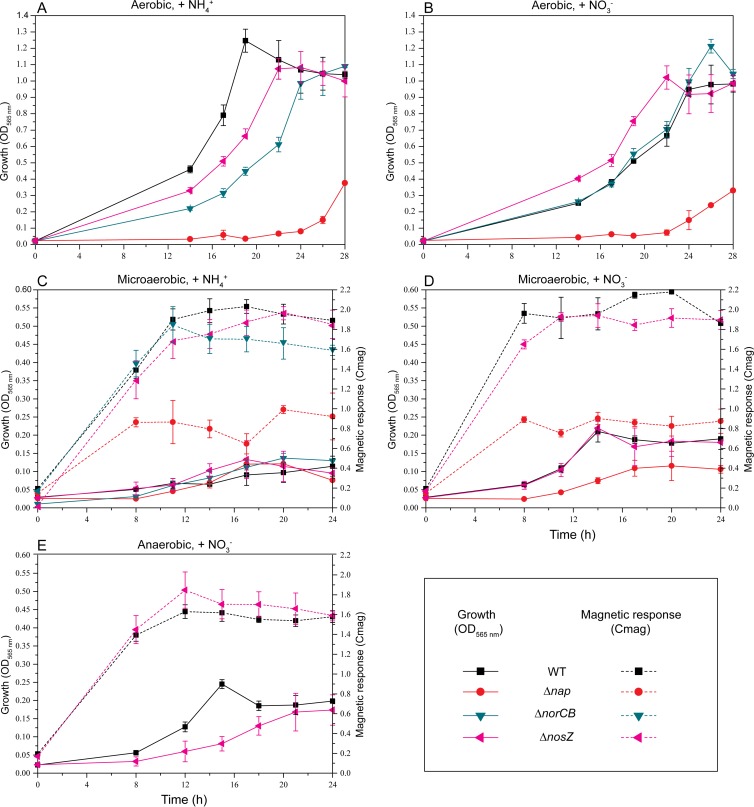
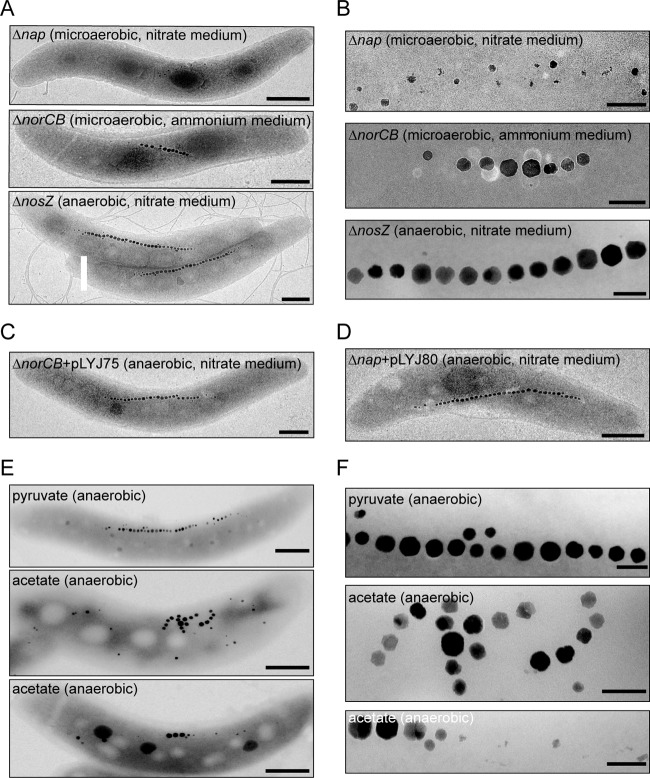
A norCB deletion strain was constructed as described in Materials and Methods. When ΔnorCB cells were incubated microaerobically in ammonium medium or aerobically in either nitrate or ammonium medium, no significant effect on growth was observed (Fig. 3A, B, and C). However, no growth occurred in the presence of nitrate under anaerobic and microaerobic conditions, probably due to the toxicity of accumulated nitric oxide, as demonstrated for AMB-1 (61). In the presence of ammonium under microaerobic conditions, the average Cmag of the ΔnorCB mutant was slightly lower than that of the WT (Fig. 3C). ΔnorCB cells were slightly shorter (3.7 ± 0.2 μm, versus 4.1 ± 0.3 μm for the WT) and contained fewer magnetosomes (14 crystals per cell, versus 23 in the WT) (Fig. 4A and B), whereas crystal size and morphology were unaffected. Complementation of ΔnorCB mutants with plasmid pLYJ75 harboring a WT norCB allele restored growth and magnetosome formation back to the WT levels (Fig. 4C; see Fig. S3A in the supplemental material). For comparison, a nosZ deletion mutant was constructed. As shown in Fig. S3B in the supplemental material, in the ΔnosZ mutant no bubble was detected in semisolid agar due to the high solubility of N2O, and ΔnosZ was complemented with plasmid pLYJ76, which restored the activity of nitrous oxide reduction to dinitrogen gas. The deletion of nosZ did not affect magnetosome formation under any tested condition (Fig. 3A, B, C, D, and E), and cells contained magnetosomes virtually identical to those of the WT with respect to crystal morphology, size, and number (Fig. 4A and B). However, loss of nosZ resulted in a slightly reduced growth under anaerobic conditions compared to that of the WT, which may result from reduced energy yields obtained by incomplete denitrification (Fig. 3E).
