Abstract
The experimental malleability and unique phylogenetic position of the sea squirt Ciona intestinalis as part of the sister group to the vertebrates have helped establish these marine chordates as model organisms for the study of developmental genetics and evolution. Here we summarize the tools, techniques, and resources available to the Ciona geneticist, citing examples of studies that employed such strategies in the elucidation of gene function in Ciona. Genetic screens, germline transgenesis, electroporation of plasmid DNA, and microinjection of morpholinos are all routinely employed, and in the near future we expect these to be complemented by targeted mutagenesis, homologous recombination, and RNAi. The genomic resources available will continue to support the design and interpretation of genetic experiments and allow for increasingly sophisticated approaches on a high-throughput, whole-genome scale.
Keywords: ascidian, development, transgenesis, electroporation
THE sea squirt Ciona intestinalis (Figure 1) has recently emerged as a powerful model organism for biological research in the postgenomic era. Ever since their days as a favorite of early developmental biologists like Laurent Chabry (Chabry 1887), Ed Conklin (Conklin 1905), and even famed geneticist Thomas Hunt Morgan (Morgan 1923), sea squirts, or ascidians, have been recognized as choice organisms for experimental embryology due to their simple embryos, rapid development, and ease of manipulation. These advantages, coupled to their amenability to genomic and systems approaches (Satoh et al. 2003), today drive the sure but steady acceptance of ascidians into the mainstream of developmental biology.
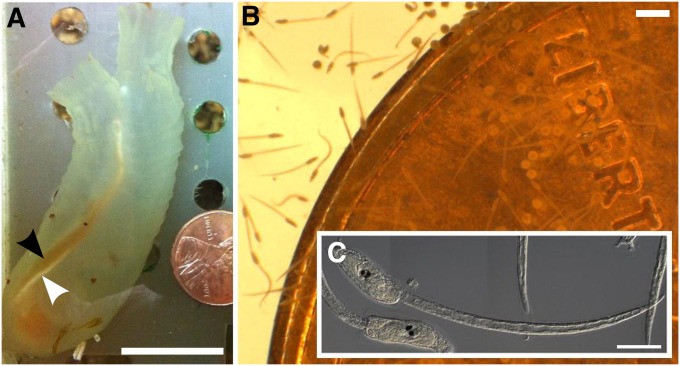
Figure 1 .
The sea squirt Ciona intestinalis. (A) Medium-sized adult type A C. intestinalis. Adults are typically 5–15 cm in length when removed from the wild. Notice egg (black arrowhead) and sperm (white arrowhead) ducts laden with gametes. An adult this size will carry >500 eggs. The animal is next to a United States one cent coin, for scale. (B) Tadpole larvae of C. intestinalis, swimming next to the same coin as in A. (C) DIC image of two C. intestinalis larvae. Bars in A, ∼2 cm; B, ∼100 µm; and C, ∼50 µm.
Ascidians are a paraphyletic group within the subphylum Tunicata in the chordate phylum. For a long time it was believed that cephalochordata as the sister group to the vertebrates. However, improved phylogenetic methods and sequencing of tunicate (also known as “urochordate”) and cephalochordate genomes clearly confirmed that the tunicates, although highly divergent in body plan and genomic architecture, are our closest living invertebrate relatives (Delsuc et al. 2006; Putnam et al. 2008). This phylogenetic position has been an important factor for the adoption of the sea squirt as a “model organisms” for the biomedical sciences.
We will not provide an in-depth discussion of all the advantages or disadvantages of using ascidians, more specifically Ciona spp., in biological research. Nor do we intend this review to be a primer on ascidian biology. These have been covered elsewhere (Christiaen et al. 2009c; Lemaire 2009, 2011). Instead, we will focus on the genetic tools available to the biologist studying C. intestinalis and related species. These include not only the traditional approaches of forward and reverse genetic screens, but also molecular genetic techniques, especially those based on transient transfection of whole embryos with plasmid DNA. The two major experimental species, C. intestinalis and C. savignyi, are weakly cross-fertile (Byrd and Lambert 2000) and are treated here almost interchangeably, referred to as simply Ciona. In fact, evidence points to the grouping of more than one cryptic species within C .intestinalis (Suzuki et al. 2005; Iannelli et al. 2007), so it would be more appropriate to talk about a Ciona species complex. Embryogenesis is virtually identical in all Ciona spp., despite neutral genomic sequence variation 10 times greater than that observed between rat and mouse (Hill et al. 2008).
Other ascidian species utilized as experimental species in the laboratory include Halocynthia roretzi, Phallusia mammilata, Molgula spp., Ascidiella aspersa, Ecteinascidia turbinata, and Botryllus schlosseri. Extreme conservation of embryonic development among these distantly related species is a general rule (Lemaire 2009), in spite of profound differences in genomic sequence (Oda-Ishii et al. 2005) or in choice of cell signaling pathway used for induction of a given cell fate (Tokuoka et al. 2007; Hudson and Yasuo 2008). This dichotomy of phenotypic constancy and molecular divergence is of great interest to those studying the evolution of development. Overall, the remarkable conservation of ascidian embryogenesis also provides some hope that most techniques described here might be readily applied to other sea squirt species.
Forward Genetics
The latest techniques and strategies for genetic screens in Ciona have recently been reviewed and outlined in exceptional detail (Veeman et al. 2011). Here we attempt to summarize basic aspects of doing forward genetics in Ciona. One particular advantage that has motivated the development of forward genetics in Ciona, especially for developmental studies, is the minimal overlap in gene functions. Due to tunicates having branched off from vertebrates before the latter underwent two whole-genome duplication events (Dehal and Boore 2005), several paralogous gene families in vertebrates are each represented by a single ortholog in Ciona. This means the requirement for these genes can be readily tested without the need for double or triple mutants to circumvent such overlap in gene function.
The generation time of Ciona is 1–3 mo. Under certain culturing conditions, sperm can be obtained in 1 mo and eggs after 2 mo (Sasakura et al. 2003). Most solitary ascidians are hermaphroditic broadcast spawners. This presents advantages as well as disadvantages to the geneticist studying them. One advantage is that Ciona adults show a modest ability to self-fertilize. This allows one to screen for recessive mutations in the progeny of selfed F0 animals. Second, their broadcast spawning strategy is tied to their large effective population sizes, which has resulted in extreme polymorphism rates (Small et al. 2007b). Genome-wide average single nucleotide polymorphism (SNP) heterozygosity is at 1.2% in C. intestinalis and reaches 4.5% in C. savignyi (Dehal et al. 2002; Kim et al. 2007; Small et al. 2007b). In other words, any two Ciona individuals from the same population are as different from one another, at the sequence level, as humans are from chimpanzees. This means that there is no shortage of SNPs to use as genetic markers, averaging a SNP or indel per 80 bp of genome. Indeed, the latest positional mapping strategies in Ciona use direct sequencing of SNPs (Veeman et al. 2011).
Unfortunately, the elevated levels of polymorphism make identifying the particular mutation underlying a phenotype difficult to pinpoint. Nevertheless, successful identification of genes underlying mutant phenotypes has been carried out. Screening for short-tailed mutants revealed essential roles for prickle and laminin α3/4/5 in notochord cell polarity and convergence (Figure 2, B–F) (Jiang et al. 2005; Veeman et al. 2008). These mutants were identified by screening progeny from self-fertilized “wild-caught” Ciona. This demonstrates how the extreme natural variation in wild Ciona populations also provides a wealth of naturally occurring recessive mutations. Additionally, N-ethyl-N-nitrosourea (ENU) has been successfully used to induce mutations (Figure 2A) (Chiba et al. 2009). Mutant strains are propagated for distribution by the Ascidian Stock Center at the University of California, Santa Barbara (http://www.ascidiancenter.ucsb.edu/). Currently, there are no isogenic inbred lines available for Ciona.
