Abstract
Next-generation sequencing technologies are accelerating gene discovery by combining multiple steps of mapping and cloning used in the traditional map-based approach into one step using DNA sequence polymorphisms existing between two different accessions/strains/backgrounds of the same species. The existing next-generation sequencing method, like the traditional one, requires the use of a segregating population from a cross of a mutant organism in one accession with a wild-type (WT) organism in a different accession. It therefore could potentially be limited by modification of mutant phenotypes in different accessions and/or by the lengthy process required to construct a particular mapping parent in a second accession. Here we present mapping and cloning of an enhancer mutation with next-generation sequencing on bulked segregants in the same accession using sequence polymorphisms induced by a chemical mutagen. This method complements the conventional cloning approach and makes forward genetics more feasible and powerful in molecularly dissecting biological processes in any organisms. The pipeline developed in this study can be used to clone causal genes in background of single mutants or higher order of mutants and in species with or without sequence information on multiple accessions.
Keywords: gene cloning, next-generation sequencing, R gene, abscisic acid, disease resistance
ISOLATING causal genes is essential for the understanding of biological processes that mutations in these genes affect. When a mutant phenotype is generated by insertion of a tag (such as a transposon or a T-DNA) in the genome, the causal gene can be identified through isolating sequences flanking the tag. When it is a result of nucleotide sequence change, deletion/insertion, or an epimodification of nucleotide, the causal genes can be identified by their chromosomal locations or maps on the basis of molecular markers linked to these mutations. In the latter case, the mutant organism in an accession/strain/background is crossed to a wild-type (WT) organism in a different accession to generate a population where both the causal mutation and the DNA sequence polymorphisms between the two accessions are segregating. By identifying polymorphisms that are tightly associated with the mutant phenotype, a map position (chromosome location) of the mutation can be defined and candidate mutations can be identified through sequencing the mutation region (Lukowitz et al. 2000; Jander et al. 2002; Peters et al. 2003). This method has been widely used in diverse organisms such as Drosophila melanogaster and Caenorhabditis elegans. The past two decades have also seen an explosion of use of such map-based cloning in plants to identify important genes in various processes that range from growth and development to biotic and abiotic responses.
Despite being a powerful approach in gene discovery, the classical map-based cloning has several limitations. First, it is dependent on the availability of molecular polymorphisms between genomes of the two accessions. Second, this method is traditionally labor intensive because many mutant plants (often hundreds if not thousands) in a segregating population need to be genotyped to identify recombinants that refine the mutation position. Third, it requires crossing plants in two different accessions to generate a mapping population. If a mutant phenotype is modified by natural variations between accessions, inferring genotype by phenotype will be complicated. Inconveniently for map-based cloning, natural modification is not uncommon in any species. For example, 5% of the Saccharomyces cerevisiae genes that are essential in one accession are not so in another accession, and two to four genes could be required for the phenotypic modification (Dowell et al. 2010). In addition, if a new mutant (such as a suppressor or enhancer) is isolated in the background of an existing mutant, the original mutation needs to be introgressed into another accession before a mapping population for the new mutation can be generated. Introgression is a long process as it is done by reisolating the mutant (usually from the F2 population) from an outcross to a second accession and repeating the process more than five times to obtain a mutant line with the majority of the chromosomes coming from the second accession.
The first two limitations are now overcome by the recent technical advances in next-generation sequencing where vast amounts of sequence information can be acquired in a fast and cost-effective way (Mardis 2008; Shendure and Ji 2008). This technology has revolutionized genomic studies and enabled our understanding of dynamics and functions of cells and population at single-base resolution (Lister et al. 2009). Interrogation of whole genome information on various accessions is becoming more affordable and accessible whether a related reference genome is available (Cao et al. 2011; Gan et al. 2011) or not (Elshire et al. 2011). Although it is possible to reveal mutations in a mutant plant by direct whole genome sequencing when a reference wild-type genome is available, it is almost impossible to identify the causal mutation because there are usually numerous base changes irrelevant to the phenotype in a particular mutant. Therefore, combining the traditional mapping with whole genome sequencing in one step becomes a powerful and effective way to clone genes. Instead of sequencing only one mutant plant (as in direct sequencing of a mutant) or analyzing one plant at a time for hundreds of plants (as in traditional map-based cloning), segregants of the same phenotype from the F2 progenies are pooled for deep sequencing to identify causal mutations in one single step. In Arabidopsis, this method has been used for identifying a number of genes. For instance, SHOREmap used 500 mutant F2 progenies from a cross between a growth defective mutant in the Columbia-0 (Col-0) accession with a wild-type plant in the Ler accession to identify the causal mutation in one next-generation sequencing run of 20× coverage of the genome (Schneeberger et al. 2009). Subsequently, use of pools of much fewer segregants was also successful in identifying genes involved in microRNA precursor processing, cell wall, and growth regulation (Cuperus et al. 2010; Austin et al. 2011; Uchida et al. 2011).