Fig 3.
Growth (OD565) and magnetic response (Cmag) of WT MSR-1 and the Δnap, ΔnorCB, and ΔnosZ mutants under different conditions. Under aerobic conditions, the Cmag values were always zero and not shown. (A) Aerobic, ammonium medium; (B) aerobic, nitrate medium; (C) microaerobic, ammonium medium; (D) microaerobic, nitrate medium; (E) anaerobic, nitrate medium. Results from representative experiments were measured in triplicate, and values are given as means and standard deviations.
Fig 4.
(A) TEM micrographs of Δnap, ΔnorCB, and ΔnosZ whole cells under the indicated conditions. Scale bars, 500 nm. (B) Close-up views of the magnetosome crystals shown in panel A. Scale bars, 100 nm. (C and D) TEM micrographs of anaerobically grown ΔnorCB (C) and Δnap (D) cells complemented with plasmids pLYJ75 and pLYJ80, respectively, harboring their WT alleles. Scale bars, 500 nm. (E) TEM micrographs of WT cells grown anaerobically on pyruvate and acetate. Scale bars, 500 nm. (F) Close-up views of the magnetosome crystals shown in panel E. Scale bars, 100 nm.
Nap functions as a nitrate reductase during anaerobic growth.
To abolish its function, we first attempted to interrupt the nap operon by insertion of a kanR cassette into napA (napA::kanR) in MSR-1 (details are shown in Fig. S4 in the supplemental material). However, this napA::kanR mutant still showed a WT-like phenotype for nitrate reduction and magnetite formation, probably due to some residual activity (data not shown). Therefore, a further deletion mutant (Δnap) was constructed by unmarked excision of the entire nap operon. Compared to the WT, the Δnap mutant displayed a markedly delayed growth (it took more than 50 h to reach stationary phase, compared to about 24 h for the WT) when cells were cultured aerobically in either nitrate or ammonium medium (Fig. 3A and B). Hardly any difference in growth was found in ammonium medium between the WT and the Δnap mutant under microaerobic conditions (Fig. 3C). However, in microaerobic nitrate medium Δnap cells did not consume and reduce nitrate and reached lower cell densities than the WT, probably due to reduced energy yields (Fig. 3D). No growth or nitrate utilization was observed for the Δnap mutant in nitrate medium under anaerobic conditions, confirming that Nap functions as the primary nitrate reductase for anaerobic respiration in MSR-1.
Deletion of nap severely affects microaerobic magnetite biomineralization.
Under microaerobic conditions, the loss of nap genes resulted in significantly decreased Cmag values (<1.0) (Fig. 3C and D) not only in ammonium medium but also nitrate medium, in which mutant cells grew only by aerobic respiration due to the absence of Nap. As shown in Fig. 4A and B, compared to ΔnosZ and ΔnorCB, deletion of nap had a much stronger effect on magnetosomes, which were significantly smaller (24.8 ± 4.5 nm, versus 41.1 ± 2.5 nm for WT crystals under microaerobic conditions in nitrate medium), present in lower numbers (9 crystals per cell, versus 25 crystals per cell in the WT), and appearing as irregularly shaped, misaligned particles. To ensure that the observed phenotypes in fact were caused by the introduced mutation, the Δnap mutant was complemented with plasmid pLYJ80, which restored the activity of nitrate reduction (data not shown), as well as WT-like growth and magnetosome formation (Fig. 4D; see Fig. S3A in the supplemental material).