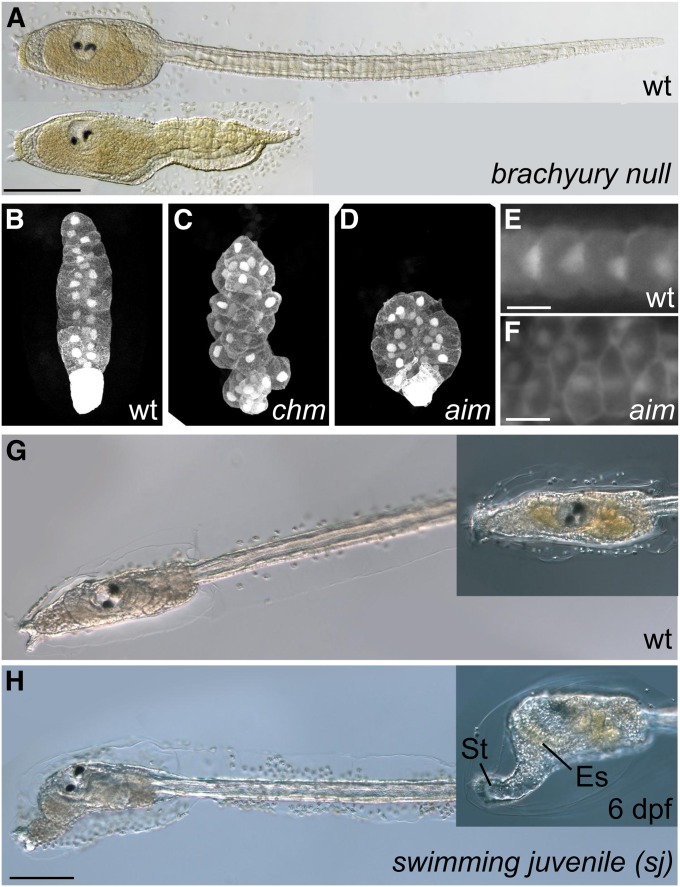
Figure 2 .
Ciona mutants. (A) Wild type (top) and ENU-induced Brachyury (Bra) homozygous null (bottom) larvae of C. savignyi. Brachyury is a transcription factor required for notochord cell specification. Bra null mutants show severe disruption of tail morphogenesis, due to improper specification of convergent-extending notochord cells. (B–D) Notochord cells of (B) wild-type and homozygous, (C) chonmague (chm), and (D) aimless (aim) mutant C. savignyi embryos. Chm and aim are naturally occurring alleles screened from progeny of self-fertilized wild animals. Chm encodes a homolog of Laminin α3/4/5, and aim encodes the planar cell polarity (PCP) pathway component Prickle. Both are downstream of Brachyury and are required for proper notochord cell convergent extension. (E and F) Besides its role in regulating mediolateral polarity during convergent extension, Prickle is involved in regulation of later PCP in the notochord. Note asymmetric localization of nuclei at the posterior end of wild-type notochord cells, contrasted to anterior or random placement of nuclei in notochord cells of an aim mutant. (G) Wild-type and (H) swimming juvenile (sj) mutant C. intestinalis larvae. sj mutation is due to a Minos retrotransposon insertion in the cellulose synthase A (CesA) gene. The synthesis of cellulose (a major component of the larval and adult tunics) is impaired in sj mutants, resulting in larvae that metamorphose without settlement on a solid substrate and without apoptosis of tail structures. Note elongated stalk (St) and endostyle (Es), hallmarks of metamorphosis, in larvae that are still swimming by day 6 after fertilization (6 dpf). Bars in A, 50 µm; E and F, 10 µm; and H, 100 µm. A is adapted from Chiba et al. 2009 and B–D, from Veeman et al. 2008 with permission of the authors and Development/The Company of Biologists. E and F are reprinted from Jiang et al. (2005) with permission of Elsevier and the authors, and G and H, from Sasakura et al. (2005), with permission of the authors.
Ciona also present unique challenges relating primarily to their captivity and husbandry. As marine filter-feeders, they require circulating sea water. Both open circulation systems utilizing a natural source of sea water and food as well as closed circulation systems utilizing cultured microplankton as a food source have been developed to rear Ciona (Hendrickson et al. 2004; Joly et al. 2007). Open systems are easier to maintain, but may be difficult to set up, depending greatly on local availability of microplankton-laden sea water. Closed systems can be set up far inland, but may encounter problems such as inadequate food supply, water fouling, pests, etc. Nonetheless, the breeding and rearing of Ciona in the laboratory is feasible and stands to be further developed as more groups begin to incorporate genetic screens and genetic engineering in their studies (see below).
Reverse Genetics
The sequencing of the C. intestinalis (Dehal et al. 2002) and C. savignyi (Vinson et al. 2005; Small et al. 2007a) genomes in the early to mid 2000s immediately made reverse genetics in Ciona quite alluring. This is especially the case for studies in developmental biology where a limited number of candidate genes identified from genomic and EST sequences are predicted to effect early embryogenesis. For instance, of ∼385 annotated transcription factor genes predicted from the genome, only 65 are zygotically expressed before gastrulation (Imai et al. 2004). That is a very short list of genes potentially controlling most of early Ciona embryogenesis, tempting many a researcher to derive specific working hypotheses that can be addressed by targeting a small number of selected genes.
Transient genetic perturbations
Reverse genetics traditionally involves the generation of mutations at known positions of the genome prior to phenotyping. For those without the resources or infrastructure to build and maintain a marine culturing system in which to raise mutants, many modern molecular techniques are available for transient genetic perturbations in wild-caught Ciona. An abundance of gravid adults can be found in natural and artificial harbors across the globe, in some places year round (Lambert and Lambert 1998). Adults are simply pried away from the substrate and brought to the laboratory where they are kept alive in tanks containing artificial sea water for up to 3 weeks without feeding. Animals can even be “seeded” on detachable substrates and left to fend for themselves until sexual maturity in such places. However, care should be taken to not introduce the species to a location where it does not already occur, as Ciona are notoriously invasive and can cause serious ecological and economic damage (Edwards and Leung 2008).
Microinjection
Thousands of synchronized embryos can be obtained by simply mixing gametes from two or three cultured or wild-caught animals. These embryos are then ready for any number of experiments. Traditionally, the most common manipulation of gene function was through microinjection of mRNA, and to a lesser extent, plasmid DNA (Hikosaka et al. 1993; Imai et al. 2000). mRNA microinjection is still recommended for examining the role of maternally expressed genes, and is the method of choice for sea squirt species for which a suitable DNA electroporation protocol has not yet been established, such as H. roretzi (Kumano et al. 2011).
Researchers studying Ciona embryogenesis have made great use of injected morpholino oligonucleotides (“morpholinos”) to inhibit mRNA translation or splicing, through hybridization to start codon or splicing signal sequences, respectively (Satou et al. 2001a; Heasman 2002). The unnatural backbones of morpholinos preclude their degradation by the cellular machinery. Thus, morpholinos are stable enough to persist throughout embryogenesis, irreversibly inhibiting bound target mRNAs.
Microinjection of Ciona eggs is a relatively labor-intensive technique primarily due to the small size of the ascidian egg (0.1 mm in diameter). Compare this to the egg size of other model chordates such as zebrafish (0.7 mm), Xenopus laevis (1–1.3 mm), and X. tropicalis (0.7–0.8 mm). The elastic nature of the oocyte cortex also makes handling and microinjection of Ciona eggs comparable in difficulty to microinjection of mammalian oocytes (Christiaen et al. 2009b). Published results typically report ∼20–60 successfully injected survivors per morpholino tested.
Despite the relative difficulty of morpholino injections, commitment and hard work can make it pay off in a big way. It was using morpholinos that the provisional gene regulatory “blueprint” of the Ciona embryo was generated. Imai and colleagues assayed the expression of 73 developmental regulators at different embryonic stages, in 25 different morphant backgrounds generated by morpholino injection. This revealed over 3000 gene regulatory connections operating in the early Ciona embryo (Imai et al. 2006). Thus, microinjection skills are, for now, indispensable for a rigorous and thorough investigation of gene function in Ciona.
Electroporation
Due to the constraints of the microinjection technique, a protocol for electroporating plasmid DNA into fertilized, single-cell zygotes was developed (Corbo et al. 1997). Although electroporation has been used to transfect cells in tissue culture or in avian embryos, as far as we know Ciona are the only animals for which electroporation is used to introduce DNA to whole embryos, en masse. For this technique, plasmid DNA is simply mixed with embryos at the one-cell stage. This mixture is placed in an electroporation cuvette, which is subjected to an electric current, much akin to electroporation of cultured mammalian cells (Chu et al. 1987). It is no exaggeration to say this revolutionized the study of Ciona embryogenesis as thousands of synchronized, transgenic embryos could be generated at the push of a button. Today, electroporation remains the main molecular genetic technique used in most Ciona laboratories worldwide (Zeller 2004).
