Abstract
Although mammals are a well-studied group of animals, making accurate field identification of small mammals is still complex because of morphological variation across developmental stages, color variation of pelages, and often damaged osteological and dental characteristics. In 2008, small mammals were collected for an epidemiological study of a spotted fever outbreak in Hainan, China. Ten species of small mammals were identified by morphological characters in the field, most using pelage color characters only. The study is extended here, in order to assess whether DNA barcoding would be suitable as an identification tool in these small mammals. Barcode clusters showed some incongruence with morphospecies, especially for some species of Rattus and Niviventer, so molecular delineation was carried out with an expanded dataset of combined cytochrome b (Cyt-b) and cytochrome c oxidase subunit I (COI) sequences. COI sequences were successfully amplified from 83% of collected mammals, but failed in all specimens of Suncus murinus, which were thus excluded in DNA barcoding analysis. Of note, ten molecular taxonomic units were found from samples of nine morphologically identified species. Accordingly, 11 species of small mammals were present in the investigated areas, including four Rattus species, three Niviventer species, Callosciurus erythraeus, Neohylomys hainanensis, Tupaia belangeri, and Suncus murinus. Based on the results of the phylogenetic and molecular delineation analyses, the systematic status of some rodent species should be redefined. R. rattus hainanicus and R. rattus sladeni are synonyms of R. andamanensis. R. losea from China and Southeast Asia comprises two independent species: R. losea and R. sakeratensis. Finally, the taxonomic status of three putative species of Niviventer should be further confirmed according to morphological, molecular and ecological characters.
Introduction
Rodents are important host animals for many zoonoses that threaten public health worldwide [1], [2]. Typically, there is a specific association between pathogens and host animals, such as the coevolutionary relationship between hantavirus and their rodent hosts [3]. Thus, gaining accurate taxonomic information on host animals is important for surveillance and epidemiological investigation of rodent-borne diseases.
Mammals rank amongst the most studied animal groups, with their taxonomy and species diversity well documented in the literature [4]. However, field identification of many small mammal species remains difficult, in large part because of morphological variation through development, and color variation of pelages (mammalian coat) between individuals. Only through analysis of internal morphology (e.g. skull and dentition) can definitive identification be made. Furthermore, molecular data from one previous study suggests the frequent occurrence of cryptic mammal species that are overlooked when using morphological characters alone [5]. Therefore, a standard molecular identification system is necessary as a complement to morphological methods, in order to reduce uncertainties in the identification of mammal species.
One standardized molecular identification approach, termed DNA barcoding [6], [7], has been extensively used in recent years. This technique can also provide genetic references to validate field identifications made by researchers with limited taxonomic background, which makes it a particularly valuable tool for conducting ecological and epidemiological surveys. Previous applications of this technique in primates and small mammals indicate that it is a valuable method for species identification [8], [9], [10], and DNA barcoding has been instrumental in reassessing the species diversity of regional faunas of small mammals and other taxa [11], [12], [13].
Three potential outcomes of using DNA barcodes in the investigation of species diversity of a specific geographic region are: 1) morphologically homogeneous specimens sharing DNA barcodes with little intraspecific variation; 2) morphologically homogeneous specimens possessing DNA barcodes divergent at a level beyond that expected for species, indicating the possibility of overlooked species [9], [10], [14]; 3) a putative new species or a new record species for the area [9]. When a cryptic species or new species are indicated, a study of their systematic position becomes necessary. However, because systematic information content of the COI barcode is limited, this fragment alone is insufficient for reliable molecular phylogenetic reconstruction and the assignment of new species [15]. Therefore, species delimitation approaches based on multi-locus phylogenies are necessary to define the species status of studied samples, and clarify the relationship with other closely related species. However, using certain phylogenetic based species delimitation methods [16], [17], [18], subjective judgment regarding morphologic and ecological traits are necessary in order to determine whether a highly supported clade should be considered an independent species. Pons et al. [19] proposed a statistical method of DNA-based species delimitation which determines the switch or threshold point of transition from species-level to population-level branching on a phylogenetic tree, giving an estimate of the number of species. This method has been successfully used in species delimitation of asexual mites [20] and rodents of the Rattini tribe from Southeast Asia [21].