Nap functions in maintaining proper redox balance for magnetosome formation.
Although nitrate was no longer utilized in Δnap cells, redundant nitrate at levels as high as 8 mM did not affect magnetite synthesis (Table 1). Furthermore, Δnap cells growing microaerobically in ammonium medium (in the absence of nitrate) also produced fewer and irregular magnetosomes, resembling those in the presence of nitrate. This indicated that besides being required for nitrate reduction, Nap might have an additional function for magnetosome formation.
On the other hand, it had been hypothesized previously by Taoka and colleagues that nitrate reduction was not essential for Fe3O4 synthesis (56), whereas reduction of nitrite was implicated in the oxidation of ferrous iron to produce the mixed-valence Fe3O4 (65). Therefore, we asked whether the observed effect of nap deletion on magnetosomes might result indirectly from a regulatory effect on nitrite reduction, which might be suppressed in the Δnap mutant. To test this possibility, aerobically grown nonmagnetic Δnap and WT cells were precultured as described in Materials and Methods. Growth experiments were then performed under microaerobic and anaerobic conditions, in which 500 μM nitrite, the product of nitrate reduction catalyzed by Nap in the WT, was added to ammonium medium. As shown in Fig. 5, under microaerobic conditions in the absence of nap, nitrite was still utilized. However, again the Cmag value was much lower than that of the WT. When Δnap cells were incubated anaerobically in the absence of nitrate but in the presence of 500 μM nitrite, nitrite was completely consumed after 20 h, although neither the WT nor the Δnap mutant obviously grew under these conditions. The Cmag was only about 0.6 in the Δnap mutant, while a Cmag value of 1.6 was found in the WT. Taken together, these results precluded effects of deregulated nitrite reduction in the Δnap mutant.
Fig 5.

Growth (OD565) and magnetic response (Cmag) of WT MSR-1 and the Δnap mutant under microaerobic conditions with 500 μM nitrite added to ammonium medium.
Alternatively, it has been shown previously that in other denitrifying bacteria, such as Paracoccus pantotrophus and R. sphaeroides, the periplasmic Nap enzyme is regulated by the oxidation state of carbon substrates, and it is thought to play a role in maintaining redox homeostasis by dissipating excess reductant during aerobic growth (10, 16, 50, 55). Furthermore, it was observed by us that there was a significant growth lag in the Δnap mutant of MSR-1 under aerobic conditions (Fig. 3A and B), which would be consistent with the suggestion by Richardson et al. (41) that excess reductant causes lower growth rates because NADH has to be reoxidized by cell maintenance reactions, as also observed in P. pantotrophus and Rhodobacter capsulatus growing on carbon sources which are more reduced than the biomass (10, 40). Therefore, we hypothesized that in MSR-1 the Nap system also might be involved in the maintenance of the intracellular redox balance, which consequently may affect the species of iron within the periplasm and magnetosome vesicles. Transcription from the nap promoter was tested in both the WT and the Δnap mutant in the presence of carbon substrates with different oxidation states (Table 3). No difference in the β-glucuronidase activity with different carbon substrates under microaerobic conditions and under aerobic conditions was found for the WT (Table 3). Only a slight increase in nap-gusA expression was observed in the WT under aerobic conditions compared to that under microaerobic conditions, which is in agreement with the observed upregulation of nap expression by oxygen (Table 2). In the Δnap mutant, no effect on expression on different carbon sources was found under microaerobic conditions, and a similar level of β-glucuronidase activity was detectable under aerobic conditions. However, when we shifted nonmagnetic cells from 21% to 0% oxygen, WT cells developed much lower Cmag values on the more reduced carbon substrate acetate than on the more oxidized substrates pyruvate and succinate (Table 3). TEM showed that magnetite biomineralization in acetate-grown cells was affected: in addition to cells with regular magnetosome chains, a variable proportion (>50%) of cells with irregular and shorter chains and with fewer (Fig. 4E and F) and smaller crystals (36.2 ± 1.3 nm, versus 42.7 ± 1.2 nm with pyruvate), as well as cells entirely devoid of magnetite crystals, was present. In contrast, Cmag values in the Δnap mutant were equally low with different carbon substrates (Table 3). Therefore, in acetate-grown cells of the WT, impaired magnetosome formation could be due to regulation of Nap activity rather than nap expression by the oxidation state of carbon sources, similar to what has been described for Nap of R. sphaeroides DSM158, in which expression of a napA-lacZ gene fusion was similar in cells grown with carbon substrates of different oxidation states, whereas Nap activity was higher on reduced carbon sources (16).
