Abstract
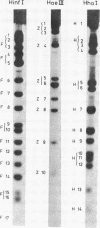
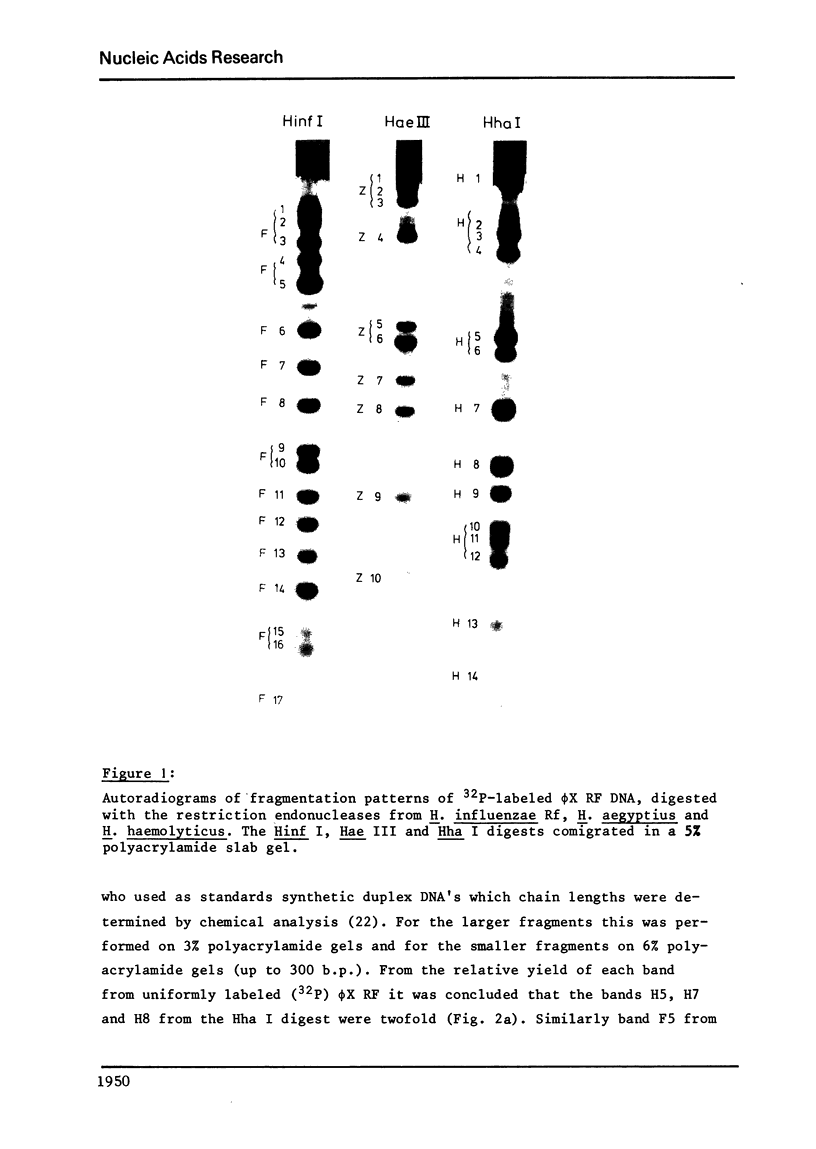
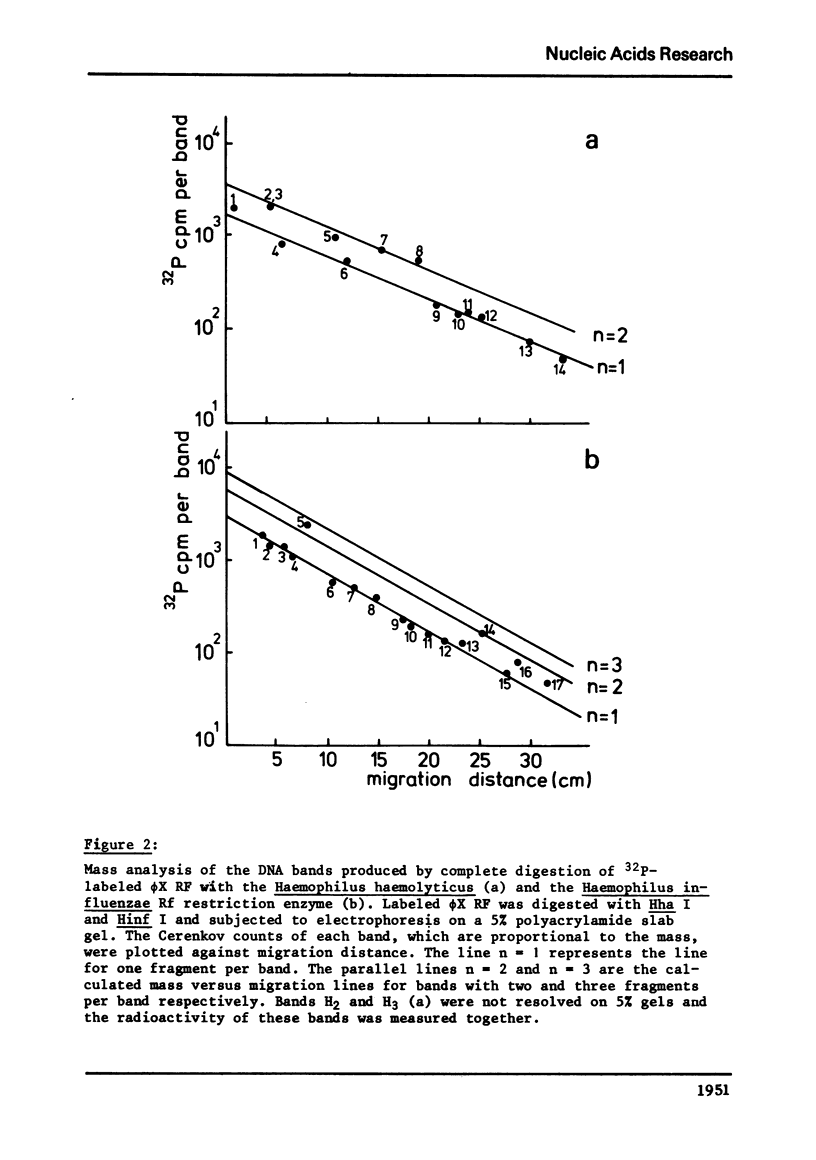
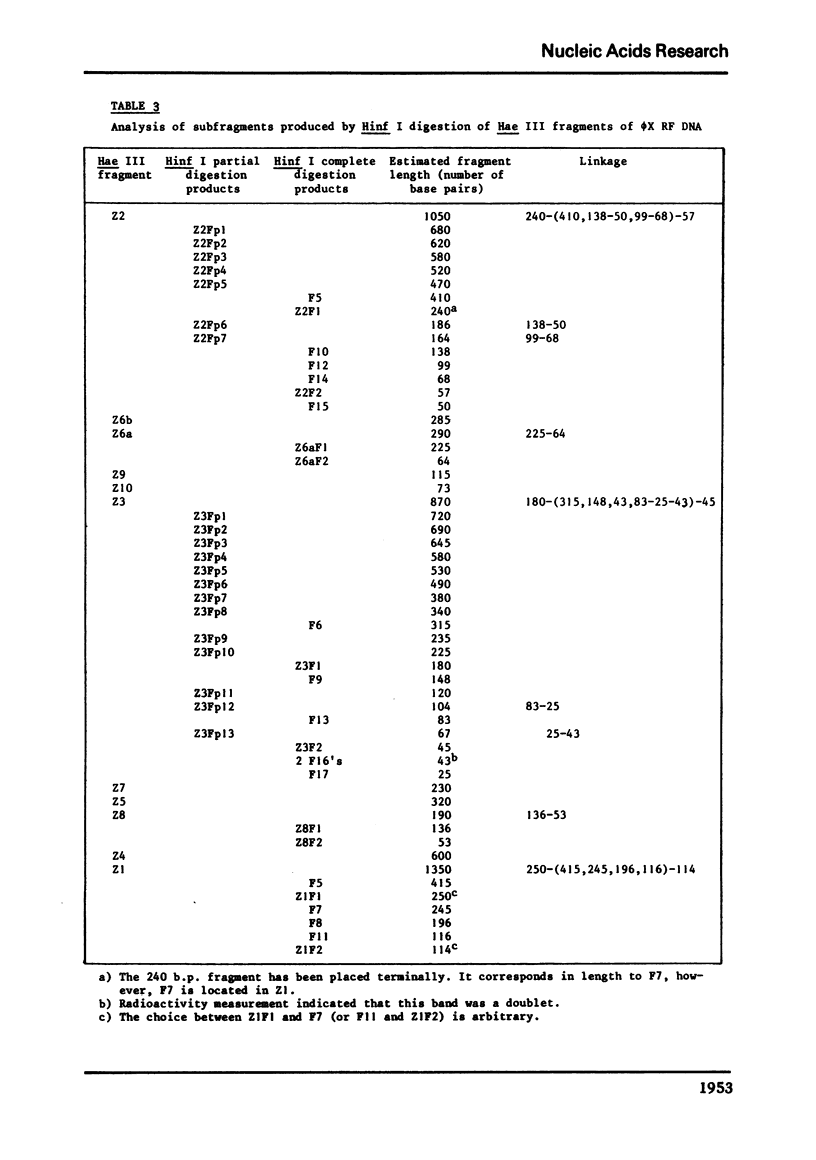
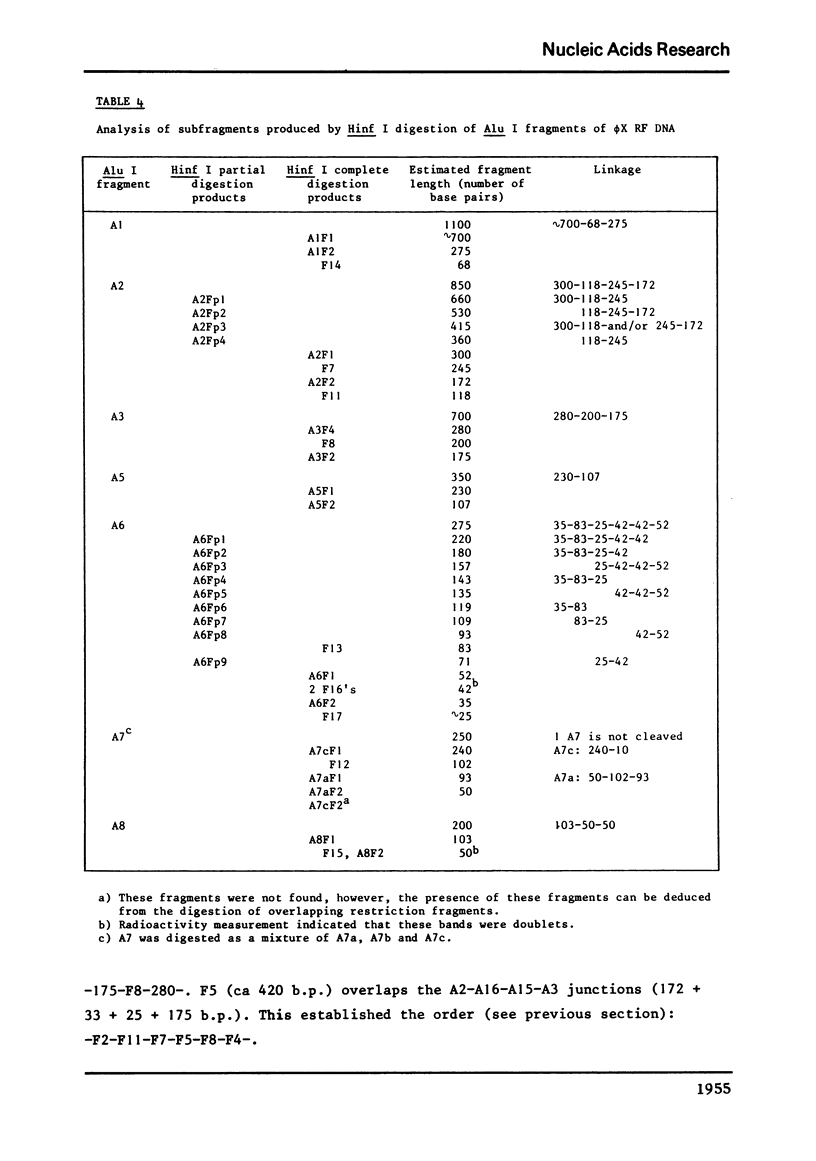
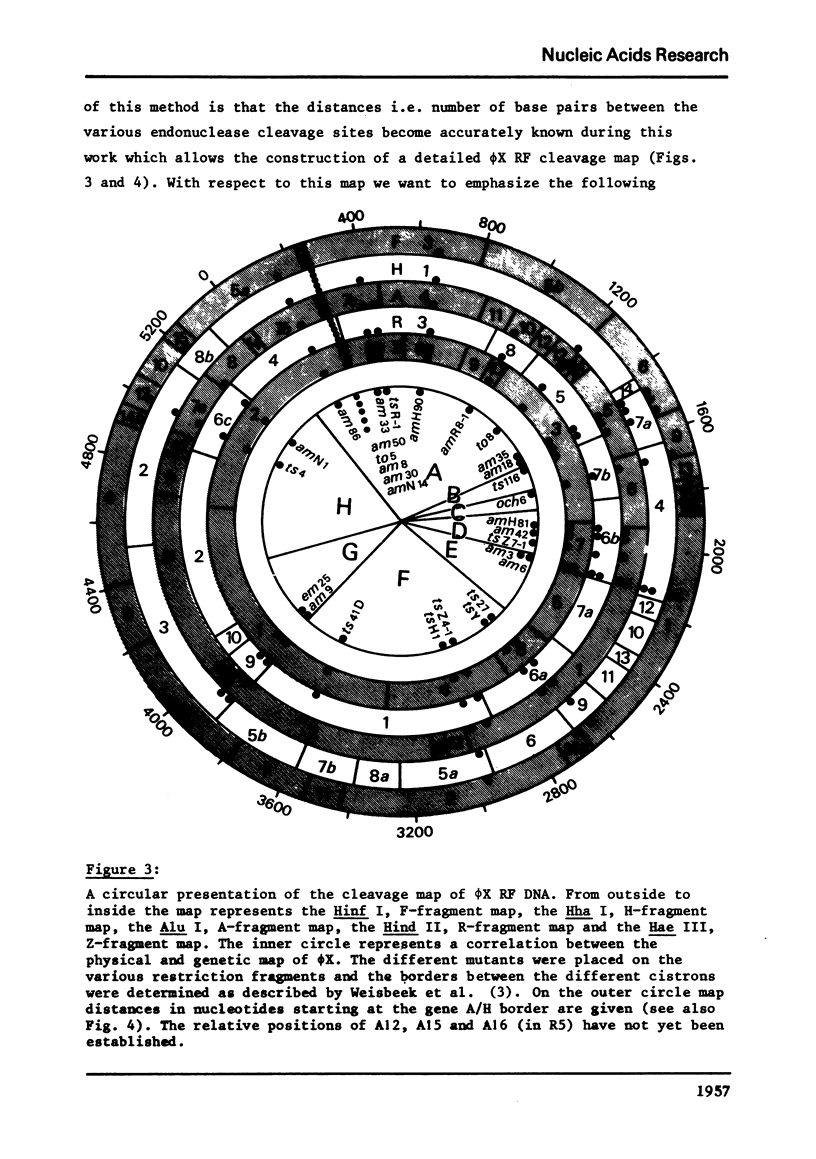
phiX RF DNA was cleaved by restriction enzymes from Haemophilus influenzae Rf (Hinf I) and Haemophilus haemolyticus (Hha. I). Twenty one fragments of approximately 25 to 730 base pairs were produced by Hinf I and seventeen fragments of approximately 40 to 1560 base pairs by Hha I. The order of these fragments has been established by digestion on Haemophilus awgyptius (Hae III) and Arthrobacter luteus (Alu I) endonuclease fragments of phiX RF with Hinf I and Hha1. By this method of reciprocal digestion a detailed cleavage map of phiX RF DNA was constructed, which includes also the previously determined Hind II, Hae III and Alu I cleavage maps of phiX 174 RF DNA (1, 2). Moreover, 28 conditional lethal mutants of bacteriophage phiX174 were placed in this map using the genetic fragment assay (3).