After cloning and preparation of plasmid DNA, the entire procedure from adult dissection to fertilization, dechorionation, and electroporation of embryos, takes ∼1 hr. (Christiaen et al. 2009a). Several plasmids can be tested in hundreds of embryos each, daily. The sheer numbers of transfected embryos generated in short time by the electroporation technique allow for robust, large-scale yet rapid and low-cost phenotyping not possible in other chordate model organisms. In one such instance, Brown and colleagues were able to assay 85,506 transgenic embryos resulting from 1237 electroporations (Brown et al. 2007).
The typical plasmid electroporated into Ciona embryos consists of a driver and a transgene. We refer to the combination of distal cis-regulatory (“enhancers”) and basal promoter elements as the “driver.” The compact genomes of Ciona species allow for easy cloning and testing of intergenic and/or intronic sequences for enhancer activity. This has been facilitated by a shared library of plasmids that allows for the Gateway method of restriction-free, modular assembly of driver/transgene combinations (Roure et al. 2007). Several lineage-, tissue- and cell-type–specific drivers (Figure 3) have been described and are constantly being discovered and added to the researcher’s toolkit. Transgenes can be reporter genes (“reporters”) and/or perturbation genes. We refer to perturbation genes as simply any sequence expected to have a biological function in Ciona. We will later discuss which types of perturbation genes are frequently used in Ciona and which are still but promising possibilities.
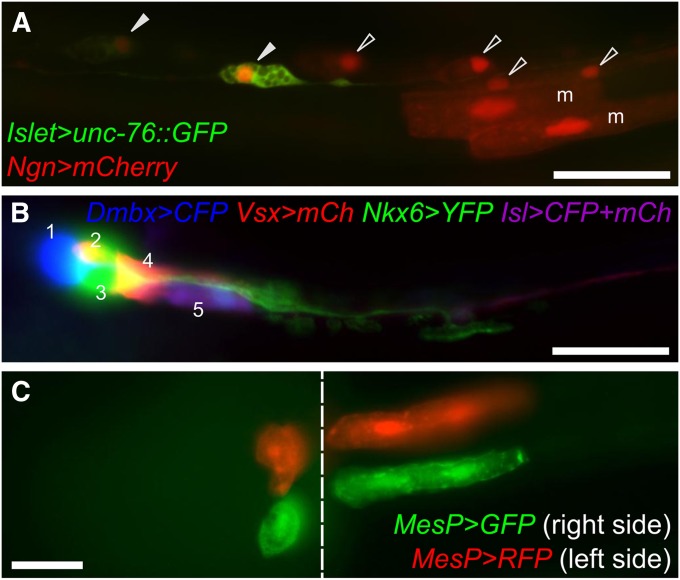
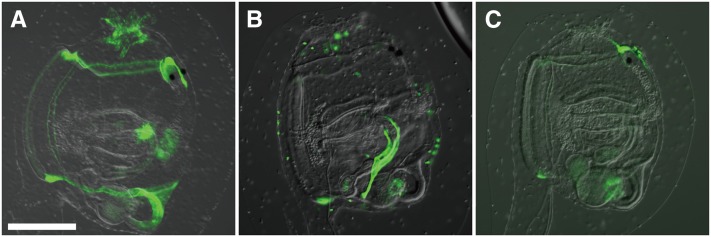
Figure 3 .
Electroporation of reporter gene plasmids. (A) Posterior region of the tail of a midtailbud-stage embryo, coelectroporated with Islet > unc-76::GFP (green) and Ngn > mCherry (red) reporter constructs. Ngn is expressed in A- and b- neural plate border cells, which gives rise to ependymal cells (open arrowheads) and bipolar neurons (solid arrowheads) of the posterior tail nerve cord, as well as secondary tail muscle cells (“m”). The Islet driver used is Islet -5915/-5396 (Stolfi et al. 2010), which only drives expression in bipolar tail neurons. This exemplifies the use of lineage- and cell-type–specific reporter plasmids to visualize cells and trace their ontogeny. Ngn > mCherry was constructed using 5 kb upstream of the endogenous Ngn start codon. (B) Motor ganglion of a C. intestinalis larva transfected with five different plasmids for a combination of four different drivers and three different fluorescent reporter genes. This mix allows for labeling of all five neuronal subtypes of the motor ganglion (numbered 1–5) in a single individual. All reporters were fused to unc-76 tag for efficient labeling of axons. mCh, mCherry; Isl, Islet. Panel adapted from Stolfi and Levine (2011). (C) Ventral view of a midtailbud-stage embryo electroporated once with MesP > GFP (green), rinsed, and electroporated with MesP > RFP (red). Due to mosaicism, this individual is expressing MesP > GFP only in cells of the right hemisphere, and MesP > RFP only in cells of the left hemisphere. Image is a composite of images taken at different focal planes of the same embryo. The seam between the two focal planes is indicated by the dashed line. Migratory heart precursors are labeled in the left panel, while tail muscles are labeled in the right panel. Both are descended from the B7.5 pair of blastomeres, which express MesP. Bars in A and B, ∼25 µm.
General strategies, caveats, and surprises of the electroporation technique
Multiple plasmids are often mixed together and “coelectroporated” into the embryos (Figure 3A). In one example, five plasmids representing four different enhancers and three different fluorescent reporters were coelectroporated to label all five neuronal subtypes in the Ciona motor ganglion (Figure 3B) (Stolfi and Levine 2011). Multiple perturbation constructs can also be used to create complex epistatic backgrounds. In one study, converting an entire cell lineage into heart through overexpression of an active form of the FGF/MAPK-signaling effector Ets1/2 while simultaneously blocking cell migration by expressing a repressor form of the transcription factor FoxF, resulted in the uncoupling of specification and directed migration of cardiac precursor cells (Beh et al. 2007).
The electroporation technique presents at least four peculiarities that can represent significant disadvantages when it is compared to stable transgenesis. They are: transience, variable dosage, mosaicism, and leakiness (Figure 4). However, simply keeping these caveats in mind when interpreting your experiments goes a long way toward minimizing their disadvantages. And, as always, it is up to the creative researcher to take these potential pitfalls and harness them to one’s advantage.
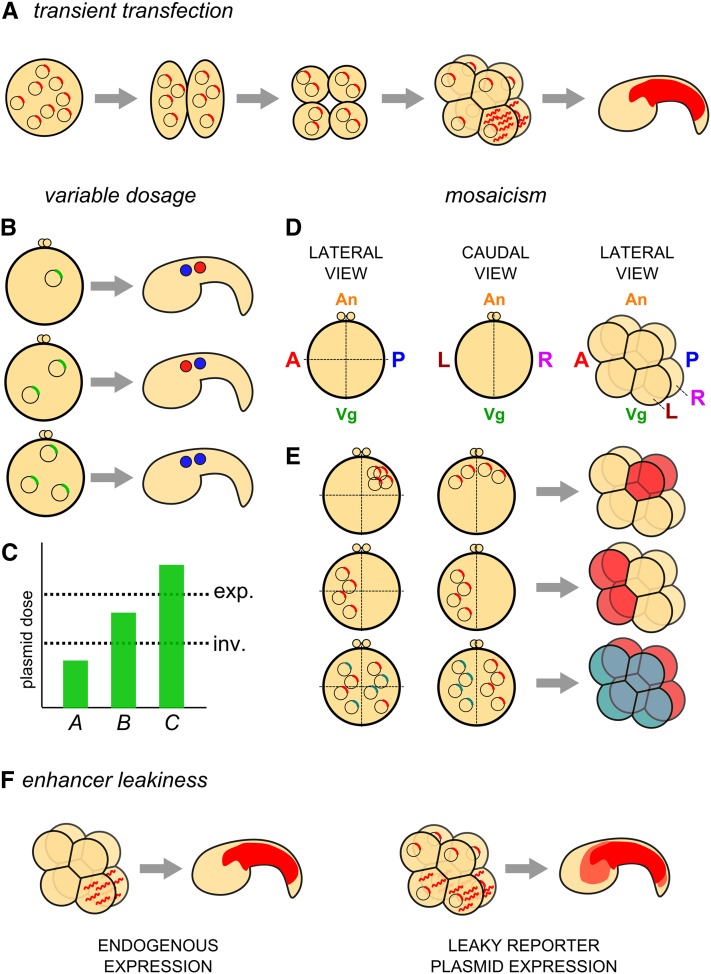
Figure 4 .
