Abstract
Changes in neural activity influence synaptic plasticity/scaling, gene expression, and epigenetic modifications. We present the first evidence that short-term and persistent changes in neural activity can alter adenosine-to-inosine (A-to-I) RNA editing, a post-transcriptional site-specific modification found in several neuron-specific transcripts. In rat cortical neuron cultures, activity-dependent changes in A-to-I RNA editing in coding exons are present after 6 hr of high potassium depolarization but not after 1 hr and require calcium entry into neurons. When treatments are extended from hours to days, we observe a negative feedback phenomenon: Chronic depolarization increases editing at many sites and chronic silencing decreases editing. We present several different modulations of neural activity that change the expression of different mRNA isoforms through editing.
Keywords: RNA, adenosine-to-inosine (A-to-I) RNA editing, neuron, neural plasticity, transcriptional modulation
CHANGES in neural activity can modulate synaptic strength at the level of individual neurons (Burrone et al. 2002) and whole networks (Turrigiano et al. 1998), influence developmental and differentiation decisions (Borodinsky et al. 2004), and bias neurons toward inclusion in novel memory formation (Zhou et al. 2009). At the mRNA level, immediate early genes (IEGs) are transcribed rapidly in response to increased neural activity and have been described as a “genomic action potential” (Clayton 2000). The promoter regions of these and other genes involved in neuronal plasticity can undergo activity-dependent alterations in their chromatin structure, such as cytosine demethylation and histone acetylation (Tsankova et al. 2004; Ma et al. 2009). In addition to the major, genome-wide epigenetic changes known to occur during early development, recent studies have found activity-driven epigenetic modifications in post-mitotic neurons during fear memory recall, exposure to drugs of abuse, and emotional stress (Renthal et al. 2007). We sought to understand if neural activity influences a post-transcriptional genetic modification, adenosine-to-inosine (A-to-I) RNA editing.
A-to-I RNA editing is an enzymatically catalyzed, site-specific nucleotide change (deamination) in pre-mRNA that changes adenosine into inosine, which reads as guanosine during translation (Bass 2002; Nishikura 2010). The best-known example is the developmentally regulated editing of the glutamate receptor subunit Gria2, where A-to-I editing at a single site controls whether GRIA2-containing AMPA receptors are permeable to Ca2+ ions (Sommer et al. 1991). In neurons, many transcripts undergo editing: Previous large-scale genomic screens in Drosophila and human cells found that synapse-related transcripts are enriched in RNA-editing sites (Paul and Bass 1998; Hoopengardner et al. 2003; Li et al. 2009). More generally, inosine is greatly enriched in brain tissue compared to other tissues (Paul and Bass 1998) and the mammalian adenosine deaminases acting on RNA (ADARs) tend to be preferentially or exclusively expressed in the brain (Melcher et al. 1996). Knockout of ADARs in mice, Drosophila, and Caenorhabditis elegans leads to aberrant neurological and behavioral phenotypes (Brusa et al. 1995; Tonkin et al. 2002; Savva et al. 2012). This strongly supports that RNA editing influences neural function, but it is not well understood whether changes in neural activity can affect RNA editing to exert control over transcript diversity and protein function. Previously, it has been shown that editing at two sites in serotonin 2C (Htr2c) pre-mRNA is regulated in a serotonin-dependent manner without any change in transcript expression levels (Gurevich et al. 2002); early life stress and the common antidepressant fluoxetine can also alter Htr2c editing (Englander et al. 2005; Bhansali et al. 2007). These studies suggest a possible role for neural activity in inducing changes in RNA editing, but this connection has not been systematically explored on a genome-wide scale with newer sequencing techniques. In this work, we characterize the effects of membrane potential depolarization and synaptic activity on RNA editing across many editing sites from the coding regions of a diverse group of transcripts.