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Air G. M., Blackburn E. H., Sanger F., Coulson A. R. The nucleotide and amino acid sequences of the N (5') terminal region of gene G of bacteriophage phiphiX 174. J Mol Biol. 1975 Aug 25;96(4):703–719. doi: 10.1016/0022-2836(75)90147-3. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Bacteriophage phiX174 DNA synthesis in a replication-deficient host: determination of the origin of phiX DNA replication. J Mol Biol. 1976 Apr 15;102(3):633–656. doi: 10.1016/0022-2836(76)90339-9. [DOI] [PubMed] [Google Scholar]
- Berkowitz S. A., Day L. A. Molecular weight of single-stranded fd bacteriophage DNA. High speed equilibrium sedimentation and light scattering measurements. Biochemistry. 1974 Nov 5;13(23):4825–4831. doi: 10.1021/bi00720a022. [DOI] [PubMed] [Google Scholar]
- Borrias W. E., Wilschut I. J., Vereijken J. M., Weisbeek P. J., van Arkel G. A. Induction and isolation of mutants in a specific region of gene A of bacteriophage phi chi 174. Virology. 1976 Mar;70(1):195–197. doi: 10.1016/0042-6822(76)90251-8. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Hutchison C. A., 3rd, Edgell M. H. Isolation and genetic localization of three phi-X174 promoter regions. Nat New Biol. 1973 Jun 20;243(129):233–236. doi: 10.1038/newbio243233a0. [DOI] [PubMed] [Google Scholar]
- Darby G., Dumas L. B., Sinsheimer R. L. The structure of the DNA of bacteriophage phiX174. VI. Pyrimidine sequences in the complementary strand of the replicative form. J Mol Biol. 1970 Sep 14;52(2):227–238. doi: 10.1016/0022-2836(70)90027-6. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Denhardt D. T. Structure of nascent phiX174 replicative form: evidence for discontinuous DNA replication. Proc Natl Acad Sci U S A. 1974 Mar;71(3):984–988. doi: 10.1073/pnas.71.3.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Denhardt D. T. The mechanism of replication of phiX174 single-stranded DNA. X. Distribution of the gaps in nascent phiX174 replicative form DNA. Biochem Biophys Res Commun. 1974 Nov 27;61(2):532–537. doi: 10.1016/0006-291x(74)90989-9. [DOI] [PubMed] [Google Scholar]
- Godson G. N. Origin and direction phiX174 double- and single-stranded DNA synthesis. J Mol Biol. 1974 Nov 25;90(1):127–141. doi: 10.1016/0022-2836(74)90261-7. [DOI] [PubMed] [Google Scholar]
