Abstract
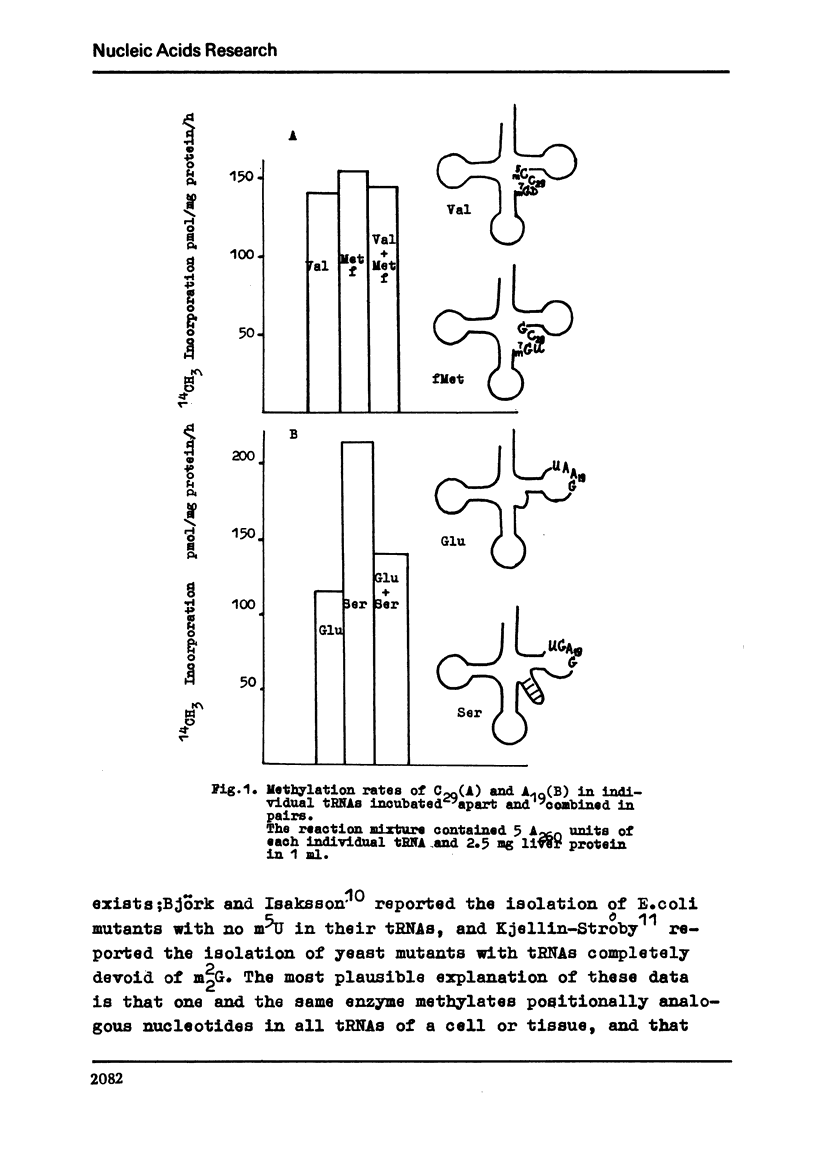
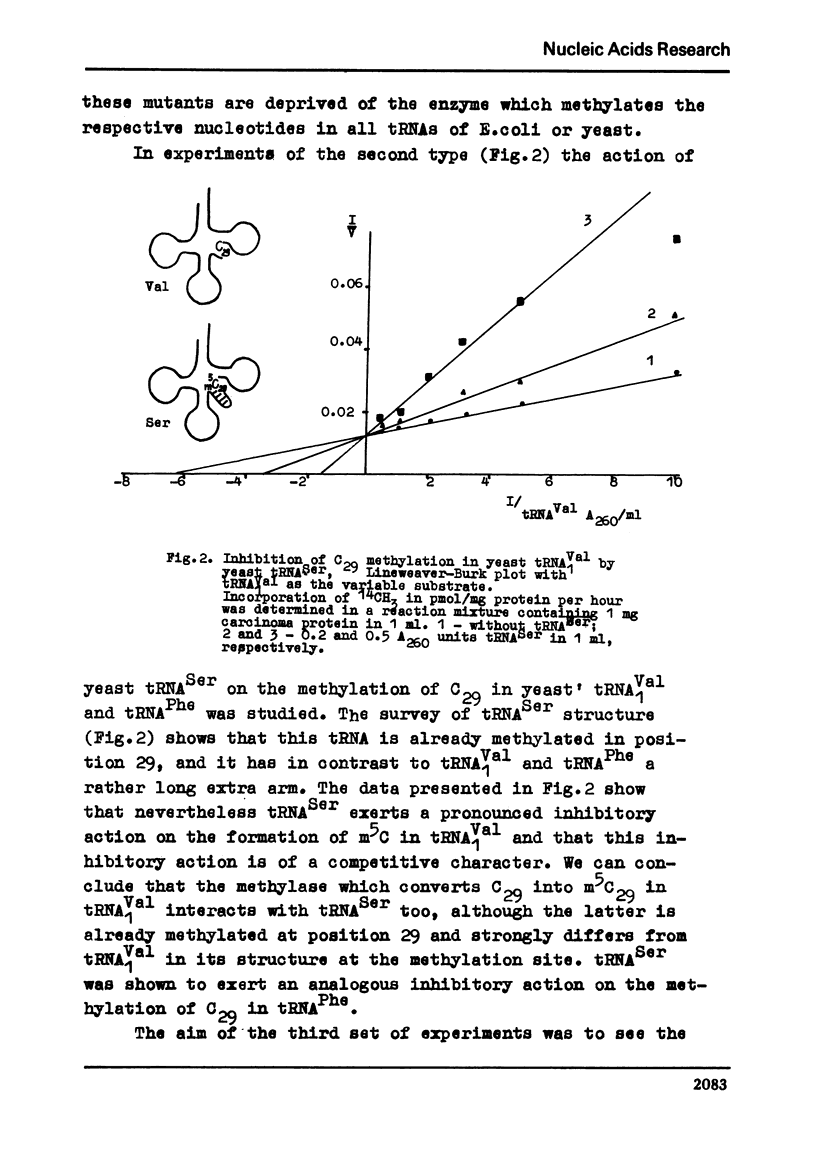
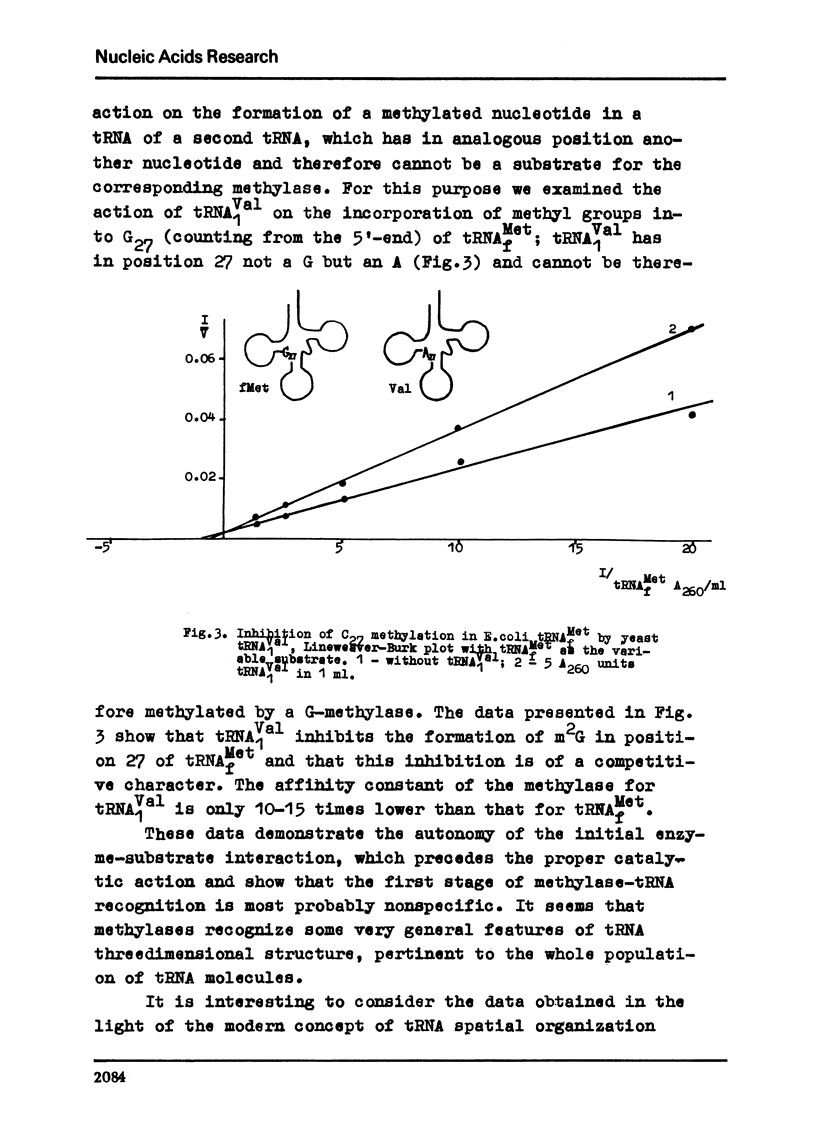
In order to further elucidate the mechanism of tRNA methylase-tRNA intreaction the methylation of some individual tRNAs separately and by pairs was performed. In conditions of tRNA excess the methylation rates of positionally analogous nucleotides in tRNA molecules are not summed up when two substrates are simultaneously present in the reaction mixture. The inhibitory action of yeast tRNASer, possessing m5c in position 29, on the methylation of C29 in other individual tRNAs was shown. Yeast tRNAVal which possesses an A residue in position 27 was shown to inhibit the methylation of G27 in E. coli tRNAMet. The data obtained confirm the suggestion that tRNA methylases recognizes the tertiary structure of tRNAs. They show also that the recognition and the proper catalytic action are two autonomous processes and that the former at least in its first stage is rather unspecific.