In 2007, a severe spotted fever case was reported in Hainan, China [22], which prompted epidemiological investigation. The investigation was carried out in three different counties in the north and central areas of the province, and focused on reservoir animals carrying the Rickettsia bacteria that are responsible for spotted fever. Investigators collected blood and/or tissue samples of livestock, pets, and small mammals (rodents, moonrats, and shrews) for the study. However, identification of the small mammals, carried out prior to collection of blood and tissue samples, was hasty and by means of external morphological characteristics only. Confirmation of the species assignments is necessary to ensure the utility of the epidemiology studies.
Although it is a well-studied group of rodents, there are still some disagreements about species classification in Rattus, such as R. rattus occupying China, and R. losea. There are four subspecies of R. rattus recorded in China: namely R. rattus rattus, R. rattus alexandrinus, R. rattus sladeni and R. rattus hainanicus [23]. Among them, R. rattus rattus and R. rattus alexandrinus were regarded as imported subspecies, while R. rattus sladeni and R. rattus hainanicus were native to southern mainland China, and Hainan Island, respectively [23]. However, Musser and Carleton [24] downgraded R. rattus sladeni to a synonym of R. tanezumi, and downgraded R. rattus hainanicus to a synonym of R. andamanensis. R. losea was described from Taiwan, and recorded in China and other countries of Southeast Asia [24]. This species is discontinuously distributed across mainland Southeast Asia and East Asia in general, and displays two regional forms in its morphology [25] and genetics [26]. Pagès et al. [21] studied the taxonomy of the Rattini tribe based on samples collected from Southeast Asia, using a phylogenetic-based species delimitation method. The authors were hesitant to name one putative species as R. losea because only specimens of Southeast Asia were included, although these specimens were all morphologically identified as R. losea. Aplin et al. [27] confirmed subsequently that R. losea from Taiwan and Southeast Asia were two independent species, and named the latter R. sakeratensis.
The genus Niviventer occurs in China and Southeast Asia, with 17 species recorded [24]. The characters traditionally used to distinguish these species are not completely unequivocal, especially amongst some closely related species. Musser examined a large number of specimens to give morphological and geographic species-limits to some morphologically homogeneous species [28], [29], [30]. However, identification remains difficult even from areas with detailed species records, for example, two putative species reported by Pagès et al. [21]. Jing et al. studied the molecular phylogeny of Niviventer species of China [31], but their species identification was later questioned [21]. The karyotype studies of different Niviventer species summarized by Li et al. [32] also suggested that there are identification problems in these taxa.
Therefore, in the present study, we use DNA barcoding to confirm the field identification assignments, and highlight rodent species prone to misidentification. After barcoding identification, we investigate the taxonomic status of some popular rodent species in Hainan further, including species of Rattus and Niviventer, using molecular species delimitation.
Materials and Methods
Ethical statement
Ethical approval for this study was obtained from the Ethical Committee of Chinese Center for Disease Control and Prevention. Small mammals were live trapped in areas which were not privately owned or protected. All small mammals involved in this paper were neither endangered nor listed as protected species. No specific permits were required for the described field studies.