Results
Acute depolarization of rat cortical neurons for 6 hr but not 1 hr alters A-to-I editing in the coding regions of diverse transcripts
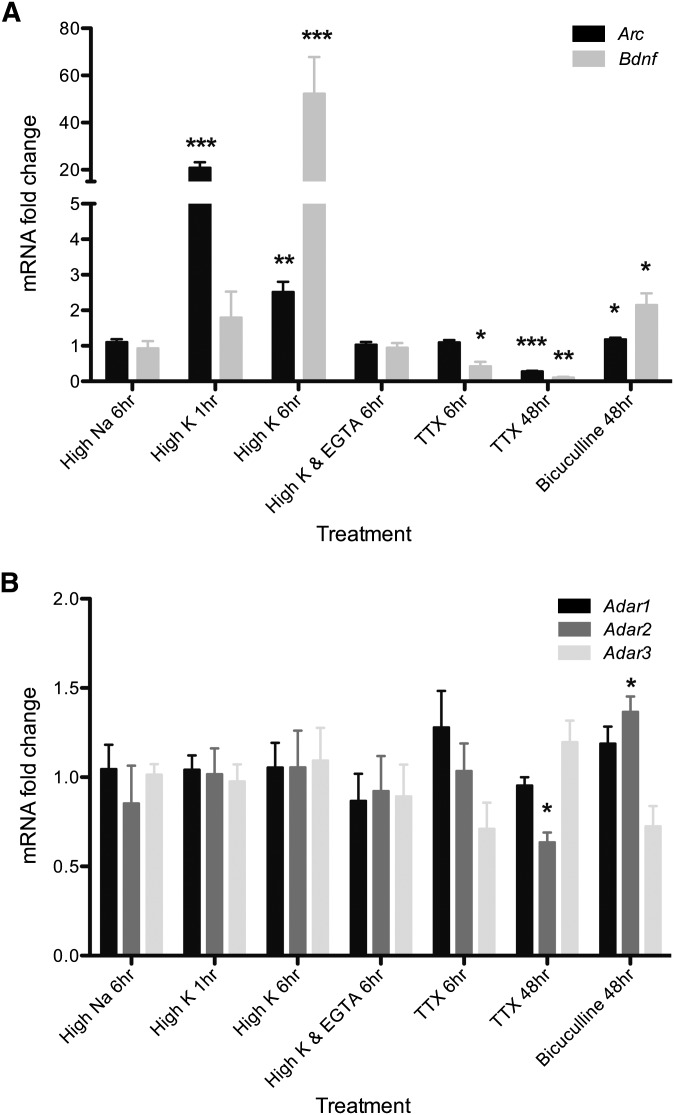
We depolarized primary cultures of neonatal rodent cortical neurons by iso-osmotically increasing potassium to 60 mM for 1 or 6 hr (see Supporting Information, File S1 for full methods). In four biological replicates, we extracted RNA and reverse-transcribed cDNA. To validate the efficacy of our activity induction, we measured the expression of two IEGs, Arc and Bdnf, and observed similar results as reported previously (Figure 1A) (Lin et al. 2008). Arc is rapidly transcribed after 1 hr depolarization (>20-fold), whereas Bdnf transcripts increase significantly only after 6 hr of depolarization (>50-fold). For all experiments, we used the expression of these two IEGs to validate the membrane potential or synaptic activity modulation, as Arc and Bdnf transcripts have previously been shown to increase after depolarization and decrease after inhibition (Shepherd et al. 2006; Aid et al. 2007; Lin et al. 2008). Depolarization for either 1 or 6 hr does not change transcript expression of any of the adenosine deaminases (Adar1-3) required for A-to-I editing (Figure 1B).
Figure 1 .
Transcription of immediate early genes after neuron activity modulation. (A) Comparison of the transcript levels of the immediate early genes Arc and Bdnf between indicated treatments and matched controls using quantitative RT-PCR (qRT-PCR). The High K treatments used 60 mM potassium depolarization buffer. The High Na treatments used a control buffer with sodium substituted for potassium. The High K & EGTA treatment consisted of pretreatment for 30 minutes with 2 mM EGTA and then 6 hours with 60 mM potassium depolarization buffer and 2 mM EGTA. Matched controls were the following: High Na 1 hr (for High Na 6 hr, High K 1 hr); High Na 6 hr (for High K 6 hr, High K and EGTA 6 hr, TTX 6 hr); Neurobasal 48 hr (for Bic 48 hr, TTX 48 hr). Asterisks indicate significant differences in expression from the matched control (*P < 0.05, **P < 0.01, ***P < 0.001; n = 4 cultures, one-sample, two-tailed t-test). Error bars indicate SEM. (B) Relative change in the transcript levels of the adenosine deaminases Adar1, Adar2, and Adar3 between indicated treatments and matched controls using qRT-PCR. Treatments and matched controls are the same as in A.