In all these examples of gene cloning with the next-generation sequencing approach, mapping and defining gene location are based on the polymorphisms between two accessions and therefore the third limitation resulting from availability of a proper mapping parent in a different accession still exists. Here, we report the use of pooled segregants from progenies of a backcross to identify causal mutations in a double mutant using next-generation sequencing. A different accession or an introgressed mutant are not needed to create the mapping population because this method relies on polymorphisms generated during mutagenesis. This method therefore has potential for identifying causal mutations in any chemical or physical mutagen-induced mutant even when no traditional mapping parents are available. While we were submitting this article, an independent study identified agronomically important loci in rice with a similar approach (Abe et al. 2012), demonstrating the powerful use of this strategy.
Materials and Methods
Plant growth and phenotype analyses
Arabidopsis growth, disease resistance tests, mapping, and protoplast transformation were carried out as previously described (Zhu et al. 2010). For EMS (ethyl methanesulfonate) mutagenesis, Arabidopsis seeds were treated with 2.5% EMS for 12 hr. Information on markers nga63, ciw12, and M59 are at TAIR (http://www.arabidopsis.org). Primer sequences for the markers JH15 and JHdcaps21 are: JH15-F 5′-CCTGTGTTGGTCATTTGCAC, JH15-R 5′-ACCAATTGCAACAATCATGC, JHdcaps21-F 5′-AACCAAAGTGACTACTAATTCC, and JHdcaps21-R 5′-CGAGAGAAGCTATCCTTCATTGAG. Amount of salicylic acid (SA) and abscisic acid (ABA) were analyzed as previously described (Pan et al. 2008).
Next-generation sequencing
F2 progenies from a cross of int70 snc1-1 and snc1-1 were grown and scored at 28°. Equal amount of leaf tissues were collected from each plant and tissues from mutant and nonmutant plants were pooled separately for genomic DNA extraction using the E.Z.N.A. plant DNA Midi kit (OMEGA Bio-tek, Inc, http://www.omegabiotek.com). DNA libraries for Hi-Seq were constructed according to manufacturer instructions (Illumina, http://www.illumina.com). The libraries were run on Illumina Hi-Seq with single reads of 51 bp.
Results
Isolation of temperature-insensitive disease resistance mutants
Plant disease resistance is modulated by temperature and an elevated temperature often renders an otherwise resistant plant susceptible to pathogen invasion (Wang et al. 2009). To investigate how temperature affects disease resistance, we carried out genetic screens for mutants that retain disease resistance at high temperature in otherwise temperature-sensitive disease resistant mutants (Zhu et al. 2010). BON1(BONZAI1) is a negative regulator of a TIR-NB-LRR type of R (Resistance) gene SNC1 (Suppressor of npr1-1, constitutive 1) and the loss of BON1 function in bon1-1 leads to activation of plant defense responses (Yang and Hua 2004). The snc1-1 (or snc1) has a missense mutation resulting in an autoactive SNC1, conferring constitutive defense responses (Zhang et al. 2003). Both bon1-1 and snc1-1 mutants exhibited a dwarf phenotype at 22° but a wild-type growth phenotype at 28° (Yang and Hua 2004) (Figure 1A). These temperature-sensitive autoimmune mutants were mutagenized with EMS and M2 plants were screened at 28° for dwarf phenotypes. Putative int (insensitive to temperature) mutants were further tested for disease resistance at 28° to identify those with heat-stable disease resistance. Our prior study on the int102 mutant finds that the TIR-NB-LRR protein SNC1 is the component that confers temperature sensitivity to disease resistance mediated by SNC1 (Zhu et al. 2010). Study of another mutant int173 reveals that mutations in the ABA2 (ABA Deficient 2) gene enhanced disease resistance mediated by SNC1 and another R gene RPS4 (Resistance to Pseudomonas Syringae 4) at high temperature (Mang et al. 2012).