Caveats of the electroporation technique. (A) Diagram of the principle of transient transfection of Ciona embryos with plasmid DNA. Plasmid molecules are nonreplicative and segregate randomly, presumably as extrachromosomal DNA “arrays.” Transcription of a transgene in a given cell (determined by the activity of the driver) will result in inheritance of mRNA and/or resulting protein by all descendants of the cell. Given the short developmental time of Ciona (16 hr from fertilization to hatching), most reporters expressed during early embryogenesis are still visible late in development. (B and C) Diagram of possible outcomes of variable penetrance of a given perturbation transgene, presumably due to variation in total molecules of plasmid DNA taken up by the embryo at electroporation. (B) An embryo transfected with n copies of a plasmid carrying a gain-of-function gene (top) might not show any phenotype when compared to an unelectroporated control. If 2n copies of the same plasmid are transfected into an embryo, a given phenotype might occur. In this example, the fates of two cells are inverted along the anterior–posterior axis (middle). In embryos transfected with 3n copies of the plasmid, a qualitatively different phenotype might occur, in this case, an expansion of the anterior cell fate (blue) and the expense of its posterior neighbor (red) (bottom). (C) These outcomes can be interpreted as being due to different thresholds of plasmid dosage required for either phenotype (inv., inversion of cell fates; exp., posterior expansion of the anterior cell fate). Remembering that, we have little control over how many copies of the plasmid go into each embryo, meaning in any given batch of electroporated embryos, one will see the whole range of plasmid dosage represented. (D) Diagram of one-cell Ciona zygotes, viewed laterally (left) or caudally (center), and an eight-cell–stage embryo viewed laterally (right). Major embryonic axes and coordinates are indicated (A, anterior pole; P, posterior pole; An, animal pole; Vg, vegetal pole; L, left hemisphere; R, right hemisphere). (E) Model for mechanism underlying mosaicism in electroporated embryos. Expected mosaic expression patterns at the eight-cell stage (right) given the localization of plasmid DNA before the first cleavage (left and center, representing lateral and caudal views of the same embryo, respectively). Top: plasmid that is taken up primarily at the posterior, animal pole on either side of the embryo results in expression (red fill) in the cells that derive from this part of the zygote. Center: plasmid taken up at the anterior pole of the left side of the embryo will result in expression in only the anterior/left-derived cells. Bottom: mutually exclusive uptake of distinct plasmids (represented by blue and red transgenes) by left and right hemispheres after sequential electroporations will result in mutually exclusive, left–right expression of these plasmids (blue and red fill, respectively; see Figure 3C). (F) Diagram of the transcription of a reporter gene by a “leaky” driver (right), that is, one that does not faithfully recapitulate the expression of the endogenous gene from which it was derived, as assayed by in situ hybridization or immunostain (left), being somewhat active in other cells not normally expressing the endogenous gene, labeling the descendants of these cells with lower (though sometimes equal or stronger) levels of reporter gene protein.
Transience:
Empirical evidence suggests electroporated plasmid DNA does not frequently integrate into the genome, but probably remains as nonreplicating, extrachromosomal arrays, like exogenous DNA in transgenic lines of the nematode Caenorhabditis elegans (Stinchcomb et al. 1985). This nonreplicative, nonintegrative mode of mitotic segregation and inheritance means that transgene expression is transient (Figure 4A), and indeed seldom have we observed de novo transgene expression beyond metamorphosis, ∼72 hr postfertilization. Due to the rapid development time of Ciona embryos (16 hr to hatching, 24 hr to onset of metamorphosis), this transience is not really an obstacle for studies focused on the embryo or larva. However, unless an irreversible modification is being carried out (e.g., targeted mutagenesis by overexpression of zinc-finger nucleases, as proposed below), plasmid electroporation is not a suitable method for genetic perturbations in juveniles or adults. This can actually be an advantage, because certain driver > perturbation combinations that might be adult lethal or sterile can be easily assayed during embryogenesis.
Variable dosage:
The uptake of DNA by electroporated cells is not well understood (Escoffre et al. 2009). What we do know is that we have an imprecise control over exactly how many copies of the plasmid go into each embryo. This means that, in any batch of electroporated embryos, one sees a gradient of transgene dosage, though there is no simple way to accurately quantify this. This is why the large numbers of transfected embryos obtained by electroporating them en masse is so useful, as it allows one to average phenotypic quantification data over large numbers of individuals.
The one advantage of this variability is the possibility of uncovering the effects of gene dosage in a particular process. In this way, the gradient of plasmid dosage seen in a batch of electroporated embryos is akin to an allelic series. Whereas different transgenic strains must be generated to uncover the phenotypes associated with variable gene copy number in organisms such as flies, in Ciona one can see the entire range of phenotypes on a single slide. In some cases, qualitatively different phenotypes can arise from different plasmid doses, if, when, or where certain regulatory thresholds are met (Figure 4, B and C).
Mosaicism:
This term relates to the spatial variation in expression of exogenous DNA, another issue inherent to the electroporation technique. One hypothesis is that mosaicism might occur by random, unequal distribution of plasmid DNA molecules into one of two daughter cells resulting from a mitotic division. Another possibility is random loss or silencing of plasmid DNA. Regardless of the underlying cause, transgene expression is frequently seen only in cells descended from one of the major “quadrants” or hemispheres of the early embryo (Figure 4, D and E). Mosaicism within a lineage is also known to occur (Zeller et al. 2006). However, with the appropriate optimization of electroporation rigs and parameters, one can increase or decrease the incidence of mosaicism at will, suggesting the underlying cause of mosaicism is closely linked to the initial distribution of plasmid DNA within the unicellular zygote (Zeller et al. 2006).
Mosaicism can be exploited to deliberately drive expression of different transgenes in equivalent cells of different lineages or on either side of the bilaterally symmetric embryo (Stolfi and Levine 2011). In embryos electroporated more than once, each time with a distinct plasmid, some proportion of embryos will randomly incorporate plasmid in one hemisphere and another plasmid in the opposite hemisphere (Figure 3C). This provides an internal control in an experiment in which cells on a genetically manipulated side of the embryo can be compared to cells on the opposite, unmanipulated side.
Leakiness:
Quite often, the expression of a transgene under the control of a given driver does not exactly mirror that of the endogenous gene from which the driver was obtained as determined by in situ hybridization or transcriptional profiling. “Leaky” expression will be seen in closely related lineages or different lineages altogether (Figure 4F). Presumably, this is because the entire complement of a given gene’s cis-regulatory sequence is not contained within the driver. Alternatively, leakiness might be due to an intrinsic refractivity of exogenous plasmid DNA to chromatin state regulatory mechanisms that might be contributing to silencing of endogenous genes (Beisel and Paro 2011). Leakiness can be problematic because it can confound interpretation of perturbation experiments. You think a certain phenotype is induced when you express gene X in cell A, but the phenotype might in fact be due to leaky expression of gene X in cell B.
Leaky reporter plasmids have been used to counterselect certain cell types in fluorescence-activated cell sorting of dissociated Ciona embryos. For instance, MesP > GFP is used to sort cells descended from the B7.5 blastomeres, which express MesP (Figure 3C). However, this plasmid shows leaky expression in mesenchyme. A second plasmid, MyoD > YFP, which is also leaky in the mesenchyme, was used to counterselect these cells in a YFP-dependent negative gate (Christiaen et al. 2008). This use of leakiness to fight leakiness is a perfect representation of the dichotomy between the simultaneously annoying and useful quirks of electroporation in Ciona.
What to electroporate?
Overexpression of dominant-interfering neomorphs (often called “dominant negatives,” much to the chagrin of the rigorous geneticist) is widely used to study transcription factors and signaling pathways in development. Overexpression of a fusion between the DNA-binding domain of a given transcriptional activator and a Groucho corepressor-recruiting WRPW motif can partially mimic its loss-of-function phenotype as established by morpholino injection (Beh et al. 2007). Perturbation of signaling pathways by overexpression of dominant-interfering or constitutively active pathway components is better established. Transmembrane receptors truncated or mutated at their intracellular side efficiently sequester ligands of the FGF, BMP, and Ephrin families. (Davidson et al. 2006; Picco et al. 2007; Christiaen et al. 2010). Similarly, a DNA-binding mutant of the Notch transcriptional coactivator Supressor of Hairless [Su(H)] can inhibit the transcriptional output of Delta/Notch signaling (Pasini et al. 2006). Self-dimerizing forms of certain receptor kinases can partially mimic activation of the wild-type receptor by their ligands (Shi and Levine 2008; Christiaen et al. 2010). Cytoskeleton dynamics have been perturbed by overexpression of dominant-interfering or constitutively active mutant forms of small GTPases such as RhoD/F and Cdc42 (Christiaen et al. 2008).