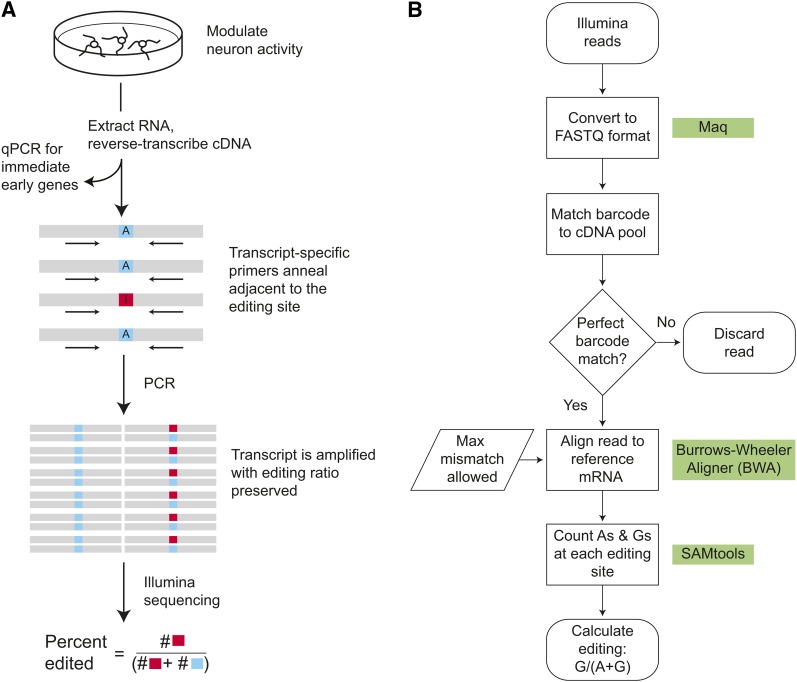
After depolarizing neurons, we quantified the level of RNA editing occurring at 25 rat exonic positions that are conserved and known to be edited in humans (Table 1). Each region was first PCR-amplified, and the amplified transcripts were pooled and sequenced using an Illumina GAIIx (Figure 2A). Compared to standard RNA sequencing (RNA-seq), this targeted deep-sequencing approach can reliably quantify small changes and do so even in rare transcripts, since all editing sites are sampled with roughly the same (large) number of reads (Figure S1). In a single Illumina flow cell, we sequenced reads from multiple experimental treatments and multiple editing sites simultaneously. As shown in Figure 2B, reads are properly identified using treatment-specific barcodes and a Burrows–Wheeler short-read aligner (Li and Durbin 2009). The average number of reads per editing site was 20,000 ± 900 reads (mean ± SEM, n = 25 sites), allowing us to quantify the editing level with <1% error (upper bound on the 95% confidence interval).
Table 1 . Comparison of RNA editing percentage with human homologs.
| Transcript and editing site in rat RefSeq | Mean editing level in Na 1-hr treatment (%, n = 4 cultures) | Standard error of the mean (%) | Previously reported human editing levels at homologous site (%) |
|---|---|---|---|
| Blcap K15R | 16.5 | 1.1 | 11–18, various brain regions (Li et al. 2009) |
| Blcap Q5R | 28.0 | 1.0 | 8–16, various brain regions (Li et al. 2009) |
| Blcap Y2C | 42.9 | 0.6 | ∼30, cerebellum (Kwak et al. 2008) |
| Cadps E1186G | 37.8 | 1.2 | 20–26, various brain regions (Li et al. 2009) |
| Cyfip2 K320E | 50.7 | 0.6 | 84, cerebellum (Kwak et al. 2008); 41–83, various brain regions (Li et al. 2009) |
| Flna Q2333R | 4.5 | 0.5 | 14, cerebellum (Kwak et al. 2008); 27–40, various brain regions (Li et al. 2009) |
| Flnb Q2272R | 20.1 | 2.8 | |
| Gabra3 I342M | 57.6 | 1.9 | 57, frontal lobe (Li et al. 2009) |
| Gria2 Q607R | 99.5 | 0.0 | 95–100, motor neurons (Kawahara et al. 2004); 57–98, various brain regions (Li et al. 2009) |
| Gria2 flip R764G | 51.8 | 1.3 | 58, gray matter (Maas et al. 2001); 17–65, various brain regions (Li et al. 2009) |
| Gria2 flop R764G | 54.7 | 3.0 | |
| Gria3 flip R769G | 83.4 | 1.3 | 48–95, various brain regions (Li et al. 2009) |
| Gria3 flop R769G | 77.0 | 4.6 | |
| Grik1 Q636R | 41.4 | 2.5 | 47, frontal lobe (Li et al. 2009); ∼50, cerebellum and corpus callosum (Barbon et al. 2003) |
| Grik2 I567V | 46.7 | 0.5 | |
| Grik2 Q621R | 45.2 | 2.2 | |
| Grik2 Y571C | 50.2 | 0.2 | |
| Htr2c A | 79.0 | 2.4 | 74–80, various brain regions (Niswender et al. 2001) |
| Htr2c B | 76.5 | 2.9 | 40–60, various brain regions (Niswender et al. 2001) |
| Htr2c E | 3.8 | 1.7 | 16–22, various brain regions (Niswender et al. 2001) |
| Htr2c C | 37.6 | 4.7 | 60–66, various brain regions (Niswender et al. 2001) |
| Htr2c D | 62.8 | 6.4 | 46–66, various brain regions (Niswender et al. 2001) |
| Kcna1 I400V | 16.5 | 2.1 | 20, cerebellum (Kwak et al. 2008); 26–59, various brain regions (Li et al. 2009) |
| Kcnma1 S41G | 2.5 | 0.7 | |
| Neil1 K242R | 1.0 | 0.1 |
Comparison of adenosine-to-inosine editing levels found in this study (rat cortical neurons) and the previously reported levels for the homologous editing sites in humans. All editing levels are given as percentage inosine (or a range of the percentage inosine).