- HALL J. B., SINSHEIMER R. L. The structure of the DNA of bacteriophage x-174. IV. Pyrimidine sequences. J Mol Biol. 1963 Feb;6:115–127. doi: 10.1016/s0022-2836(63)80127-8. [DOI] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Fragment maps of phiX-174 replicative DNA produced by restriction enzymes from haemophilus aphirophilus and haemophilus influenzae H-I. J Virol. 1974 Nov;14(5):1142–1151. doi: 10.1128/jvi.14.5.1142-1151.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. G., Sanders L., Slocombe P. M. A restriction cleavage map of phiX 174 DNA by pulse-chase labelling using E. coli DNA polymerase. Nucleic Acids Res. 1976 May;3(5):1323–1329. doi: 10.1093/nar/3.5.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. H., Sinsheimer R. L. Structure of an intermediate in the replication of bacteriophage phiX174 deoxyribonucleic acid: the initiation site for DNA replication. J Mol Biol. 1974 Feb 15;83(1):47–61. doi: 10.1016/0022-2836(74)90423-9. [DOI] [PubMed] [Google Scholar]
- Khorana H. G., Agarwal K. L., Büchi H., Caruthers M. H., Gupta N. K., Kleppe K., Kumar A., Otsuka E., RajBhandary U. L., Van de Sande J. H. Studies on polynucleotides. 103. Total synthesis of the structural gene for an alanine transfer ribonucleic acid from yeast. J Mol Biol. 1972 Dec 28;72(2):209–217. doi: 10.1016/0022-2836(72)90146-5. [DOI] [PubMed] [Google Scholar]
- Kiger J. A., Jr, Sinsheimer R. L. Vegetative lambda DNA. IV. Fractionation of replicating lambda DNA on benzoylated-naphthoylated DEAE cellulose. J Mol Biol. 1969 Mar 28;40(3):467–490. doi: 10.1016/0022-2836(69)90166-1. [DOI] [PubMed] [Google Scholar]
- Lee A. S., Sinsheimer R. L. A cleavage map of bacteriophage phiX174 genome. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2882–2886. doi: 10.1073/pnas.71.7.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. S., Sinsheimer R. L. A continuous electroelution method for the recovery of DNA restriction enzyme fragments. Anal Biochem. 1974 Aug;60(2):640–644. doi: 10.1016/0003-2697(74)90279-6. [DOI] [PubMed] [Google Scholar]