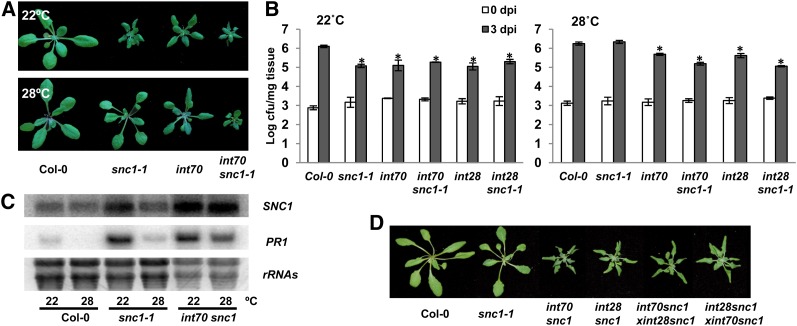
Figure 1 .
The int70 and int28 mutations conferred enhanced disease resistance to snc1-1 mutants at high temperature. (A) The int70 mutation confers a dwarf phenotype to snc1-1 at 28°. Shown are the wild-type Col-0, int70, snc1-1, and int70 snc1-1 plants grown at 22° and 28°. (B) The int70 snc1 and int28 snc1 double mutants displayed enhanced disease resistance to the virulent pathogen Pseudomonas syringae pv tomato (Pst) DC3000 at high temperature. Shown are pathogen amounts in Col-0, snc1-1, int70, int70 snc1-1, int28, and int28 snc1-1 grown at 22° (top) and 28° (bottom) at day 0 and day 3 postinnoculation (dpi). Values represent mean ± SD (n = 3). The asterisks indicate a significant difference from the wild-type Col-0 as determined by Student’s t-test (P ≤ 0.05). Similar results were observed in three independent experiments. (C) Expression of SNC1 and PR1 was increased in int70 snc1-1 at 28°. Shown is the RNA blot of total RNAs from the wild-type Col-0, snc1-1, and int70 snc1-1 plants grown at 22° and 28°. rRNA’s stained with ethidium bromide were used as a loading control. (D) The int70 and int28 mutations are allelic. Shown are the 28° grown plants of the wild-type Col-0, snc1-1, int70 snc1, int28 snc1, a F1 plant from int70 snc1 (female) cross to int28 snc1 (male), and a F1 plant of a reciprocal cross. The F1 plants had a similar growth phenotype to those of the two double mutant parents.
To further investigate the regulation of plant immunity by temperature, we characterized two additional int mutants: int70 and int28. Unlike the snc1-1 plant that is dwarf at 22° but wild type at 28°, the int70 snc1-1 double mutant showed a dwarf phenotype at both 22° and 28° (Figure 1A) and a similar growth phenotype was found in int28 snc1-1. At 22°, both the int70 snc1-1 and the int28 snc1-1 double mutants exhibited an enhanced disease resistance to the virulent pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 similarly to the snc1-1 single mutant when compared to the wild-type Col-0 (Figure 1B). However, both double mutants exhibited an increased resistance to Pst DC3000 at 28° compared to snc1-1, which was as susceptible as the wild-type Col-0 at this temperature (Figure 1B). Correlated with the growth and disease resistance phenotype, expression of the defense response marker gene PR1 as well as the SA-induced SNC1 gene were upregulated in the int70 snc1-1 mutant at 28° compared to snc1-1 (Figure 1C). Because int70 snc1-1 and int28 snc1-1 had a similar phenotype and were mapped to a similar region (see below), we tested whether they have lesions in the same gene. The F1 plants of a cross between these two double mutants exhibited at 28° a dwarf phenotype similar to those of the single mutants, indicating that they are indeed allelic (Figure 1D).