Animal collection
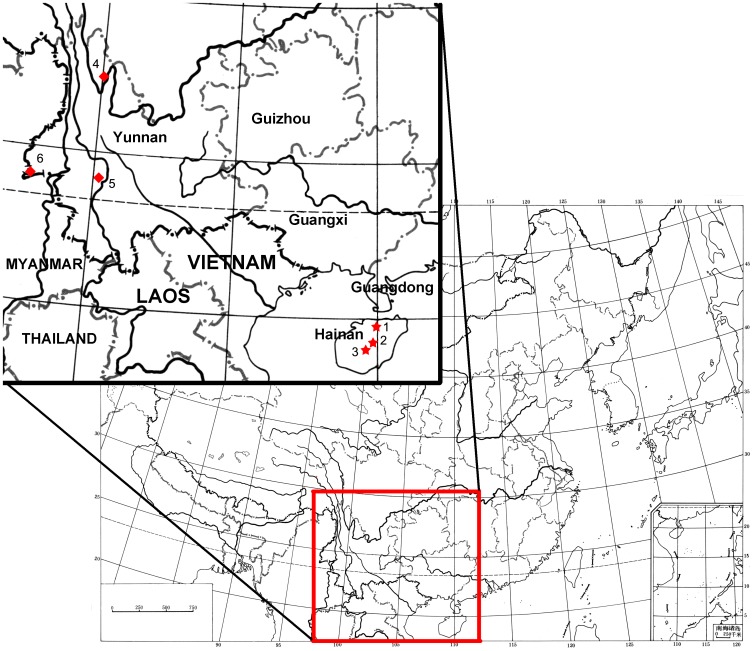
Host animals were collected from three counties of Hainan, including Chengmai, Qiongzhong, and Wuzhishan. Small mammals were live trapped indoors, on farmlands and forests, around villages in these counties (Figure 1). Trapped small mammals were euthanized with CO2 before species identification and sample collection. After standard morphological identification (based on color of pelage, and length of body and tail) [23], blood, liver and spleen samples were collected, stored in liquid nitrogen in the field, and maintained at −80°C in the laboratory until used for experiments. Some material from specimens in good condition were kept as voucher specimens, including skull and pelages. All voucher specimens were identified by taxonomic experts after the field work. Field sample identifications and locality information are listed in Table 1.
Figure 1. Sample locations of the small mammal collected in Hainan, and Rattus rattus sladeni caught from Yunnan.
1: Chengmai, 2: Qiongzhong, 3: Wuzhishan, 4: Deqin, 5: Lincang and 6: Ruili.
Table 1. Information of small mammals collected from Hainan, including field identification results, number of individuals, collection site, and accession numbers of COI and Cyt-b sequences submitted to Genbank.
| Field identification | Number of individuals | Locality | Voucher specimens | COI | Cyt-b |
| Rattus losea | 28 | Qiongzhong | HM031871–HM031896 | HM031709–HM031721 | |
| 5 | Chengmai | ||||
| Rattus rattus hainanicus | 5 | Qiongzhong | HN115 | HM031790–HM031836 | HM031722–HM031763 |
| 6 | Wuzhishan | ||||
| 42 | Chengmai | ||||
| Rattus norvegicus | 2 | Qiongzhong | HM031897–HM031910 | HM031676–HM031682 | |
| 17 | Chengmai | ||||
| Rattus tanezumi | 33 | Chengmai | HM031837–HM031870 | HM031683–HM031708 | |
| 1 | Qiongzhong | ||||
| Niviventer confucianus | 3 | Qiongzhong | HN113 | HM031911–HM031913 | JF714939, JF714941 |
| 1 | Wuzhishan | ||||
| 4 | Chengmai | ||||
| Niviventer fulvescens | 6 | Wuzhishan | HN131, HN162 | HM031914–HM031931 | HM031673–HM031675 |
| 13 | Chengmai | JF714932–JF714938 | |||
| 3 | Qiongzhong | JF 714940, JF714942 | |||
| Callosciurus erythraeus | 4 | Qiongzhong | HN120 | HM031932–HM031935 | N/A |
| Suncus murinus | 4 | Wuzhishan | – | N/A | N/A |
| Neohylomys hainanensis | 2 | Qiongzhong | HN122 | HM031764–HM031766 | N/A |
| 1 | Wuzhishan | ||||
| Tupaia belangeri | 25 | Chengmai | – | HM031767–HM031789 | N/A |
Sequence acquisition
Total genomic DNA of small mammals was isolated using the Qiagen DNAeasy blood and tissue DNA isolation kit (Qiagen China, Pudong, Shanghai) according to the manufacturer's instructions. To amplify 650 bp of the cytochrome c oxidase subunit 1 gene (COI), universal primers BatL5310, R6036R and related amplification conditions were used according to Robins et al. [33]. If amplification with the universal primers failed, the cocktail primer sets were used instead [34]. Although the cocktail primer set was initially designed for fish DNA barcoding, it was successfully used in barcoding of bats [9], and pikas and shrews in author's laboratory. Conditions for the cocktail primer sets were: 94°C for 1 min, five cycles of 94°C for 30 s, 50°C for 40 s, and 72°C for 1 min, followed by 35 cycles of 94°C for 30 s, 54°C for 40 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. For samples from species of Rattus and Niviventer, 1200 bp of the mitochondrial cytochrome b gene (Cyt-b) were also amplified using the primers L14724 and H15915 of Irwin et al. [35]. Each PCR cycle consisted of 93°C for 1 min, 50°C for 1 min and 72°C for 2 min. The cycle was repeated 35 times with a final extension at 72°C for 10 min. All amplicons were directly sequenced in both directions with the ABi 3100 automatic sequencer (Perkin–Elmer, Waltham, MA) using the ABi PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA). COI and Cyt-b sequences of Rattus and Niviventer obtained by Pagès et al. [21] showed substantial overlap with the fragments used here, and so were downloaded from GenBank and added to the dataset. COI and Cyt-b sequences of R. rattus sladeni from Yunnan (Figure 1) were also included in the dataset to confirm the status of the subspecies. Information of sequences of Pagès et al. [21] and R. rattus sladeni were listed in Table 2.