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Björk G. R., Svensson I. Analysis of methylated constituents from RNA by thin-layer chromatography. Biochim Biophys Acta. 1967 Apr 18;138(2):430–432. doi: 10.1016/0005-2787(67)90504-7. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Nishimura S. Methylation of Escherichia coli transfer ribonucleic acids by adenylate residue-specific transfer ribonucleic acid methylase from rat liver. Biochemistry. 1974 Aug 27;13(18):3683–3688. doi: 10.1021/bi00715a010. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Seno T., Nishimura S. Fragmented E. coli methionine tRNA f as methyl acceptor for rat liver tRNA methylase: alteration of the site of methylation by the conformational change of tRNA structure resulting from fragmentation. Biochem Biophys Res Commun. 1971 May 7;43(3):476–483. doi: 10.1016/0006-291x(71)90638-3. [DOI] [PubMed] [Google Scholar]
- Pegg A. E. The effects of diamines and polyamines on enzymic methylation of nucleic acid. Biochim Biophys Acta. 1971 Apr 8;232(4):630–642. doi: 10.1016/0005-2787(71)90755-6. [DOI] [PubMed] [Google Scholar]
- Phillips J. H., Kjellin-Stråby K. Studies on microbial ribonucleic acid. IV. Two mutants of Saccharomyces cerevisiae lacking N-2-dimethylguanine in soluble ribonucleic acid. J Mol Biol. 1967 Jun 28;26(3):509–518. doi: 10.1016/0022-2836(67)90318-x. [DOI] [PubMed] [Google Scholar]
- Rich A. Transfer RNA and protein synthesis. Biochimie. 1974;56(11-12):1441–1449. doi: 10.1016/s0300-9084(75)80265-3. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Shershneva L. P., Venkstern T. V., Bayev A. A. A study of tRNA methylases by the dissected molecule method. FEBS Lett. 1971 May 20;14(5):297–298. doi: 10.1016/0014-5793(71)80283-1. [DOI] [PubMed] [Google Scholar]
- Shershneva L. P., Venkstern T. V., Bayev A. A. A study of transfer RNA methylation. Biochim Biophys Acta. 1973 Jan 19;294(2):250–262. [PubMed] [Google Scholar]
- Turchinsky M. F., Shershneva L. P. A simple method of quantitative recovery of nucleotides from thin layers. Anal Biochem. 1973 Aug;54(2):315–318. doi: 10.1016/0003-2697(73)90358-8. [DOI] [PubMed] [Google Scholar]