Isolation of the INT70 gene through next-generation sequencing on bulked segregants from a backcross
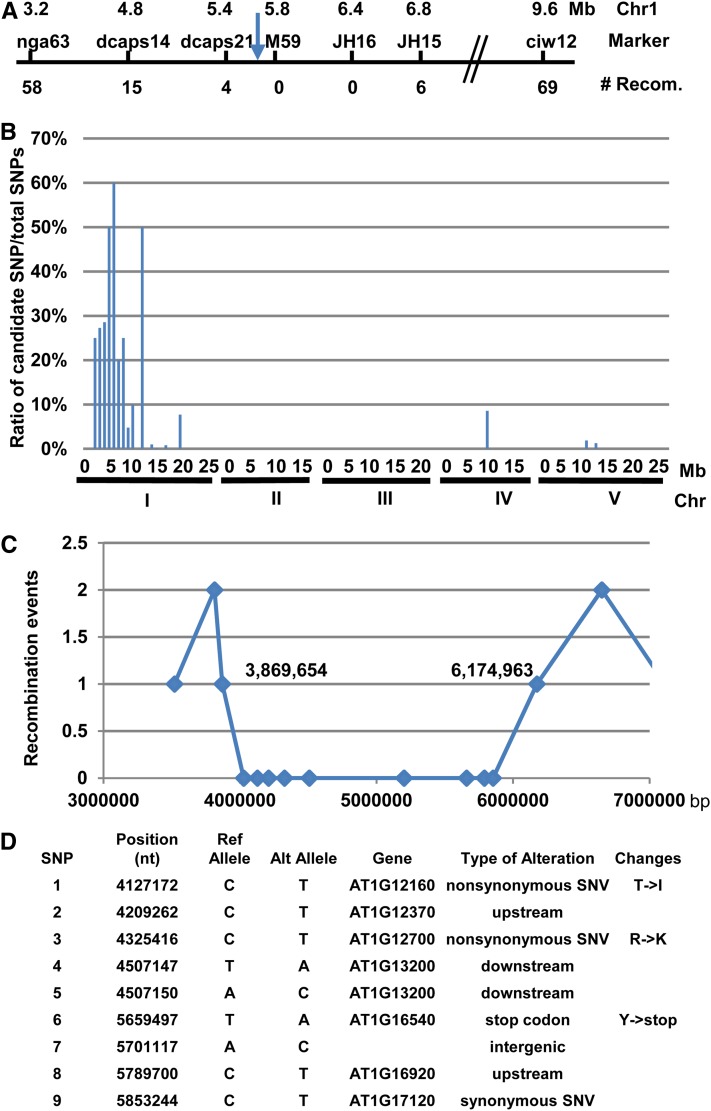
To identify the molecular bases of enhanced disease resistance by the int70 mutation at high temperature, we carried out a traditional map-based cloning to identify the causal mutation. The int70 snc1-1 double mutant in the Col-0 accession was crossed to the wild-type Wassilewskija (Ws) accession. F2 populations were grown at 28° and the “int-” looking plants (segregated at ∼1/16) were selected for mapping using SSLP or CAPS markers between Col-0 and Ws (Lukowitz et al. 2000; Zhu et al. 2010). With bulked segregants of ∼50 int-looking plants, the int70 mutation was mapped to a region on chromosome I flanked by markers nga63 and ciw12. Using a total of 498 mutant plants, the mutation was refined to a region between markers JHdcaps21 and JH15, but no further recombinants could be identified for inner markers M59 or JH16 (Figure 2A). A second mapping cross was made with between int70 snc1-1 and the wild-type Ler plants, but again no further recombinants could be identified internal to JHdcaps21 and JH15 from hundreds of int-looking segregants. Thus, there appears to be a recombination suppression in the region of 1.4 Mb between JHdcaps21 and JH15.
Figure 2 .
Mapping and cloning of the int70 mutation. (A) Traditional mapping of the int70 mutation. Shown are the molecular markers and their positions on chromosome I around the int70 region. Close to 500 mutant plants from the progenies of a cross of int70 snc1-1 (in Col-0) with the wild-type Ws were used to define the position of int70. The number of plants with recombination events at these markers is indicated. The arrow indicates the int70 mutation position. (B) Plot of normalized candidate SNP counts along the five chromosomes. Shown is the ratio (in percentage) of number of candidate SNPs vs. total number of the SNPs in a 1-Mb window. Candidate SNPs are defined as having the nonreference allele being >90% in the mutant pool and <50% in the nonmutant pool. (C) Recombination events detected near the peak of candidate SNPs. X-axis is the nucleotide position on chromosome I and the Y-axis is the number of reference alleles found in the mutant pool. Each positive SNP is represented by a square. (D) Annotation of all nine candidate SNPs in the defined int70 region. Shown are the base position, sequence of the reference allele, sequence of the nonreference allele, its associated gene, type of alteration, and predicted amino acid change for each SNP.
The recombination suppression might have been due to a number of reasons such as a high level of DNA sequence polymorphisms between accessions or large inversions induced by EMS. We thus decided to try using bulked segregants from F2 progenies of a backcross in the same accession background combined with next-generation sequencing for mapping and gene identification. The int70 snc1-1 double mutant in Col-0 was crossed to the original snc1-1 mutant in Col-0, and the int70 snc1-1 double mutant segregated at one out of four in the F2 progenies grown at 28°. Approximately 25 int-looking plants and 50 wild-type–looking plants were collected and pooled, respectively. Genomic DNAs were extracted and purified from these mutant and nonmutant pools and libraries constructed from these DNAs were run in a single lane on the Illumina Hi-Seq machine with single-end reads of 51 bp.