Table 3.
Effects of different carbon sources on magnetic response and transcriptional expression of nap-gusA
| Carbon substrate (concn, mM) | Avg oxidation no. of carbons |
Cmag |
β-Glucuronidase activity (U)a |
||||
|---|---|---|---|---|---|---|---|
| WT, anaerobic | Δnap mutant, anaerobic | WT |
Δnap mutant |
||||
| Microaerobic | Aerobic | Microaerobic | Aerobic | ||||
| Pyruvate (27) | +0.66 | 1.7 ± 0.1 | 0.6 ± 0.1 | 31.7 ± 1.0 | 49.9 ± 4.4 | 72.3 ± 7.9 | 77.3 ± 0.1 |
| Succinate (27) | +0.50 | 1.6 ± 0.1 | 0.5 ± 0.0 | 33.4 ± 1.0 | 54.2 ± 2.2 | 77.5 ± 4.0 | 83.7 ± 6.3 |
| Acetate (27) | +0.00 | 0.8 ± 0.0 | 0.5 ± 0.0 | 31.2 ± 7.7 | 42.7 ± 1.0 | 75.3 ± 6.8 | 78.5 ± 0.2 |
Values are averages and standard deviations for at least replicate cultures.
DISCUSSION
Except for nap, the maximum expression of all identified denitrification genes (nir, nor, and nos) coincided with conditions of highest magnetite synthesis, i.e., the presence of nitrate and low concentrations or absence of oxygen. This pattern is similar to that for other nonmagnetic bacteria, such as P. stutzeri (29), R. sphaeroides 2.4.3 (1, 57), and Bradyrhizobium japonicum (5). However, under microaerobic conditions in the absence of nitrate, where expression of denitrification genes nirS, nor, and nosZ was reduced, magnetite biomineralization of WT cells was independent of the presence of nitrate, suggesting that denitrification and oxygen respiration may have overlapping functions under microaerobic conditions. Whereas deletion of nosZ only had a weak effect on anaerobic growth and biomineralization, we were unable to detect any growth in our ΔnorCB mutant of MSR-1 under anaerobic and microaerobic conditions in the presence of nitrate, probably due to the toxicity of the accumulated intermediate nitric oxide, since growth could be rescued by additional deletion of nirS in the ΔnorCB background (Y. Li and D. Schüler, unpublished data). In contrast, in a previous study Wang and colleagues reported some growth for an AMB-1 ΔnorB mutant under anaerobic conditions (61). The discrepancy between nor mutants of MSR-1 (no growth) and AMB-1 (poor growth) might be due to different genotypes, since in the AMB-1 mutant only norB (located downstream of norC) was interrupted by transposon insertion, thereby possibly retaining some residual activity of the nitric oxide reductase enzyme. The slightly reduced magnetosome number of the MSR-1 ΔnorCB mutant is unlikely to result from limited energy yields during denitrification as speculated by Wang et al. (61), since oxygen respiration in ammonium medium is unlikely to be affected by loss of denitrification proteins. On the other hand, the comparatively high expression levels of our norCB-gusA fusion suggest that in the absence of nitrate under microaerobic and aerobic conditions, NorCB may be involved in further, yet-unknown functions directly or indirectly linked to magnetosome formation. For example, it has been shown that Nor from Paracoccus denitrificans is capable of reducing oxygen to water in vitro (13, 15), which also might be a possible function for Nor in MSR-1 during microaerobic respiration and magnetite biomineralization.