Figure 2 .
Targeted deep-sequencing assay for RNA editing. (A) Schematic of RNA editing percentage quantification. Primary cortical neuron cultures are exposed to various activity-increasing and -decreasing drug treatments, followed by RNA extraction and reverse transcription to cDNA. PCR was used to check for gDNA contamination of the cDNA (Figure S2 and Table S1). To validate activity-modulating treatments, immediate early gene expression is measured using qPCR (Table S2). For each transcript in each condition/replicate, a separate PCR is done to amplify the transcript using primers that anneal a few bases before the editing site(s) of interest (Table S3). Only one transcript and one editing site are shown in the schematic; our experiments included 25 editing sites in 14 transcripts. Blue squares indicate a transcript that is unedited (adenosine at editing site) and red squares indicate an edited transcript (inosine at editing site, which is read as guanosine). A separate PCR is used to attach barcodes and sequencing primers (Table S4 and Table S5), and then all transcripts are sequenced with Illumina. Reads are aligned, and the ratio of reads with guanosine at the editing site over the total reads (adenosine and guanosine) gives the editing percentage. This can be assayed even when the transcript has very few copies in the original cDNA library. (B) Flowchart of data-processing steps to compute the editing percentage at each editing site from Illumina reads. Green squares indicate steps where existing open-source tools were used. Briefly, all Illumina reads were first converted to the standard FASTQ format. Then barcodes were examined to assign each of the (multiplexed) reads back to their respective experimental conditions; barcodes were removed from the read. Any read without an exact-match barcode in the first four bases of the read was discarded entirely. Then reads were aligned to a database of reference sequences surrounding each editing site with the maximum allowable mismatch between the read sequence and the reference sequence determined by the maximum number of editing sites in a single read. Pileup statistics were computed at each editing site for all aligned reads, allowing direct calculation of the editing percentage.
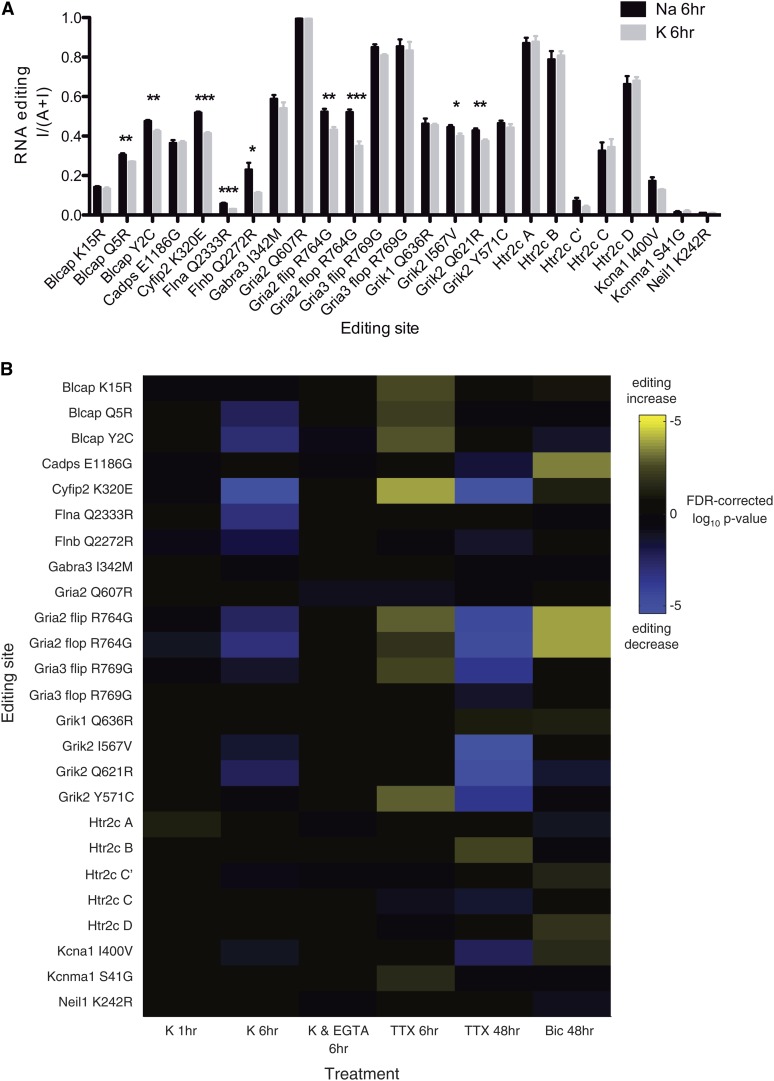
After 1 hr depolarization, we found no differences in editing levels at any of the sites when compared to neurons treated with a control buffer with sodium substituted for potassium. The editing levels in rat cortical neuron culture were generally in good agreement with the corresponding editing levels reported in humans (Table 1). However, after 6 hr of high potassium depolarization, we found small but significant decreases at 9 of the 25 editing sites (P < 0.05, n = 4 cultures, two-sample, unpaired t-test, Benjamini–Hochberg false-discovery rate correction) (Figure 3A). Editing levels changed in a diverse group of transcripts, including AMPA- and kainate-type glutamate receptor subunits (Gria2, Grik2), a fragile X mental retardation 1 interacting protein (Cyfip2), actin-interacting cytoskeletal proteins (Flna, Flnb), and a tumor suppressor protein (Blcap).