We obtained 40,584,342 reads from the mutant pool and 52,614,692 reads from the nonmutant pool, which are on average 11× and 14× coverage of the Arabidopsis genome, respectively (Supporting Information, Figure S1A). The sequence reads from the two samples were aligned to the Col-0 reference genome (TAIR v10) by the software BWA (Li and Durbin 2009), a fast program to map short reads on a reference genome. SAMtools-mpileup (Li et al. 2009) was used to identify potential SNPs at each position along the five chromosomes. For most of the chromosome regions, 5–10 single nucleotide polymorphisms (SNPs) or small insertions and deletions (INDELs) were identified per megabase and the medium SNP/INDEL count is eight per megabase, indicating one nucleotide sequence difference every 125 kb between int70 snc1 and the wild-type Col-0 (Figure S1A). Several chromosome regions exhibited an extremely high number of SNP/INDELs, and they mostly resided in regions with more sequence read coverage (Figure S1B). Very likely these regions contain repetitive sequences, and misalignment of sequence reads could result in false positive SNP/INDELs. The total number of SNP/INDELs is thus estimated to be ∼950 on the basis of eight SNP/INDELs per megabase and 119 Mb per genome. Almost all the polymorphisms should have been induced during mutagenesis on snc1-1 by EMS (12 hr at 2.5%) because most of the background mutations in the starting strain snc1-1 should have been cleaned by backcrosses. This mutation number is consistent with the earlier TILLING study where ∼1000 mutations were found to be induced by EMS per genome (Colbert et al. 2001).
We mapped the causal mutation on the basis of the frequency of the nonreference (different from the wild-type Col-0 reference) allele of a SNP/INDEL in the mutant pool and the nonmutant pool. This frequency should be 50% in both pools for most of the SNP/INDELs. However, if the nonreference allele of a SNP/INDLEL is the causal mutation, its frequency should be 100% in the mutant pool and close to 33% in the nonmutant pool. In addition, SNP/INDELs linked to the causal gene should also show a high frequency of nonreference alleles in the mutant pool. After empirically testing frequency setting to maximize detection sensitivity, we defined candidate SNP/INDELs (causal and its linked ones) as having the nonreference allele >90% frequency in the mutant pool and <50% in the nonmutant pool. A PERL script was developed to quantify nonreference allele frequency at each polymorphic site and to filter SNPs on the basis of allele frequency. Count of candidate SNP/INDELs was normalized by count of total number of SNP/INDELs in the same region, and the ratio was plotted along the five chromosomes using a 1-Mb sliding window. This normalization effectively reduced noises from false positive SNPs in the repetitive genomic region. This count ratio plot readily revealed one major peak on chromosome I, indicating the position of the int70 mutation, which is consistent with the result from the traditional mapping (Figure 2B).
Closer inspection of SNP/INDELs near the peak revealed recombination events that refined the position of int70 mutation on chromosome I (Figure 2C). A total of nine SNPs in that region were found to have the nonreference allele 100% in the mutant pool and <50% in the nonmutant pool. Flanking these nine SNPs and at positions 3,869,654 bp and 6,174,963 bp, respectively, two SNPs had one reference allele in the mutant pool, indicating recombination events between int70 snc1-1 and snc1-1 at these two sites. On the basis of annotation by ANNOVAR (Wang et al. 2010), nonreference alleles of five of nine SNPs in the int70 region were alterations in the intergenic or noncoding regions, three caused nonsynonymous mutations, and one at position 5,659,497 presumably caused a stop codon that made it a good candidate for int70 mutation (Figure 2, A and D). The reference sequence of this SNP in the wild-type Col-0 is T, and all four reads in the control pool are T, while all nine reads in the mutant pool are A. This mutation resides at nucleotide 33 (translation start being 1) in the first exon of At1g16540, leading to a predicted change of Tyr11 to a stop codon (Figure 3A).
Figure 3 .