Table 2. Information of sequences from previous studies and R. rattus sladeni included in the dataset.
| Sample number | Locality | Field Identification | Phylogenetic species* | Reference papers | COI | Cyt b |
| R2953 | Kanchanaburi (Thailand) | Rattus tanezumi | Rattus andamanensis | [21] | HM217525 | HM217396 |
| R3087 | Kanchanaburi (Thailand) | Rattus andamanensis | Rattus andamanensis | [21] | HM217533 | HM217403 |
| CB0001 | Veal Renh (Cambodia) | Rattus argentiventer | Rattus argentiventer | [21] | HM217484 | HM217362 |
| CB0104 | Veal Renh (Cambodia) | Rattus argentiventer | Rattus argentiventer | [21] | HM217486 | HM217364 |
| R0284 | Ratchaburi (Thailand) | Rattus exulans | Rattus exulans | [21] | HM217508 | HM217377 |
| R0856 | Nakhon Pathom (Thailand) | Bandicota indica | Rattus exulans | [21] | HM217510 | HM217379 |
| R2795 | Ratchaburi (Thailand) | Rattus exulans | Rattus exulans | [21] | HM217527 | HM217395 |
| R3520 | Sakhon Nakhon (Thailand) | Rattus exulans | Rattus exulans | [21] | HM217553 | HM217424 |
| R4004 | Kalasin (Thailand) | Rattus exulans | Rattus exulans | [21] | HM217564 | HM217437 |
| R5349 | Nan (Thailand) | Rattus exulans | Rattus exulans | [21] | HM217595 | HM217470 |
| R5447 | Nan (Thailand) | Rattus exulans | Rattus exulans | [21] | HM217596 | HM217472 |
| L0010 | Luang Prabang (LPDR) | Rattus sp. | Rattus nitidus | [21] | HM217488 | HM217474 |
| L0180 | Luang Prabang (LPDR) | Rattus nitidus | Rattus nitidus | [21] | HM217492 | HM217478 |
| L0192 | Luang Prabang (LPDR) | Rattus nitidus | Rattus nitidus | [21] | HM217493 | HM217479 |
| R0115 | Ratchaburi (Thailand) | Rattus norvegicus | Rattus norvegicus | [21] | HM217501 | HM217370 |
| R0223 | Ratchaburi (Thailand) | Rattus norvegicus | Rattus norvegicus | [21] | HM217504 | HM217373 |
| MDZ10Mada | Madagascar | Rattus rattus | Rattus rattus | [21] | hM217495 | HM217368 |
| ratcosR12 | Oman | Rattus rattus | Rattus rattus | [21] | HM217496 | HM217366 |
| ratcosT820 | India | Rattus rattus | Rattus rattus | [21] | HM217498 | HM217367 |
| ratcosTE4264 | Tanzania | Rattus rattus | Rattus rattus | [21] | HM217497 | HM217365 |
| R0237 | Ratchaburi (Thailand) | Rattus losea | Rattus sakeratensis | [21], [27] | HM217505 | HM217374 |
| R0238 | Ratchaburi (Thailand) | Rattus losea | Rattus sakeratensis | [21], [27] | HM217506 | HM217375 |
| R1015 | Nakhon Ratchasima (Thailand) | Rattus losea | Rattus sakeratensis | [21], [27] | HM217512 | HM217381 |
| R3484 | Loei (Thailand) | Rattus losea | Rattus sakeratensis | [21], [27] | HM217550 | HM217421 |