Since only the loss of nap genes and not that of other denitrification genes directly abolished growth and since intermediates such as nitrite supported only very weak anaerobic growth of the WT and Δnap strains, the reduction of nitrate to nitrite catalyzed by the periplasmic Nap enzyme, and not the subsequent reduction steps, is the primary energy-generating process of denitrification. Deletion of the entire nap cluster not only abolished anaerobic growth but also severely impaired magnetite synthesis under microaerobic conditions in the presence of either nitrate or ammonium, resulting in fewer, smaller, and misshapen magnetosome crystals. This is in agreement with the finding that the abolishment of nitrate reductase activity by deprivation of molybdenum, an essential cofactor of the periplasmic nitrate reductase, resulted in an approximately 60% decrease of iron content of cells from strain MS-1 (56). Unlike growth, magnetite synthesis in the WT showed no dependency on the electron acceptor (i.e., nitrate) concentration, suggesting that this effect was not primarily due to energy limitation of cells. The Nap enzymes of other nonmagnetic bacteria were implicated in redox balancing using nitrate as an ancillary oxidant to dissipate excess reductant (40). Consistent with the suggestion that lower growth rates may be caused by excess reductant (41), we observed a significant lag of growth in Δnap cells under aerobic conditions. In addition, although the oxidation state of carbon sources did not affect transcription of nap, WT cells contained shorter and irregular crystal chains on the more reduced substrate acetate than on more oxidized substrate pyruvate, while in the Δnap mutant magnetite synthesis was equally low on different carbon sources. Similar to the case for Nap in other bacteria, such as P. pantotrophus Pd1222 (50) and various strains of R. sphaeroides (16, 18, 55), our data are consistent with a role of Nap of MSR-1 in the maintenance of the intracellular redox balance, thus posing an optimum redox potential for magnetite synthesis. The lower Cmag in acetate-grown cells of even the WT might indicate that Nap activity is still insufficient to dispel all excess reductant originating from reduced carbon substrates. Moreover, the lack of a difference in magnetosome biomineralization in Δnap cells grown on different carbon sources might be explained by excess reductant in vivo even in medium with the oxidized substrate pyruvate.
Unlike that of other denitrification genes, transcription of the nap operon was induced by oxygen but unaffected by nitrate. This regulation pattern is different from that in E. coli, in which nap gene expression is induced by anaerobiosis and nitrate limitation (9, 54), and in P. pantotrophus, where nap genes are expressed only during aerobiosis (51). However, nap regulation in MSR-1 resembles that in Ralstonia eutropha and R. sphaeroides DSM158, in which nap systems show a higher expression level under aerobic conditions and are not induced by nitrate (16, 52). Furthermore, a consensus Fnr (fumarate and nitrate reduction regulatory protein) box (TTGAN6TCAA) (39) is located about 80 bp upstream of the putative translation start of napF of MSR-1, which is also consistent with nap regulation by oxygen. Taken together, these data indicate that Nap likely functions also during aerobic respiration, which is in agreement with its speculated role in dissipation of intracellular reductant.
Overall, we demonstrated that magnetite biomineralization in MSR-1 in fact is closely linked to nitrate reduction catalyzed by periplasmic nitrate reductase Nap, which participates in redox reactions required for magnetite biomineralization in addition to its role in anaerobic respiration. While the absence of nir and nosZ genes in other MTB such as MC-1 is consistent with their reported inability to grow and respire by denitrification (14, 46), this indicates that a complete denitrification pathway is not absolutely required for magnetosome formation. Interestingly, the presence of a nap cluster and nor genes even in the MC-1 genome (46) agrees with our observation that these genes are important for magnetosome formation also during aerobic respiration, suggesting that they may have functions in magnetite biomineralization which are distinct from their roles as merely respiratory enzymes. Denitrification genes absent in other MTB might be replaced by genes for other redox enzymes. For example, in the magnetotactic marine vibrio strain MV-1, which can respire anaerobically with N2O as an electron acceptor, the N-terminal sequence determined from a purified periplasmic, copper-containing Fe(II) oxidase displays homology to the putative N2O reductase from MS-1 (4), which indicates that magnetosome formation may be linked to respiration by other, unknown functions. Recently, Nishida and Silver have shown that synthesis of magnetic mineral particles is also possible in nonmagnetotactic yeast Saccharomyces cerevisiae, which confirms that intracellular redox control through carbon metabolism and iron supply is an important factor for magnetite biomineralization (38). Finally, our study provides evidence that in MSR-1 genes located outside the genomic magnetosome island are also required for synthesis of magnetosomes that are fully functional with respect to their numbers, sizes, and shapes to serve properly as navigational devices.
Supplementary Material
ACKNOWLEDGMENT
We thank the China Scholarship Council (CSC) for financial support.