Wild-type proteins can also serve as perturbation genes, when misexpressed in a cell lineage or cell type in which they are not normally expressed. Such misexpression has been used to show the inductive role of certain signaling ligands (Hudson et al. 2007) or to demonstrate the sufficiency of certain transcription factors in activating or repressing a given regulatory network (Stolfi et al. 2010; Kratsios et al. 2012).
RNAi
RNA interference (RNAi) holds great promise, but has not been developed to the point of widespread, routine use. Plasmid-based RNAi in particular is extremely useful as it allows for electroporation-based protein knockdown experiments, supplanting difficult morpholino injections. In C. intestinalis, plasmid-based RNAi against tyrosinase, the enzyme responsible for melanin synthesis, has been reported (Nishiyama and Fujiwara 2008). RNAi also was used in elucidating a role for the basement membrane-associated proteoglycan gene Leprecan in structural integrity of the notochordal sheath (Dunn and Di Gregorio 2009). However, in both cases RNAi was mediated by short hairpin RNAs (shRNAs) transcribed by RNApolIII in a constitutive and ubiquitous manner. The gold standard would be to use microRNA (miRNA)-like shRNA cassettes (“shmiR”), which can be driven by RNApolII promoters linked to cell-specific enhancers (Chang et al. 2006; Haley et al. 2010). This would allow for cell-specific knockdown of target mRNAs, by cooption of the endogenous miRNA biogenesis pathway, which is fully functional during Ciona embryogenesis (Chen et al. 2011).
Such shmiR constructs, although already used in other metazoans, have yet to be successfully adapted to Ciona. Most attempts have relied on shRNA design principles gleaned from experiments on mammals, nematodes, and insects. However, as a marine invertebrate with an optimal developmental temperature of 19°, perhaps some tweaking of hairpin stability and folding algorithms will be needed. As far as we know, targeted gene knockdown by miRNA-like RNAi constructs has not been reported in a marine invertebrate. The preponderance of microRNA-offset RNAs (moRs) in Ciona and the proclivity of Ciona Drosha to produce such moRs from heterologously expressed in Drosophila pri-miRs suggests certain peculiarities of small RNA biogenesis in Ciona might also have to be taken into account (Shi et al. 2009).
In the near future?
Other genetically encoded tools have not been explored, though in theory they could be easily adapted to Ciona. Of particular note are those based on the DNA-binding domains of zinc-finger (ZF) and transcription activator-like (TAL) transcription factors (Beerli et al. 1998, 2000; Christian et al. 2010; Morbitzer et al. 2010). Fusions of DNA endonucleases to custom-designed ZFs or TALs (termed ZFNs and TALENs, respectively) have been used to promote targeted genetic deletions or homologous recombination in systems where such events were traditionally difficult to induce (Urnov et al. 2005; Wood et al. 2011; Young et al. 2011; see note added in proof).
More sophisticated tools for precise control of gene function would include optically controlled proteins (Riggsbee and Deiters 2010), such as light-activated G protein-coupled receptors like Channelrhodopsin-2 (Nagel et al. 2003), or plant phytochrome, or phototropin fusions (Shimizu-Sato et al. 2002; Strickland et al. 2008). Chemical control of protein function is also a burgeoning field (Ouyang and Chen 2010). Several genetically encoded protein domains that interact with small molecules can be fused to other proteins. These fusions are then active, inactive, or interact with other components depending on the presence (or absence) of the small molecule. For instance, a tamoxifen-induced mutated ER ligand binding domain-Cre recombinase fusion is routinely used for small molecule-dependent DNA recombination in mouse (Feil et al. 1997). Such methods could allow for another layer of precision on top of the spatiotemporal control of gene expression already afforded by the use of cell-specific drivers, taking the electroporation technique to new heights.
Stable transgenesis and reverse genetic screens
Stable transgenesis in Ciona is a relatively new development that holds great promise in expanding the scope of genetic analysis of ascidian biology. Plasmid DNA introduced into Ciona eggs rarely integrates into the genome (Matsuoka et al. 2005). Two methods have been developed for increasing the odds of germline transmission of transgenes, one based on co-injection of I-SceI meganuclease (Deschet et al. 2003) and one based on Tc1/mariner superfamily transposable elements (Sasakura et al. 2003).
I-SceI meganuclease is an endonuclease from yeast that recognizes an 18-bp target sequence (Monteilhet et al. 1990). Co-injection of I-SceI enzyme into eggs can increase the rate of genome integration and germline transmission of transgenes flanked by two I-SceI sites (Thermes et al. 2002). It is thought that I-SceI will cause double stranded breaks flanking the transgene, which is then inserted randomly into a native chromosome by the DNA repair machinery. Deschet et al. (2003) used the same strategy to generate stable C. savignyi transgenics carrying copies of GFP under the control of a Brachyury driver from C. intestinalis.
Transposable “P” elements reinvigorated the field of fly genetics, permitting stable transgenesis, insertion/deletion mutagenesis, and enhancer and gene trapping (Bellen et al. 1989). In Ciona, the Tc1/mariner-family member Minos allows for similar applications. Germline integration of transgenes is achieved by co-injection or coelectroporation of Minos transposase mRNA and plasmid carrying the transgene flanked by Minos terminal repeats (Sasakura et al. 2003; Matsuoka et al. 2004, 2005). Using this method, reporter lines were generated using previously characterized drivers. A slightly different application has been the generation of enhancer trap lines, using the same Minos transposable element. By introducing a Minos vector carrying GFP downstream of a minimal promoter without an enhancer, random integration of the transgene resulted in lines showing distinct GFP expression patterns, depending on the enhancers found near each integration site (Figure 5) (Awazu et al. 2004, 2007). High-throughput remobilization of integration events by expression of transposase in sperm (Sasakura et al. 2008) or egg (Hozumi et al. 2010) has since expanded the collection of reporter lines. This has been useful to identify novel enhancers, though a paucity of embryonic reporter lines has been observed, perhaps owing to the choice of minimal promoter used or the rarity of embryonic enhancers in the genome. All told, the Ciona intestinalis Transgenic Line Resource center (CITRES, http://marinebio.nbrp.jp/ciona/) has 41 enhancer trap lines and several other Minos transgenic lines, which are all available to the community. With the growing availability of transgenic lines, perhaps Cre/Lox- (Sauer and Henderson 1988) or GAL4/UAS-like (Brand and Perrimon 1993) systems for modular, conditional genetic loss or gain of function could be adapted to Ciona.
Figure 5 .
Enhancer trap transgenic lines. (A–C) Postmetamorphic juveniles from different Minos-based enhancer trap transgenic lines from the Bioresource Project in Japan (http://marinebio.nbrp.jp/ciona/). (A) GFP expression seen in the endostyle, retropharyngeal band, esophagus, central nervous system, oral siphon, and atrial siphon. (B) GFP expressed in the pyloric gland. (C) GFP is expressed in the central nervous system. Bar, ∼100 µm. All images unpublished, courtesy of Y. Sasakura.
The high-throughput remobilization of single integration events also holds great promise for generating mutants for reverse genetic screens. It has already been shown that a Minos transgene can disrupt the endogenous gene at the site of its integration. For instance, swimming juvenile mutants contain a Minos enhancer trap insertion in the cellulose synthase A (CesA) gene, leading to premature body plan reorganization prior to settlement of larvae (Figure 2G) (Sasakura et al. 2005). The unique sequence of the transgene can be used to easily locate the insertion site.
Genomics
The Ciona genomes have been sequenced at draft-level coverage, and are both ∼150–170 Mb in length, coding for ∼16,000 genes. (Simmen et al. 1998; Dehal et al. 2002; Vinson et al. 2005; Small et al. 2007a). They are the smallest chordate genomes sequenced to date, with the exception of the larvacean tunicate Oikopleura dioica (Seo et al. 2001). The C. intestinalis genome has been sequenced and assembled de novo twice, once in 2002 and once in 2005. A more accurate assembly, based on the first sequence, was published in 2008 and covers ∼75% of the genome (Satou et al. 2008). A total of 65% of this has been mapped to the 14 pairs of chromosomes of C. intestinalis (Shoguchi et al. 2006). The two species’ genome sequences have been aligned against each other, and the evolutionary divergence between the two means that highly conserved noncoding sequences are usually imbued with functional significance, greatly aiding in the search for enhancers and other cis-regulatory elements. The alignment can easily be navigated on the VISTA browser (http://pipeline.lbl.gov/cgi-bin/gateway2?bg=Cioin2&selector=vistapoint) (Frazer et al. 2004).