Figure 3 .
Activity-dependent changes in RNA editing at 6 and 48 hr. (A) Editing levels (fraction inosine) for High Na 6 hr (control) and High K 6 hr (depolarizing) treatments at each editing site. Asterisks indicate significant differences between the two treatments at that editing site (*P < 0.05, **P < 0.01, ***P < 0.001; n = 4 cultures, two-sample unpaired t-test, Benjamini–Hochberg false-discovery rate correction). Error bars indicate SEM. (B) Heat map showing significance (P) value for differential editing (change in percentage of inosine) at each editing site. Yellow indicates significant increases in editing percentage and blue indicates significant decreases. Intensity denotes log significance. Editing sites are labeled Gene XNY, where Gene denotes the transcript name, X is the amino acid specified if the site is not edited, N is the amino acid position in the rat RefSeq protein record, and Y is the amino acid specified if the site is edited. Differential editing is calculated between each treatment and matched controls (see Figure 1). The difference in editing levels for each site in each condition is presented in Table S6.
We observed that, within a single transcript, not all editing sites show activity-based modulation: For example, the fraction of inosine-containing transcripts at the Gria2 Q/R site is unchanged whereas its R/G site shows a decrease in editing (Figure 3A). We speculate that such decreases in editing within a single transcript cannot be attributed to a simple increase in transcript levels due to the different effects of activity at the two editing sites. However, without additional quantification of transcript levels, a transient uncompensated change in transcript levels cannot be ruled out. For Gria2, editing at the Q/R site has been shown to be a prerequisite for splicing and transcript maturation (Higuchi et al. 2000), which could explain the differential editing. Alternatively, since the Q/R site is edited solely by ADAR2 whereas the R/G site is edited by both ADAR1 and ADAR2, it is possible that activity-induced changes in Gria2 editing are mediated solely by ADAR1. Even when the same ADAR deaminates multiple sites, it is likely that the local double-stranded RNA (dsRNA) structure at each site or other sequence-specific differences can result in different ADAR-binding affinity and degrees of editing. Also, different ADAR isoforms, such as an alternatively spliced ADAR2 truncation with no deaminase activity and a SUMOylated ADAR1 with reduced deaminase activity, and other dsRNA-binding proteins may also influence editing in a site-selective manner through competitive inhibition (Wahlstedt and Ohman 2011).