The amount of ABA affects SNC1-mediated disease resistance. (A) Mutations in the ABA3 gene in int70 and int28. Shown is the diagram of the gene structure of ABA3. Boxes represent coding sequences and lines represent introns or 5′ and 3′ untranslated regions of the ABA3 gene. Nucleotide changes and the predicted amino acid changes in int70 and int28 are indicated. Mutation positions of earlier aba3 alleles (los5-1, los5-2, aba3-1, and aba3-2) were also indicated for comparison. (B) Mutation of aba3-1 confers int phenotype to snc1-1. Shown are wild-type Col-0, aba3-1, snc1-1, and aba3-1snc1-1 plants grown at 22° and 28°. (C) The int70 mutant is deficient in ABA. Shown are ABA levels in the wild-type Col-0, snc1-1, int70, and int70 snc1 plants before bolting grown at 22° and 28°. (D) SA is accumulated to a high level in the aba3-21 snc1-1 mutant. Shown is the amount of SA in the wild-type Col-0, snc1-1, int70, and int70 snc1 plants grown at 22° and 28°. (E) Expression of GFP fusions of various SNC1 forms at 22° and 28° in protoplasts isolated from the wild-type Col-0 and aba3-21. Shown are GFP images from representative protoplasts. At 28°, all forms of SNC1:GFP (including SNC1WT, SNC1-1, SNC1-3, and SNC1-4) proteins had a very strong nuclear accumulation in aba3-21 while only SNC1-3 and SNC1-4 had a strong nuclear accumulation in the wild-type Col-0. (F) ABA spray partially rescued the growth defect of snc1-1 at 22°. Plant on the right was sprayed with 10 μM ABA twice a day for 3 days at week 4 while plant on the left was sprayed with buffer. Shown are their growth phenotypes at 6 weeks old.
Confirmation of ABA3 as the INT70 gene
This gene At1g16540 was previously identified as LOS5 or ABA3 essential for ABA biosynthesis (Xiong et al. 2001). It encodes a molybdenum cofactor sulfurase that catalyzes the generation of the sulfurylated form of MoCo, a cofactor required by aldehyde oxidase that functions in the last step of ABA biosynthesis (Bittner et al. 2001). If INT70 is ABA3, int28 should also contain a mutation in the ABA3 gene. Sequencing the ABA3 gene in the int28 snc1 mutant indeed revealed a G nucleotide (at 1415 position of cDNA) in exon 12 substituted by an A nucleotide leading to a missense mutation of arginine 472 to lysine (Figure 3B). This R472K mutation is close to the G469E mutation found in aba3-1.
To further verify that mutations in ABA3 are indeed the causal mutations of int70 and int28, we crossed aba3-1 with snc1-1 and isolated the aba3-1 snc1-1 double mutant. This double mutant had a dwarf phenotype at 28° similar to those of the double mutants int70 snc1-1 and int28 snc1-1 (Figure 3B). We thus conclude that ABA3 is the INT70/INT28 gene and renamed int70 as aba3-21 and int28 as aba3-22.
The aba3-21 mutant is presumably a null mutant, due to a predicted stop codon in the very beginning of the ABA3 protein. We analyzed the ABA amount in the aba3-21 single and aba3-21 snc1-1 mutant and found that the ABA level is ∼30% that of the wild-type Col-0 (Figure 3C). The amount of SA is found to be higher in the aba3-21 snc1-1 double mutant at 28° compared to the wild-type Col-0 or the snc1-1 single mutant (Figure 3D), which is correlated with an enhanced disease resistance in the aba3 snc1 mutants at high temperature (Figure 1C). Interestingly, SA amount is higher in aba3-21 than in Col-0 at both temperatures (Figure 3D), which might account for an enhanced disease resistance to Pst DC3000 in the aba3 single mutants aba3-21 (int70) and aba3-22 (int28) compared to the wild-type Col-0 (Figure 1B).
Nuclear accumulation of SNC1 proteins in aba3-21
High temperature inhibits nuclear accumulation of the SNC1 protein and therefore inhibits defense responses (Zhu et al. 2010). Enhanced nuclear accumulation of the SNC1 protein at high temperature was found to confer int phenotypes caused by a missense mutation in SNC1 and an ABA-deficient mutant aba2 (Zhu et al. 2010; Mang et al. 2012). We therefore analyzed the subcellular distribution of various forms of SNC1:GFP fusions in the aba3-21 mutant. Protoplasts were isolated from mesophyll cells of the wild-type Col-0 and the aba3-21 seedlings, and GFP fusions of various SNC1 forms under the control of the strong CaMV 35S promoter were expressed in these protoplasts incubated at 22° and 28°, respectively. The SNC1 forms include the WT, temperature-sensitive autoactive (SNC1-1), and temperature-insensitive autoactive (SNC1-3 and SNC1-4). As reported earlier, SNC1 WT and SNC1-1 had a very strong nuclear accumulation at 22° but not 28°, while SNC1-3 and SNC1-4 had a strong nuclear accumulation at both temperatures (Zhu et al. 2010) (Figure 3E). Strikingly, in aba3-21, all SNC1 forms had a high nuclear accumulation at both 22° and 28° (Figure 3E), correlating with an enhanced disease resistance in the aba3 mutants. This further supports the notion that ABA deficiency enhances nuclear accumulation of the R protein SNC1 and consequently confers heat-stable disease resistance.