Having a sequenced genome places Ciona into a select category of “postgenomic” model organisms. Not only does it allow for researchers to more easily clone genomic and cDNA sequences as experimental tools, but also allows for genome-wide observational studies that might generate testable hypotheses. For instance, chromatin immunoprecipitation combined with genome tiling chips (ChIP-chip) revealed the genome-wide binding profile for 11 transcription factors during development (Kubo et al. 2010). In other studies, a combination of high-throughput mRNA sequencing and computational methods have allowed for detection and mapping of Ciona microRNA loci (Hendrix et al. 2010; Keshavan et al. 2010). These are but some examples of genome-scale studies performed in Ciona.
On-line resources
Various databases and online resources serving the Ciona research community are spread around the world. Reflecting the long tradition of ascidian biology in Japan, extensive EST sequencing data (Satou et al. 2001b, 2002a) and cDNA collections (Satou et al. 2002b) are available to researchers as part of the GHOST portal (http://ghost.zool.kyoto-u.ac.jp/cgi-bin/gb2/gbrowse/kh/) (Satou et al. 2005), which also harbors the most recent and accurate assembly and gene annotation for the C. intestinalis genome (“KyotoHoya” or “KH” gene models) (Satou et al. 2008). For more protein-oriented research, there is CIPRO (http://cipro.ibio.jp/current/) (Endo et al. 2011), which is another genomic and gene expression database that integrates 2D-PAGE and mass-spectrometry data from Ciona embryos.
Another ascidian research hotspot is France, which hosts the ANISEED portal (http://www.aniseed.cnrs.fr/) (Tassy et al. 2010). This database is more explicitly oriented toward developmental genetics, with a large curated collection of in situ hybridization images and other gene expression, gene model, and anatomical data. Unlike GHOST or CIPRO, ANISEED is also meant to cover other experimental ascidian genera, such as Phallusia and Halocynthia.
Recently, a greater effort toward integration of the various databases and resources has been made, with leaders responsible for the individual portals agreeing to cross-reference their information and delineate their “spheres of influence” in such a way that redundancies are slowly eliminated. The First International Workshop on Tunicate Information Systems, held in November 2010, served to outline these goals (Inaba et al. 2011). It was preliminarily agreed that genomic and sequence data would be curated and presented by GHOST, expression data by ANISEED, protein data by CIPRO, and cellular and anatomical data by a fourth database, Four dimensional Ascidian Body Atlas (http://chordate.bpni.bio.keio.ac.jp/faba/1.4/top.html) (Hotta et al. 2007). Although the information found at each website would not be strictly limited to one type of data, the missions of each have been altered to emphasize these broad divisions while reinforcing their interconnectivity. With this, a more seamless integration is hoped to be achieved, without sacrificing the independence of each database.
Conclusion
In sum, Ciona spp. are versatile organisms amenable to diverse genetic perturbation strategies (Table 1). Various reverse genetic approaches, as well as traditional forward genetics, have been successfully used to study gene function in Ciona. Transient transfection experiments using the electroporation technique are now being complemented by stable, germ-line transgenesis. The Ciona community is also on the cusp of harnessing targeted loss-of-function techniques such as ZFNs, TALENs, and RNAi. In the near future, we anticipate that three major approaches—genetic screens, electroporation, and stable transgenesis—will be increasingly integrated in comprehensive functional studies. The growing stable of genomic resources for Ciona, as well as the suitability of Ciona embryos for medium- to high-throughput strategies, will be key to their continued employ as model organisms, especially in emerging fields such as systems biology. We believe that the upcoming years will be an exciting time for the study of Ciona genetics, and look forward to the continued growth of the Ciona community.
Table 1 . Genetic toolkit of Ciona intestinalis.
| Technique | Published? | Routine? | Reference |
|---|---|---|---|
| Forward genetic screens | Y | Y | Jiang et al. (2005) |
| Germline transgenesis | Y | Y | Deschet et al. (2003); Sasakura et al. (2003) |
| Transient transgenesis | Y | Y | Corbo et al. (1997) |
| Morpholinos | Y | Y | Satou et al. (2001a) |
| Retrotransposon remobilization | Y | Y | Awazu et al. (2007) |
| Reverse genetic screens of retrotransposon insertions | Y | N | Sasakura et al. (2005) |
| RNAi | Y | N | Nishiyama and Fujiwara (2008) |
| Targeted mutagenesis | Y | N | Kawai et al. (2012) |
| Homologous recombination | N | — | — |
| Cre/Lox or Flp/FRT recombination | N | — | — |
| GAL4-UAS heterologous expression system | N | — | — |
Genetic perturbation techniques available in Ciona. Techniques count as published if appearing at least once in a peer-reviewed journal article. Techniques are considered “routine” if three or more articles have been published using the technique. Reference given is the first (to our knowledge) published account of its use in Ciona. Y = yes, N = no. Techniques with no published instance of use in Ciona are considered to be on the “wish list,” with some currently in the process of being adapted to Ciona.
Acknowledgments
We are grateful to Robert Zeller for comments on the manuscript and to Bill Smith, Michael Veeman, and Yasunori Sasakura for sharing images. We thank Florian Razy-Krajka for reading the manuscript and for the larva picture shown in Figure 1C. Our work is supported by grants 10SDG4310061 from the American Heart Association; R01GM096032 from the National Institute of General Medical Sciences/National Institutes of Health (NIH); R01HL108643 from the National Heart, Lung, and Blood Institute/NIH; the New York Cardiac Center; and New York University College of Arts and Sciences.
Note added in proof: While this manuscript was in revision, the first report of targeted mutagenesis in Ciona was published by Kawai, N., H. Ochiai, T. Sakuma, L. Yamada, H. Sawada et al., 2012 Efficient targeted mutagenesis of the chordate Ciona intestinalis genome with zinc-finger nucleases. Dev. Growth Differ. 54: 535–545.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Awazu S., Sasaki A., Matsuoka T., Satoh N., Sasakura Y., 2004. An enhancer trap in the ascidian Ciona intestinalis identifies enhancers of its Musashi orthologous gene. Dev. Biol. 275: 459–472 [DOI] [PubMed] [Google Scholar]
- Awazu S., Matsuoka T., Inaba K., Satoh N., Sasakura Y., 2007. High-throughput enhancer trap by remobilization of transposon Minos in Ciona intestinalis. Genesis 45: 307–317 [DOI] [PubMed]
- Beerli R. R., Segal D. J., Dreier B., Barbas C. F., 1998. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 95: 14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli R. R., Dreier B., Barbas C. F., 2000. Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl. Acad. Sci. USA 97: 1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh J., Shi W., Levine M., Davidson B., Christiaen L., 2007. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development 134: 3297–3305 [DOI] [PubMed] [Google Scholar]
- Beisel C., Paro R., 2011. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 12: 123–135 [DOI] [PubMed] [Google Scholar]
- Bellen H. J., O’Kane C. J., Wilson C., Grossniklaus U., Pearson R. K., et al. , 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3: 1288. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Brown C. D., Johnson D. S., Sidow A., 2007. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science 317: 1557. [DOI] [PubMed] [Google Scholar]
- Byrd J., Lambert C. C., 2000. Mechanism of the block to hybridization and selfing between the sympatric ascidians Ciona intestinalis and Ciona savignyi. Mol. Reprod. Dev. 55: 109–116 [DOI] [PubMed] [Google Scholar]
- Chabry L., 1887. Contribution to the normal and teratological embryology of solitary ascidians. F. Alcan; (translated from French) [Google Scholar]
- Chang K., Elledge S. J., Hannon G. J., 2006. Lessons from nature: microRNA-based shRNA libraries. Nat. Methods 3: 707–714 [DOI] [PubMed] [Google Scholar]
- Chen J. S., San Pedro M., Zeller R. W., 2011. miR-124 function during Ciona intestinalis neuronal development includes extensive interaction with the Notch signaling pathway. Development 138: 4943–4953 [DOI] [PubMed] [Google Scholar]