Neuron activity-dependent A-to-I editing requires calcium entry
Many activity-regulated changes in neurons, such as IEG expression and LTP, depend on calcium entry through N-methyl-D-aspartate (NMDA) and voltage-gated calcium channels (Turrigiano et al. 1998; Guzowski et al. 2000; Greer and Greenberg 2008; Lin et al. 2008). To determine if activity-dependent RNA editing also requires calcium entry, we blocked extracellular calcium by pretreating cultures for 30 min with the calcium chelator ethylene glycol-bis-(2-aminoethyl)-N,N,N′, N′-tetraacetic acid (EGTA) before placing them in high-potassium depolarizing buffer that also contained EGTA for 6 hr. Previous work has shown that blocking calcium entry prevents immediate early gene transcription (Greer and Greenberg 2008; Lin et al. 2008), and, similarly, we found that neither Arc (P = 0.77) nor Bdnf (P = 0.61) expression was significantly different from that of the control. When blocking calcium and depolarizing, none of the 25 editing sites were significantly changed, indicating that entry of extracellular calcium is necessary for activity-dependent RNA editing (Figure 3B).
Blockade of neuron activity induces changes in A-to-I editing in the opposite direction from those induced by depolarization
Since increasing activity alters editing, we wondered if reducing activity could also influence editing. We placed cultured neurons in 2 μM tetrodotoxin (TTX) to block all action potentials for 6 hr and again assayed IEG expression and RNA-editing levels. We found a decrease in Bdnf expression (0.43 ± 0.12, P = 0.04) compared to control cultures (Figure 1A), reflecting the decrease in activity. TTX treatment for 6 hr increased editing at nine different editing sites in six transcripts (Figure 3B). Compared to high-potassium depolarization, we observed RNA-editing changes in the opposite direction—increased editing—at five of the same sites. Editing at these five sites (in Blcap, Cyfip2, and Gria2 transcripts) is differentially modulated depending on whether neural activity increases or decreases, indicating that some editing sites have bidirectional plasticity in their activity-dependent editing changes. As with high-potassium depolarization, we did not observe any changes in Adar1-3 transcript levels (Figure 1B) after TTX blockade of action potentials.
Chronic activity modulation produces distinct changes in A-to-I editing that are largely opposite to those induced by acute changes in neuron activity
Although changes in RNA editing are present after just 6 hr of activity modulation, we also chronically decreased or increased activity for 48 hr using 2 μM TTX or 40 μM GABA-A antagonist bicuculline, respectively, to see if different changes in RNA editing occurred after longer periods of activity modulation. When using TTX to block action potentials for 48 hr, Arc and Bdnf expression was reduced to 0.28 ± 0.02 (P = 4 × 10−4) and 0.11 ± 0.02 (P = 2 × 10−3) of control, respectively (Figure 1A). With chronic TTX, significant changes in editing occurred at 12 sites, more than in any other treatment, and, unlike the acute TTX treatment, all except 1 of the 12 sites reflected decreases in editing from control (Figure 3B).
When using bicuculline to block inhibitory synaptic currents and thus increase membrane depolarization for 48 hr, there were small elevations in Arc (1.18 ± 0.05, P = 0.03) and Bdnf (2.15 ± 0.33, P = 0.02) expression (Figure 1A). As in the case of chronic activity blockade, the changes after 48 hr of increased excitability were in the opposite direction of those seen with acute (6 hr) activity increase (Figure 3B). Of the sites with decreased editing after 6 hr of high-potassium depolarization, only one of them, Grik2 Q621R, had a similar decrease after the 48-hr bicuculline treatment. Both splice isoforms of Gria2 had increases instead of decreases in editing at the R/G site. In Htr2c, chronic bicuculline induced editing at two different editing sites (D and E) from chronic TTX (B and C), suggesting possible opposing signaling roles for these editing sites.
Unlike the acute activity modulations, chronic depolarization and inhibition also induce modest changes in Adar2 transcription (but not in Adar1 or Adar3) that are complementary to the majority of changes in editing seen in each condition (Figure 1B). After chronic TTX treatment, Adar2 transcript is reduced to 0.63 ± 0.06 (P = 7 × 10−3) and, after chronic bicuculline, Adar2 increases to 1.37 ± 0.09 (P = 0.02). ADAR2 deaminates several of the sites with corresponding decreases in editing after chronic TTX and increases in editing after chronic bicuculline, such as Cyfip2 K320E, Kcna1 I400V, and Gria2 R764G. The only site that shows increased editing after chronic TTX, Htr2c B, is deaminated exclusively by ADAR1 and thus would not be expected to display reduced editing in response to decreased ADAR2.