Footnotes
Published ahead of print 22 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Bartnikas TB, Tosques IE, Laratta WP, Shi J, Shapleigh JP. 1997. Characterization of the nitric oxide reductase-encoding region in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 179:3534–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bazylinski DA, Blakemore RP. 1983. Denitrification and assimilatory nitrate reduction in Aquaspirillum magnetotacticum. Appl. Environ. Microbiol. 46:1118–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bazylinski DA, Frankel R, Jannasch HW. 1988. Anaerobic magnetite production by a marine magnetotactic bacterium. Nature 334:518–519 [Google Scholar]
- 4. Bazylinski DA, Williams T. 2006. Ecophysiology of magnetotactic bacteria, p 64 In Schüler D. (ed), Magnetoreception and magnetosomes in bacteria. Springer Verlag, Heidelberg, Germany [Google Scholar]
- 5. Bedmar EJ, Robles EF, Delgado MJ. 2005. The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem. Soc. Trans. 33:141–144 [DOI] [PubMed] [Google Scholar]
- 6. Blakemore RP, Short KA, Bazylinski DA, Rosenblatt C, Frankel R. 1985. Microaerobic conditions are required for magnetite formation within Aquaspirillum magnetotacticum. Geomicrobiol. J. 4:53–71 [Google Scholar]
- 7. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 8. Cuypers H, Berghofer J, Zumft WG. 1995. Multiple nosZ promoters and anaerobic expression of nos genes necessary for Pseudomonas stutzeri nitrous oxide reductase and assembly of its copper centers. Biochim. Biophys. Acta 1264:183–190 [DOI] [PubMed] [Google Scholar]
- 9. Darwin AJ, Ziegelhoffer EC, Kiley PJ, Stewart V. 1998. Fnr, NarP, and NarL regulation of Escherichia coli K-12 napF (periplasmic nitrate reductase) operon transcription in vitro. J. Bacteriol. 180:4192–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ellington MJ, Bhakoo KK, Sawers G, Richardson DJ, Ferguson SJ. 2002. Hierarchy of carbon source selection in Paracoccus pantotrophus: strict correlation between reduction state of the carbon substrate and aerobic expression of the nap operon. J. Bacteriol. 184:4767–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faivre D, et al. 2004. Mineralogical and isotopic properties of inorganic nanocrystalline magnetites. Geochim. Cosmochim. Acta 68:4395–4403 [Google Scholar]
- 12. Faivre D, Böttger LH, Matzanke BF, Schüler D. 2007. Intracellular magnetite biomineralization in bacteria proceeds by a distinct pathway involving membrane-bound ferritin and an iron(II) species. Angew. Chem. Int. Ed. Engl. 46:8495–8499 [DOI] [PubMed] [Google Scholar]
- 13. Flock U, Watmough NJ, Adelroth P. 2005. Electron/proton coupling in bacterial nitric oxide reductase during reduction of oxygen. Biochemistry 44:10711–10719 [DOI] [PubMed] [Google Scholar]
- 14. Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73:994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujiwara T, Fukumori Y. 1996. Cytochrome cb-type nitric oxide reductase with cytochrome c oxidase activity from Paracoccus denitrificans ATCC 35512. J. Bacteriol. 178:1866–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gavira M, Roldan MD, Castillo F, Moreno-Vivian C. 2002. Regulation of nap gene expression and periplasmic nitrate reductase activity in the phototrophic bacterium Rhodobacter sphaeroides DSM158. J. Bacteriol. 184:1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez PJ, Correia C, Moura I, Brondino CD, Moura JJ. 2006. Bacterial nitrate reductases: molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 100:1015–1023 [DOI] [PubMed] [Google Scholar]
- 18. Hartsock A, Shapleigh JP. 2011. Physiological roles for two periplasmic nitrate reductases in Rhodobacter sphaeroides 2.4.3 (ATCC 17025). J. Bacteriol. 193:6483–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heyen U, Schüler D. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536–544 [DOI] [PubMed] [Google Scholar]
- 20. Holloway P, McCormick W, Watson RJ, Chan YK. 1996. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J. Bacteriol. 178:1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jepson BJ, et al. 2006. Evolution of the soluble nitrate reductase: defining the monomeric periplasmic nitrate reductase subgroup. Biochem. Soc. Trans. 34:122–126 [DOI] [PubMed] [Google Scholar]
- 22. Jogler C, Schüler D. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63:501–521 [DOI] [PubMed] [Google Scholar]