- Chiba S., Jiang D., Satoh N., Smith W. C., 2009. Brachyury null mutant-induced defects in juvenile ascidian endodermal organs. Development 136: 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen L., Davidson B., Kawashima T., Powell W., Nolla H., et al. , 2008. The transcription/migration interface in heart precursors of Ciona intestinalis. Science 320: 1349. [DOI] [PubMed] [Google Scholar]
- Christiaen L., Wagner E., Shi W., Levine M., 2009a. Electroporation of transgenic DNAs in the sea squirt Ciona. Cold Spring Harbor Protocols, doi: 10.1101/pdf.prot5345 [DOI] [PubMed] [Google Scholar]
- Christiaen L., Wagner E., Shi W., Levine M., 2009b. Microinjection of morpholino oligos and RNAs in sea squirt (Ciona) embryos. Cold Spring Harbor Protocols, doi: 10.1101/pdb. prot5347 [DOI] [PubMed] [Google Scholar]
- Christiaen L., Wagner E., Shi W., Levine M., 2009c. The sea squirt Ciona intestinalis. Cold Spring Harbor Protocols, doi: 10.1101/pdb. emo138 [DOI] [PubMed] [Google Scholar]
- Christiaen L., Stolfi A., Levine M., 2010. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev. Biol. 340: 179–187 [DOI] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., et al. , 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Hayakawa H., Berg P., 1987. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 15: 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin E. G., 1905. Organ-forming substances in the eggs of ascidians. Biol. Bull. 8: 205 [Google Scholar]
- Corbo J. C., Levine M., Zeller R. W., 1997. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124: 589–602 [DOI] [PubMed] [Google Scholar]
- Davidson B., Shi W., Beh J., Christiaen L., Levine M., 2006. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 20: 2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P., Boore J. L., 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P., Satou Y., Campbell R. K., Chapman J., Degnan B., et al. , 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298: 2157. [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H., 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439: 965–968 [DOI] [PubMed] [Google Scholar]
- Deschet K., Nakatani Y., Smith W. C., 2003. Generation of Ci-Brachyury-GFP stable transgenic lines in the ascidian Ciona savignyi. Genesis 35: 248–259 [DOI] [PubMed] [Google Scholar]
- Dunn M. P., Di Gregorio A., 2009. The evolutionarily conserved leprecan gene: its regulation by Brachyury and its role in the developing Ciona notochord. Dev. Biol. 328: 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards P. K., Leung B., 2008. Re-evaluating eradication of nuisance species: invasion of the tunicate, Ciona intestinalis. Front. Ecol. Environ 7: 326–332 [Google Scholar]
- Endo T., Ueno K., Yonezawa K., Mineta K., Hotta K., et al. , 2011. CIPRO 2.5: Ciona intestinalis protein database, a unique integrated repository of large-scale omics data, bioinformatic analyses and curated annotation, with user rating and reviewing functionality. Nucleic Acids Res. 39: D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoffre J. M., Portet T., Wasungu L., Teissié J., Dean D., et al. , 2009. What is (still not) known of the mechanism by which electroporation mediates gene transfer and expression in cells and tissues. Mol. Biotechnol. 41: 286–295 [DOI] [PubMed] [Google Scholar]
- Feil R., Wagner J., Metzger D., Chambon P., 1997. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 237: 752–757 [DOI] [PubMed] [Google Scholar]
- Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I., 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B., Foys B., Levine M., 2010. Vectors and parameters that enhance the efficacy of RNAi-mediated gene disruption in transgenic Drosophila. Proc. Natl. Acad. Sci. USA 107: 11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J., 2002. Morpholino oligos: Making sense of antisense? Dev. Biol. 243: 209–214 [DOI] [PubMed] [Google Scholar]
- Hendrickson C., Christiaen L., Deschet K., Jiang D., Joly J. S., et al. , 2004. Culture of adult ascidians and ascidian genetics. Methods Cell Biol. 74: 143–170 [DOI] [PubMed] [Google Scholar]
- Hendrix D., Levine M., Shi W., 2010. Method miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol. 11: R39 [Google Scholar]
- Hikosaka A., Satoh N., Makabe K. W., 1993. Regulated spatial expression of fusion gene constructs with the 5′ upstream region of Halocynthia roretzi muscle actin gene in Ciona savignyi embryos. Dev. Genes Evol. 203: 104–112 [DOI] [PubMed] [Google Scholar]
- Hill M. M., Broman K. W., Stupka E., Smith W. C., Jiang D., et al. , 2008. The C. savignyi genetic map and its integration with the reference sequence facilitates insights into chordate genome evolution. Genome Res. 18: 1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K., Mitsuhara K., Takahashi H., Inaba K., Oka K., et al. , 2007. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 236: 1790–1805 [DOI] [PubMed] [Google Scholar]
- Hozumi A., Kawai N., Yoshida R., Ogura Y., Ohta N., et al. , 2010. Efficient transposition of a single Minos transposon copy in the genome of the ascidian Ciona intestinalis with a transgenic line expressing transposase in eggs. Dev. Dyn. 239: 1076–1088 [DOI] [PubMed] [Google Scholar]
- Hudson C., Yasuo H., 2008. Similarity and diversity in mechanisms of muscle fate induction between ascidian species. Biol. Cell 100: 265–277 [DOI] [PubMed] [Google Scholar]
- Hudson C., Lotito S., Yasuo H., 2007. Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development 134: 3527–3537 [DOI] [PubMed] [Google Scholar]
- Iannelli F., Pesole G., Sordino P., Gissi C., 2007. Mitogenomics reveals two cryptic species in Ciona intestinalis. Trends Genet. 23: 419–422 [DOI] [PubMed] [Google Scholar]
- Imai K., Takada N., Satoh N., Satou Y., 2000. (beta)-catenin mediates the specification of endoderm cells in ascidian embryos. Development 127: 3009–3020 [DOI] [PubMed] [Google Scholar]
- Imai K. S., Hino K., Yagi K., Satoh N., Satou Y., 2004. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131: 4047–4058 [DOI] [PubMed] [Google Scholar]
- Imai K. S., Levine M., Satoh N., Satou Y., 2006. Regulatory blueprint for a chordate embryo. Science 312: 1183. [DOI] [PubMed] [Google Scholar]
- Inaba K., Endo T., Satou Y., Hotta K., Nishida H., et al. , 2011. Tunicate databases: toward a comprehensive informative basis at molecular and cellular level for tunicate community. 6th International Tunicate Meeting, McGill University, Montreal, Canada
- Jiang D., Munro E. M., Smith W. C., 2005. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr. Biol. 15: 79–85 [DOI] [PubMed] [Google Scholar]
- Joly J. S., Kano S., Matsuoka T., Auger H., Hirayama K., et al. , 2007. Culture of Ciona intestinalis in closed systems. Dev. Dyn. 236: 1832–1840 [DOI] [PubMed] [Google Scholar]
- Kawai N., Ochiai H., Sakuma T., Yamada L., Sawada H., et al. , 2012. Efficient targeted mutagenesis of the chordate Ciona intestinalis genome with zinc-finger nucleases. Dev. Growth Differ. 54: 535–545 [DOI] [PubMed] [Google Scholar]
- Keshavan R., Virata M., Keshavan A., Zeller R. W., 2010. Computational identification of Ciona intestinalis MicroRNAs. Zoolog. Sci. 27: 162–170 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Waterman M. S., Li L. M., 2007. Diploid genome reconstruction of Ciona intestinalis and comparative analysis with Ciona savignyi. Genome Res. 17: 1101–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P., Stolfi A., Levine M., Hobert O., 2012. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15: 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Suzuki N., Yuan X., Nakai K., Satoh N., et al. , 2010. Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development 137: 1613–1623 [DOI] [PubMed] [Google Scholar]
- Kumano G., Takatori N., Negishi T., Takada T., Nishida H., 2011. A maternal factor unique to ascidians silences the germline via binding to P-TEFb and RNAP II regulation. Curr. Biol. 21: 1308–1313 [DOI] [PubMed] [Google Scholar]
- Lambert C. C., Lambert G., 1998. Non-indigenous ascidians in southern California harbors and marinas. Mar. Biol. 130: 675–688 [Google Scholar]
- Lemaire P., 2009. Unfolding a chordate developmental program, one cell at a time: invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev. Biol. 332: 48–60 [DOI] [PubMed] [Google Scholar]
- Lemaire P., 2011. Evolutionary crossroads in developmental biology: the tunicates. Development 138: 2143. [DOI] [PubMed] [Google Scholar]
- Matsuoka T., Awazu S., Satoh N., Sasakura Y., 2004. Minos transposon causes germline transgenesis of the ascidian Ciona savignyi. Dev. Growth Differ. 46: 249–255 [DOI] [PubMed] [Google Scholar]