Discussion
Although changes in A-to-I RNA editing were previously known to influence neural function (Hoopengardner et al. 2003; Hideyama et al. 2010), this study is the first demonstration that changes in membrane potential and synaptic receptor activity can modulate RNA editing at many different sites in protein-coding regions. In this study, we focused on characterizing the scope of the activity-induced editing using a targeted deep sequencing approach, in which the absolute abundance (or lack thereof) of a particular transcript is not a bottleneck for accurate quantification of editing. Compared to standard RNA-seq, this helpful dissociation allows examination of editing at many sites simultaneously and requires virtually no optimization to examine new loci. Recently, others have also used a targeted sequencing approach for even quantification of editing at transcripts of variable abundance, such as in mice during development (Wahlstedt et al. 2009) and after antidepressant treatment (Abbas et al. 2010; Morabito et al. 2010) and in human patients with schizophrenia and bipolar disorder (Silberberg et al. 2011). For many of the sites, this is the first report of their editing in rats, and we find that the level of editing at these sites is largely conserved with the corresponding human orthologs (Table 1).
For some of the transcripts that undergo activity-dependent editing, such as glutamate and serotonin receptor subunits, defects in editing have been implicated in common nervous system disorders, including epilepsy, amyotrophic lateral sclerosis, and depression (Maas et al. 2006; Hideyama et al. 2010), yet the individual functional changes resulting from editing, e.g., biochemical, electrophysiological, etc., are known for only a few sites. Since the changes in editing that we observe are often small in magnitude, it is vital to understand the functional impact—if any—of small changes in transcript editing on the nervous system. With the recent expansion of newly identified coding and noncoding editing sites (Li et al. 2009) and the development of new sequencing and analysis techniques (Ramaswami et al. 2012), it is now possible to understand how the ensemble of editing changes allows neurons to adapt to changes in network activity. This initial characterization of how activity can alter RNA editing increases the rich diversity of mechanisms that neurons can use to control the expression of different protein isoforms.
Supplementary Material
Acknowledgments
We thank G. Church and H. S. Seung for support. We also thank J. Foley and H. Sullivan for preparing primary neuron cultures and J. Aach, M. Zody, and T. Sharpe for bioinformatics guidance. K. Ramamoorthi and Y. Lin provided helpful feedback on the neuron activity manipulations and quantitative RT-PCR. E. Wang and Z. Ouyang provided statistics advice. N.E.S. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship. E.Y.L. was supported by the Israel Science Foundation (grant no. 1466/10) and by a Marie Curie Reintegration grant. J.B.L. is supported by a Stanford University Startup Package.
Footnotes
Communicating editor: S. Piali
Literature Cited
- Abbas A. I., Urban D. J., Jensen N. H., Farrell M. S., Kroeze W. K., et al. , 2010. Assessing serotonin receptor mRNA editing frequency by a novel ultra high-throughput sequencing method. Nucleic Acids Res. 38: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aid T., Kazantseva A., Piirsoo M., Palm K., Timmusk T., 2007. Mouse and rat BDNF gene structure and expression revisited. J. Neurosci. Res. 85: 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbon A., Vallini I., La Via L., Marchina E., Barlati S., 2003. Glutamate receptor RNA editing: a molecular analysis of GluR2, GluR5 and GluR6 in human brain tissues and in NT2 cells following in vitro neural differentiation. Brain Res. Mol. Brain Res. 117: 168–178 [DOI] [PubMed] [Google Scholar]
- Bass B. L., 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhansali P., Dunning J., Singer S. E., David L., Schmauss C., 2007. Early life stress alters adult serotonin 2C receptor pre-mRNA editing and expression of the alpha subunit of the heterotrimeric G-protein G q. J. Neurosci. 27: 1467–1473 [DOI] [PMC free article] [PubMed]
- Borodinsky L. N., Root C. M., Cronin J. A., Sann S. B., Gu X., et al. , 2004. Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature 429: 523–530 [DOI] [PubMed] [Google Scholar]
- Brusa R., Zimmermann F., Koh D. S., Feldmeyer D., Gass P., et al. , 1995. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270: 1677–1680 [DOI] [PubMed] [Google Scholar]
- Burrone J., O’Byrne M., Murthy V. N., 2002. Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature 420: 414–418 [DOI] [PubMed] [Google Scholar]
- Clayton D. F., 2000. The genomic action potential. Neurobiol. Learn. Mem. 74: 185–216 [DOI] [PubMed] [Google Scholar]
- Englander M. T., Dulawa S. C., Bhansali P., Schmauss C., 2005. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J. Neurosci. 25: 648–651 [DOI] [PMC free article] [PubMed]