- 23. Jogler C, et al. 2011. Conservation of proteobacterial magnetosome genes and structures in an uncultivated member of the deep-branching Nitrospira phylum. Proc. Natl. Acad. Sci. U. S. A. 108:1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D. 2010. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol. Microbiol. 77:208–224 [DOI] [PubMed] [Google Scholar]
- 25. Katzmann E, et al. 2011. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol. Microbiol. 82:1316–1329 [DOI] [PubMed] [Google Scholar]
- 26. Kolinko I, Jogler C, Katzmann E, Schüler D. 2011. Frequent mutations within the genomic magnetosome island of Magnetospirillum gryphiswaldense are mediated by RecA. J. Bacteriol. 193:5328–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245 [DOI] [PubMed] [Google Scholar]
- 28. Komeili A. 2007. Molecular mechanisms of magnetosome formation. Annu. Rev. Biochem. 76:351–366 [DOI] [PubMed] [Google Scholar]
- 29. Korner H, Zumft WG. 1989. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl. Environ. Microbiol. 55:1670–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kraft B, Strous M, Tegetmeyer HE. 2011. Microbial nitrate respiration—genes, enzymes and environmental distribution. J. Biotechnol. 155:104–117 [DOI] [PubMed] [Google Scholar]
- 31. Lohsse A, et al. 2011. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One 6:e25561 doi:10.1371/journal.pone.0025561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mandernack KW, Bazylinski DA, Shanks WC, Bullen TD. 1999. Oxygen and iron isotope studies of magnetite produced by magnetotactic bacteria. Science 285:1892–1896 [DOI] [PubMed] [Google Scholar]
- 33. Mann S, Sparks NHC, Board RG. 1990. Magnetotactic bacteria: microbiology, biomineralization, palaeomagnetism and biotechnology. Adv. Microb. Physiol. 31:125–181 [DOI] [PubMed] [Google Scholar]
- 34. Marx C, Lidstrom M. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in Gram-negative bacteria. Biotechniques 33:1062–1067 [DOI] [PubMed] [Google Scholar]
- 35. Matsunaga T, Sakaguchi T, Tadokoro F. 1991. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl. Microbiol. Biotechnol. 35:651–655 [Google Scholar]
- 36. Matsunaga T, Tsujimura N. 1993. Respiratory inhibitors of a magnetic bacterium Magnetospirillum sp. AMB-1 capable of growing anaerobically. Appl. Microbiol. Biotechnol. 39:368–371 [Google Scholar]
- 37. Murat D, Quinlan A, Vali H, Komeili A. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. U. S. A. 107:5593–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishida K, Silver PA. 2012. Induction of biogenic magnetization and redox control by a component of the target of rapamycin complex 1 signaling pathway. PLoS Biol. 10:1–10 doi:10.1371/journal.pbio.1001269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ouchane S, Picaud M, Therizols P, Reiss-Husson F, Astier C. 2007. Global regulation of photosynthesis and respiration by FnrL: the first two targets in the tetrapyrrole pathway. J. Biol. Chem. 282:7690–7699 [DOI] [PubMed] [Google Scholar]
- 40. Richardson DJ, et al. 1988. The role of auxiliary oxidants in maintaining redox balance during phototrophic growth of Rhodobacter capsulatus on propionate or butyrate. Arch. Microbiol. 150:131–137 [Google Scholar]
- 41. Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ. 2001. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. 58:165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sakaguchi T, Arakaki A, Matsunaga T. 2002. Desulfovibrio magneticus sp. nov., a novel sulfate-reducing bacterium that produces intracellular single-domain-sized magnetite particles. Int. J. Syst. Evol. Microbiol. 52:215–221 [DOI] [PubMed] [Google Scholar]
- 43. Sambrook J, Russel D. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Habor, NY [Google Scholar]
- 44. Scheffel A, et al. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114 [DOI] [PubMed] [Google Scholar]
- 45. Schübbe S, et al. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schübbe S, et al. 2009. Complete genome sequence of the chemolithoautotrophic marine Magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 75:4835–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schüler D, Baeuerlein E. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180:159–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schüler D. 2008. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol. Rev. 32:654–672 [DOI] [PubMed] [Google Scholar]