- Matsuoka T., Awazu S., Shoguchi E., Satoh N., Sasakura Y., 2005. Germline transgenesis of the ascidian Ciona intestinalis by electroporation. Genesis 41: 67–72 [DOI] [PubMed] [Google Scholar]
- Monteilhet C., Perrin A., Thierry A., Colleaux L., Dujon B., 1990. Purification and characterization of the in vitro activity of I-Sce I, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 18: 1407–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbitzer R., Römer P., Boch J., Lahaye T., 2010. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl. Acad. Sci. USA 107: 21617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T., 1923. Removal of the block to self-fertilization in the ascidian Ciona. Proc. Natl. Acad. Sci. USA 9: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., et al. , 2003. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 100: 13940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Fujiwara S., 2008. RNA interference by expressing short hairpin RNA in the Ciona intestinalis embryo. Dev. Growth Differ. 50: 521–529 [DOI] [PubMed] [Google Scholar]
- Oda-Ishii I., Bertrand V., Matsuo I., Lemaire P., Saiga H., 2005. Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development 132: 1663–1674 [DOI] [PubMed] [Google Scholar]
- Ouyang X., Chen J. K., 2010. Synthetic strategies for studying embryonic development. Chem. Biol. 17: 590–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini A., Amiel A., Rothbächer U., Roure A., Lemaire P., et al. , 2006. Formation of the ascidian epidermal sensory neurons: insights into the origin of the chordate peripheral nervous system. PLoS Biol. 4: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco V., Hudson C., Yasuo H., 2007. Ephrin-Eph signalling drives the asymmetric division of notochord/neural precursors in Ciona embryos. Development 134: 1491–1497 [DOI] [PubMed] [Google Scholar]
- Putnam N. H., Butts T., Ferrier D. E. K., Furlong R. F., Hellsten U., et al. , 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071 [DOI] [PubMed] [Google Scholar]
- Riggsbee C. W., Deiters A., 2010. Recent advances in the photochemical control of protein function. Trends Biotechnol. 28: 468–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roure A., Rothbächer U., Robin F., Kalmar E., Ferone G., et al. , 2007. A multicassette Gateway vector set for high throughput and comparative analyses in Ciona and vertebrate embryos. PLoS ONE 2: e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura Y., Awazu S., Chiba S., Satoh N., 2003. Germ-line transgenesis of the Tc1/mariner superfamily transposon Minos in Ciona intestinalis. Proc. Natl. Acad. Sci. USA 100: 7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura Y., Nakashima K., Awazu S., Matsuoka T., Nakayama A., et al. , 2005. Transposon-mediated insertional mutagenesis revealed the functions of animal cellulose synthase in the ascidian Ciona intestinalis. Proc. Natl. Acad. Sci. USA 102: 15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura Y., Konno A., Mizuno K., Satoh N., Inaba K., 2008. Enhancer detection in the ascidian Ciona intestinalis with transposase-expressing lines of Minos. Dev. Dyn. 237: 39–50 [DOI] [PubMed] [Google Scholar]
- Satoh N., Satou Y., Davidson B., Levine M., 2003. Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet. 19: 376–381 [DOI] [PubMed] [Google Scholar]
- Satou Y., Imai K. S., Satoh N., 2001a. Action of morpholinos in Ciona embryos. Genesis 30: 103–106. [DOI] [PubMed] [Google Scholar]
- Satou Y., Takatori N., Yamada L., Mochizuki Y., Hamaguchi M., et al. , 2001b. Gene expression profiles in Ciona intestinalis tailbud embryos. Development 128: 2893–2904 [DOI] [PubMed] [Google Scholar]
- Satou Y., Takatori N., Fujiwara S., Nishikata T., Saiga H., et al. , 2002a. Ciona intestinalis cDNA projects: expressed sequence tag analyses and gene expression profiles during embryogenesis. Gene 287: 83–96 [DOI] [PubMed] [Google Scholar]
- Satou Y., Yamada L., Mochizuki Y., Takatori N., Kawashima T., et al. , 2002b. A cDNA resource from the basal chordate Ciona intestinalis. Genesis 33: 153–154 [Google Scholar]
- Satou Y., Kawashima T., Shoguchi E., Nakayama A., Satoh N., 2005. An integrated database of the ascidian, Ciona intestinalis: towards functional genomics. Zoolog. Sci. 22: 837–843 [DOI] [PubMed] [Google Scholar]
- Satou Y., Mineta K., Ogasawara M., Sasakura Y., Shoguchi E., et al. , 2008. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol. 9: R152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., Henderson N., 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 85: 5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H. C., Kube M., Edvardsen R. B., Jensen M. F., Beck A., et al. , 2001. Miniature genome in the marine chordate Oikopleura dioica. Science 294: 2506. [DOI] [PubMed] [Google Scholar]
- Shi W., Levine M., 2008. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development 135: 931–940 [DOI] [PubMed] [Google Scholar]
- Shi W., Hendrix D., Levine M., Haley B., 2009. A distinct class of small RNAs arises from pre-miRNA–proximal regions in a simple chordate. Nat. Struct. Mol. Biol. 16: 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J. M., Quail P. H., 2002. A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Shoguchi E., Kawashima T., Satou Y., Hamaguchi M., Sin T., et al. , 2006. Chromosomal mapping of 170 BAC clones in the ascidian Ciona intestinalis. Genome Res. 16: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen M. W., Leitgeb S., Clark V. H., Jones S. J. M., Bird A., 1998. Gene number in an invertebrate chordate, Ciona intestinalis. Proc. Natl. Acad. Sci. USA 95: 4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K., Brudno M., Hill M., Sidow A., 2007a. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 8: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K. S., Brudno M., Hill M. M., Sidow A., 2007b. Extreme genomic variation in a natural population. Proc. Natl. Acad. Sci. USA 104: 5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D., Shaw J., Carr S. H., Hirsh D., 1985. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 5: 3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A., Levine M., 2011. Neuronal subtype specification in the spinal cord of a protovertebrate. Development 138: 995. [DOI] [PubMed] [Google Scholar]
- Stolfi A., Gainous T. B., Young J. J., Mori A., Levine M., et al. , 2010. Early chordate origins of the vertebrate second heart field. Science 329: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D., Moffat K., Sosnick T. R., 2008. Light-activated DNA binding in a designed allosteric protein. Proc. Natl. Acad. Sci. USA 105: 10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. M., Nishikawa T., Bird A., 2005. Genomic approaches reveal unexpected genetic divergence within Ciona intestinalis. J. Mol. Evol. 61: 627–635 [DOI] [PubMed] [Google Scholar]
- Tassy O., Dauga D., Daian F., Sobral D., Robin F., et al. , 2010. The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res. 20: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermes V., Grabher C., Ristoratore F., Bourrat F., Choulika A., et al. , 2002. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 118: 91–98 [DOI] [PubMed] [Google Scholar]
- Tokuoka M., Kumano G., Nishida H., 2007. FGF9/16/20 and Wnt-5α signals are involved in specification of secondary muscle fate in embryos of the ascidian, Halocynthia roretzi. Dev. Genes Evol. 217: 515–527 [DOI] [PubMed] [Google Scholar]
- Urnov F. D., Miller J. C., Lee Y. L., Beausejour C. M., Rock J. M., et al. , 2005. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- Veeman M. T., Nakatani Y., Hendrickson C., Ericson V., Lin C., et al. , 2008. Chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development 135: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M. T., Chiba S., Smith W. C., 2011. Ciona genetics. Methods Mol. Biol. 770: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson J. P., Jaffe D. B., O’Neill K., Karlsson E. K., Stange-Thomann N., et al. , 2005. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 15: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. J., Lo T. W., Zeitler B., Pickle C. S., Ralston E. J., et al. , 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. J., Cherone J. M., Doyon Y., Ankoudinova I., Faraji F. M., et al. , 2011. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 108: 7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R. W., 2004. Generation and use of transgenic ascidian embryos. Methods Cell Biol. 74: 713–730 [DOI] [PubMed] [Google Scholar]
- Zeller R. W., Virata M. J., Cone A. C., 2006. Predictable mosaic transgene expression in ascidian embryos produced with a simple electroporation device. Dev. Dyn. 235: 1921–1932 [DOI] [PubMed] [Google Scholar]