While ABA deficiency enhances disease resistance mediated by SNC1, application of ABA apparently suppressed defense responses induced by the autoactive snc1-1 mutant gene. When the snc1-1 plants grown at 22° were sprayed with 10 μM of ABA twice a day for 3 days at 3 weeks old, these plants were much larger 2 weeks later than the snc1-1 sprayed only with buffer control (Figure 3F). This effect is consistent with the earlier finding that ABA application decreased nuclear accumulation of SNC1 proteins (Mang et al. 2012).
Discussion
Here we describe the use of bulked backcross segregants for next-generation sequencing to map and identify causal mutations in a simple and cost-effective way. As diagrammed in Figure 4, the general strategy is to first backcross a mutant isolated from a mutagenesis to the original starting strain (whether it be wild-type or already containing one or multiple mutations) to generate a segregating F2 population. A small number of mutant and nonmutant plants are pooled separately and analyzed by next-generation sequencing. DNA sequence polymorphisms induced by mutagens will be identified from sequence alignment to the reference genome, and ratios of nonreference vs. total SNP/INDELs in the mutant pool will be plotted along the chromosomes. The causal mutation and its associated nonreference SNP/INDELs should appear as a peak in this plot. Inspection of SNP/INDELs around the region in the mutant and nonmutant pools will identify recombination events that define the location of the causal mutation. The identity of causative genes will be confirmed by complementation test and/or characterizing additional mutant alleles.
Figure 4 .
Diagram of mapping and cloning strategy with bulked segregants in backcross progenies. A mutant of interest will be backcrossed to its original starting strain and F2 progenies will be scored for mutant phenotypes. Mutant plants and nonmutant plants will be separately pooled and their DNAs will be subject to next generation sequencing. The sequence reads will be aligned to reference genome, and SNP/INDEL will be called and filtered. SNP ratios will be plotted and mutation locations will be identified as peaks. Candidate SNPs will be annotated and the candidate genes will be tested and confirmed.
This approach differs from the previously described methods using next-generation sequencing on bulked segregants in that it does not rely on polymorphisms existing between two different accessions for mapping. It complements those strategies by offering advantages in several scenarios. First, a mutant line does not need to be crossed to a different accession to generate a mapping population, which eliminates the potential confounding effects of accession differences. Second, for cloning of suppressor or enhancer mutations of an existing mutant, a mapping parent does not need to be created in a different accession. Rather, the starting strain (wild type or with the existing mutation) can be used for backcrosses. This method is also advantageous over direct sequencing of a mutant line or a backcrossed line. Although there was a case of gene cloning by directly comparing the whole genome sequencing of an EMS-induced mutant line to the reference genome (Zuryn et al. 2010), the success is dependent on a complete elimination of random mutations induced by EMS, which may or may not be achieved through multiple backcrosses. By sequencing pools of mutant and nonmutant segregants simultaneously in this approach, unassociated mutations will be identified as having a similar distribution in both pools, while the causal mutation and its linked mutations will be more prevalent in the mutant pool. In addition, even when the mutation is an epigenetic modification, the map position of the mutation can be defined on the basis of its linked SNP/INDELs.
We envision that this approach has a broad application especially in species where genome information for the second accession is not available (such as nontraditional model plants) and in cloning genes from double or even higher order of mutants. Nevertheless, this approach has several limitations. First, SNPs identified from this approach might be false positive due to misalignment of short reads onto the reference genome, while SNPs between accessions are usually validated by more than one approach or sample. Therefore it is necessary to inspect allele frequencies in two pools to identify false positive SNPs. Second, polymorphisms generated by mutagens such as EMS are likely much fewer than those between accessions. EMS induces about a 1-bp change in every 125 kb at the concentration used in mutagenesis, while there is one SNP every 3.3 kb between Col-0 and Ler accessions of Arabidopsis thaliana. Therefore using polymorphisms between accessions has a higher statistic power in mapping than using EMS-induced polymorphisms. However, to fully take advantage of the high polymorphism density between accessions, a much larger number of segregants and a higher genome coverage are needed. For instance, to have recombination between the causal mutation and the nearest SNP that is 3.3 kb away, a total of 7000 plants would need to be fully sequenced assuming 200 kb per centimorgan in Arabidopsis. In this study, the mutant pool was sequenced to 11× coverage, potentially give a mapping resolution of 9 cM or 1.8–2.7 Mb, which was what we found in genome sequencing. The int70 mutation was defined to a 2.3-Mb fragment and there was no recombination of the nine SNPs spanning the 1.6-Mb region. On the basis of the frequency of EMS mutations, the most cost-effective way would have been to sequence a pool of 50 mutant plants and a pool of 50 nonmutant plants each to a 50× genome coverage, as the resolution will be 2 cM or 400–600 kb, which would point to a smaller number of candidate SNPs.