- 49. Schultheiss D, Schüler D. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89–94 [DOI] [PubMed] [Google Scholar]
- 50. Sears HJ, Spiro S, Richardi J. 1997. Effect of carbon substrate and aeration on nitrate reduction and expression of the periplasmic and membrane-bound nitrate reductases in carbon-limited continuous cultures of Paracoccus denitrificans Pd1222. Microbiology 143:3767–3774 [DOI] [PubMed] [Google Scholar]
- 51. Sears HJ, Sawers G, Berks BC, Ferguson SJ, Richardson DJ. 2000. Control of periplasmic nitrate reductase gene expression (napEDABC) from Paracoccus pantotrophus in response to oxygen and carbon substrates. Microbiology 146:2977–2985 [DOI] [PubMed] [Google Scholar]
- 52. Siddiqui RA, et al. 1993. Structure and function of a periplasmic nitrate reductase in Alcaligenes eutrophus H16. J. Bacteriol. 175:5867–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simpson PJ, Richardson DJ, Codd R. 2010. The periplasmic nitrate reductase in Shewanella: the resolution, distribution and functional implications of two NAP isoforms, NapEDABC and NapDAGHB. Microbiology 156:302–312 [DOI] [PubMed] [Google Scholar]
- 54. Stewart V, Bledsoe PJ, Chen LL, Cai A. 2009. Catabolite repression control of napF (periplasmic nitrate reductase) operon expression in Escherichia coli K-12. J. Bacteriol. 191:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tabata A, Yamamoto I, Matsuzaki M, Satoh T. 2005. Differential regulation of periplasmic nitrate reductase gene (napKEFDABC) expression between aerobiosis and anaerobiosis with nitrate in a denitrifying phototroph Rhodobacter sphaeroides f. sp. denitrificans. Arch. Microbiol. 184:108–116 [DOI] [PubMed] [Google Scholar]
- 56. Taoka A, Yoshimatsu K, Kanemori M, Fukumori Y. 2003. Nitrate reductase from the magnetotactic bacterium Magnetospirillum magnetotacticum MS-1: purification and sequence analyses. Can. J. Microbiol. 49:197–206 [DOI] [PubMed] [Google Scholar]
- 57. Tosques IE, Kwiatkowski AV, Shi J, Shapleigh JP. 1997. Characterization and regulation of the gene encoding nitrite reductase in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 179:1090–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Uebe R, et al. 2010. Deletion of a fur-like gene affects iron homeostasis and magnetosome formation in Magnetospirillum gryphiswaldense. J. Bacteriol. 192:4192–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 187:7176–7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ullrich S, Schüler D. 2010. Cre-lox-based method for generation of large deletions within the genomic magnetosome island of Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 76:2349–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang K, et al. 2011. Interruption of the denitrification pathway influences cell growth and magnetosome formation in Magnetospirillum magneticum AMB-1. Lett. Appl. Microbiol. 53:55–62 [DOI] [PubMed] [Google Scholar]
- 62. Watmough NJ, Field SJ, Hughes RJ, Richardson DJ. 2009. The bacterial respiratory nitric oxide reductase. Biochem. Soc. Trans. 37:392–399 [DOI] [PubMed] [Google Scholar]
- 63. Weiss BP, et al. 2004. Magnetic tests for magnetosome chains in Martian meteorite ALH84001. Proc. Natl. Acad. Sci. U. S. A. 101:8281–8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wilson KJ, Hughes SG, Jefferson RA. 1992. The Escherichia coli gus operon: induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria, p 7–22 In Gallagher SR. (ed), GUS protocols. Using the GUS gene as reporter of gene expression. Academic Press Inc., San Diego, CA [Google Scholar]
- 65. Yamazaki T, Oyanagi H, Fujiwara T, Fukumori Y. 1995. Nitrite reductase from the magnetotactic bacterium Magnetospirillum magnetotacticum—a novel cytochrome cd1 with Fe(II)-nitrite oxidoreductase activity. Eur. J. Biochem. 233:665–671 [DOI] [PubMed] [Google Scholar]
- 66. Yang CD, Takeyama H, Tanaka T, Matsunaga T. 2001. Effects of growth medium composition, iron sources and atmospheric oxygen concentrations on production of luciferase-bacterial magnetic particle complex by a recombinant Magnetospirillum magneticum AMB-1. Enzyme Microb. Technol. 29:13–19 [DOI] [PubMed] [Google Scholar]
- 67. Zumft W. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.