- Lee A. S., Sinsheimer R. L. Location of the 5-methylcytosine group on the bacteriophage phi X174 genome. J Virol. 1974 Oct;14(4):872–877. doi: 10.1128/jvi.14.4.872-877.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Arthrobacter luteus. J Mol Biol. 1976 Mar 25;102(1):157–165. doi: 10.1016/0022-2836(76)90079-6. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Haemophilus haemolyticus. J Mol Biol. 1976 May 5;103(1):199–208. doi: 10.1016/0022-2836(76)90060-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Takanami M., Kojo H. Cleavage site specificity of an endonuclease prepared from Heamophilus influenzae strain H-I. FEBS Lett. 1973 Feb 1;29(3):267–270. doi: 10.1016/0014-5793(73)80035-3. [DOI] [PubMed] [Google Scholar]
- Takanami M. Specific cleavage of coliphage fd DNA by five different restriction endonucleases from Haemophilus genus. FEBS Lett. 1973 Aug 15;34(2):318–322. doi: 10.1016/0014-5793(73)80821-x. [DOI] [PubMed] [Google Scholar]
- Vereijken J. M., Van Mansfeld A. D., Baas P. D., Jansz H. S. Arthrobacter luteus restriction endonuclease cleavage map of phi chi 174 RF DNA. Virology. 1975 Nov;68(1):221–233. doi: 10.1016/0042-6822(75)90163-4. [DOI] [PubMed] [Google Scholar]