We achieved a similar mapping resolution with a much smaller number of plants through next-generation sequencing as using 500 segregants in a traditional mapping population. Because we did not have a higher coverage of the genome or a larger number of mutant segregants, we were not able to resolve whether the apparent recombination suppression between crosses of two accessions would also occur in the backcross within the same accession. Further data mining using a different alignment method might reveal whether there is an inversion event in the int70 mutant.
This study further supports our earlier findings that ABA deficiency enhances disease resistance and that it does so through affecting nuclear accumulation of NBS-LRR type of proteins. The five int mutants we have isolated so far are snc1-4, bon1-6, aba2-21, aba3-21, and aba3-22 (Zhu et al. 2010; Gou et al. 2011; Mang et al. 2012). The prevalence of ABA-deficient mutants in the int mutants indicates a critical regulation of ABA on TIR-NB-LRR R protein-mediated resistance especially at high temperatures. Decreasing and increasing the ABA amount have opposite effects on SNC1-mediated disease resistance (Figures 1 and 3), which most likely through its effect on the subcellular distribution of SNC1 with ABA levels inversely correlated with nuclear accumulation of SNC1 as observed in this study (Figure 3E) and an earlier study (Mang et al. 2012). Future study on the effect of ABA on the nuclear accumulation of R proteins will likely reveal novel intersecting mechanisms between biotic and abiotic stress responses.
Supplementary Material
Acknowledgments
We thank Xin Li for snc1-1 seeds and the Arabidopsis Biological Resource Center for mutant seeds of aba3-1. This work is supported by National Science Foundation IOS-0919914 to J.H. and the National Science Foundation of China 31170254 to Y.Z.
Footnotes
Communicating editor: S. R. Poethig
Literature Cited
- Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., et al. , 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178 [DOI] [PubMed] [Google Scholar]
- Austin R. S., Vidaurre D., Stamatiou G., Breit R., Provart N. J., et al. , 2011. Next-generation mapping of Arabidopsis genes. Plant J. 67: 715–725 [DOI] [PubMed] [Google Scholar]
- Bittner F., Oreb M., Mendel R. R., 2001. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 276: 40381–40384 [DOI] [PubMed] [Google Scholar]
- Cao J., Schneeberger K., Ossowski S., Gunther T., Bender S., et al. , 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Colbert T., Till B. J., Tompa R., Reynolds S., Steine M. N., et al. , 2001. High-throughput screening for induced point mutations. Plant Physiol. 126: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J. T., Montgomery T. A., Fahlgren N., Burke R. T., Townsend T., et al. , 2010. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. USA 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell R. D., Ryan O., Jansen A., Cheung D., Agarwala S., et al. , 2010. Genotype to phenotype: a complex problem. Science 328: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X., Stegle O., Behr J., Steffen J. G., Drewe P., et al. , 2011. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M., Shi Z., Zhu Y., Bao Z., Wang G., et al. , 2011. The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 69: 411–420 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S. R., Rounsley S. D., Bush D. F., Levin I. M., et al. , 2002. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Gregory B. D., Ecker J. R., 2009. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr. Opin. Plant Biol. 12: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C. S., Scheible W. R., 2000. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang H. G., Qian W., Zhu Y., Qian J., Kang H. G., et al. , 2012. Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of Resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell 24: 1271–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E. R., 2008. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9: 387–402 [DOI] [PubMed] [Google Scholar]
- Pan X., Welti R., Wang X., 2008. Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 1773–1781 [DOI] [PubMed] [Google Scholar]
- Peters J. L., Cnudde F., Gerats T., 2003. Forward genetics and map-based cloning approaches. Trends Plant Sci. 8: 484–491 [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A. H., et al. , 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Shendure J., Ji H., 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Uchida N., Sakamoto T., Kurata T., Tasaka M., 2011. Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 52: 716–722 [DOI] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bao Z., Zhu Y., Hua J., 2009. Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant Microbe Interact. 22: 498–506 [DOI] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhu J. K., 2001. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Hua J., 2004. A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16: 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Goritschnig S., Dong X., Li X., 2003. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1–1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Qian W., Hua J., 2010. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 6: e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S., Le Gras S., Jamet K., Jarriault S., 2010. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics 186: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.