- Greer P. L., Greenberg M. E., 2008. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59: 846–860 [DOI] [PubMed] [Google Scholar]
- Gurevich I., Englander M. T., Adlersberg M., Siegal N. B., Schmauss C., 2002. Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J. Neurosci. 22: 10529–10532 [DOI] [PMC free article] [PubMed]
- Guzowski J. F., Lyford G. L., Stevenson G. D., Houston F. P., McGaugh J. L., et al. , 2000. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J. Neurosci. 20: 3993–4001 [DOI] [PMC free article] [PubMed]
- Hideyama T., Yamashita T., Suzuki T., Tsuji S., Higuchi M., et al. , 2010. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J. Neurosci. 30: 11917–11925 [DOI] [PMC free article] [PubMed]
- Higuchi M., Maas S., Single F. N., Hartner J., Rozov A., et al. , 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81 [DOI] [PubMed] [Google Scholar]
- Hoopengardner B., Bhalla T., Staber C., Reenan R., 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832–836 [DOI] [PubMed] [Google Scholar]
- Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., et al. , 2004. Glutamate receptors: RNA editing and death of motor neurons. Nature 427: 801. [DOI] [PubMed] [Google Scholar]
- Kwak S., Nishimoto Y., Yamashita T., 2008. Newly identified ADAR-mediated A-to-I editing positions as a tool for ALS research. RNA Biol. 5: 193–197 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. B., Levanon E. Y., Yoon J. K., Aach J., Xie B., et al. , 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Lin Y., Bloodgood B. L., Hauser J. L., Lapan A. D., Koon A. C., et al. , 2008. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455: 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D. K., Jang M. H., Guo J. U., Kitabatake Y., Chang M. L., et al. , 2009. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323: 1074–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S., Patt S., Schrey M., Rich A., 2001. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. USA 98: 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S., Kawahara Y., Tamburro K. M., Nishikura K., 2006. A-to-I RNA editing and human disease. RNA Biol. 3: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher T., Maas S., Herb A., Sprengel R., Higuchi M., et al. , 1996. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 271: 31795–31798 [DOI] [PubMed] [Google Scholar]
- Morabito M. V., Ulbricht R. J., O’Neil R. T., Airey D. C., Lu P., et al. , 2010. High-throughput multiplexed transcript analysis yields enhanced resolution of 5-hydroxytryptamine 2C receptor mRNA editing profiles. Mol. Pharmacol. 77: 895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79: 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender C. M., Herrick-Davis K., Dilley G. E., Meltzer H. Y., Overholser J. C., et al. , 2001. RNA editing of the human serotonin 5–HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology 24: 478–491 [DOI] [PubMed]
- Paul M. S., Bass B. L., 1998. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 17: 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., Lin W., Piskol R., Tan M. H., Davis C., et al. , 2012. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods 9: 579–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W., Maze I., Krishnan V., Covington H. E., III, Xiao G., et al. , 2007. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56: 517–529 [DOI] [PubMed] [Google Scholar]
- Savva Y. A., Jepson J. E., Sahin A., Sugden A. U., Dorsky J. S., et al. , 2012. Auto-regulatory RNA editing fine-tunes mRNA re-coding and complex behaviour in Drosophila. Nat. Commun. 3: 790
- Shepherd J. D., Rumbaugh G., Wu J., Chowdhury S., Plath N., et al. , 2006. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52: 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg G., Lundin D., Navon R., Ohman M., 2011. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum. Mol. Genet. 21: 311–321 [DOI] [PubMed] [Google Scholar]
- Sommer B., Kohler M., Sprengel R., Seeburg P. H., 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19 [DOI] [PubMed] [Google Scholar]
- Tonkin L. A., Saccomanno L., Morse D. P., Brodigan T., Krause M., et al. , 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 21: 6025–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N. M., Kumar A., Nestler E. J., 2004. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J. Neurosci. 24: 5603–5610
- Turrigiano G. G., Leslie K. R., Desai N. S., Rutherford L. C., Nelson S. B., 1998. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature 391: 892–896 [DOI] [PubMed] [Google Scholar]
- Wahlstedt H., Ohman M., 2011. Site-selective vs. promiscuous A-to-I editing. Wiley Interdiscip. Rev. RNA 2: 761–771 [DOI] [PubMed] [Google Scholar]
- Wahlstedt H., Daniel C., Enstero M., Ohman M., 2009. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 19: 978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Won J., Karlsson M. G., Zhou M., Rogerson T., et al. , 2009. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat. Neurosci. 12: 1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.