| R3510 | Phrae (Thailand) | Rattus losea | Rattus sakeratensis | [21], [27] | HM217552 | HM217423 |
| R4203 | Phrae (Thailand) | Rattus losea | Rattus sakeratensis | [21], [27] | HM217570 | HM217443 |
| R4230 | Loei (Thailand) | Rattus losea | Rattus sakeratensis | [21], [27] | HM217573 | HM217446 |
| R4402 | Loei (Thailand) | Rattus losea | Rattus sakeratensis | [21], [27] | HM217581 | HM217454 |
| CB0028 | Veal Renh (Cambodia) | Rattus tanezumi | Rattus sp. | [21] | HM217485 | HM217363 |
| R0169 | Ratchaburi (Thailand) | Rattus tanezumi | Rattus sp. | [21] | HM217503 | HM217372 |
| R1818 | Prachinburi (Thailand) | Rattus tanezumi | Rattus sp. | [21] | HM217520 | HM217389 |
| R2976 | Nakhon Pathom (Thailand) | Rattus andamanensis | Rattus sp. | [21] | HM217528 | HM217397 |
| R3029 | Bangkok (Thailand) | Rattus tanezumi | Rattus sp. | [21] | HM217530 | HM217399 |
| R4188 | Phrae (Thailand) | Rattus sp. | Rattus sp. | [21] | HM217569 | HM217442 |
| L0100 | Luang Prabang (LPDR) | Rattus tanezumi | Rattus tanezumi | [21] | HM217489 | HM217475 |
| L0194 | Luang Prabang (LPDR) | Rattus tanezumi | Rattus tanezumi | [21] | HM217494 | HM217480 |
| R3122 | Kanchanaburi (Thailand) | Rattus tanezumi | Rattus tanezumi | [21] | HM217537 | HM217407 |
| R3214 | Kanchanaburi (Thailand) | Rattus tanezumi | Rattus tanezumi | [21] | HM217540 | HM217410 |
| R3548 | Phrae (Thailand) | Rattus andamanensis | Rattus tanezumi | [21] | HM217555 | HM217426 |
| R3573 | Nakhon Pathom (Thailand) | Rattus tanezumi | Rattus tanezumi | [21] | HM217558 | HM217430 |
| R4003 | Kalasin (Thailand) | Rattus tanezumi | Rattus tanezumi | [21] | HM217563 | HM217436 |
| R4377 | Loei (Thailand) | Rattus andamanensis | Rattus tanezumi | [21] | HM217579 | HM217452 |
| R4424 | Phrae (Thailand) | Rattus tanezumi | Rattus tanezumi | [21] | HM217582 | HM217456 |
| R4436 | Phrae (Thailand) | Rattus tanezumi | Rattus tanezumi | [21] | HM217583 | HM217457 |
| R4481 | Phrae (Thailand) | Rattus andamanensis | Rattus tanezumi | [21] | HM217584 | HM217458 |
| R5294 | Nan (Thailand) | Rattus tanezumi | Rattus tanezumi | [21] | HM217592 | HM217466 |
| R5296 | Nan (Thailand) | Rattus tanezumi | Rattus tanezumi | [21] | HM217593 | HM217467 |
| R1833 | Nakhon Sri Thammarat (Thailand) | Rattus tanezumi | Rattus tiomanicus | [21] | HM217522 | HM217391 |
| R3427 | Kanchanaburi (Thailand) | Niviventer sp. | Niviventer fulvescens | [21] | HM217545 | HM217416 |
| R3429 | Loei (Thailand) | Niviventer sp. | Niviventer fulvescens | [21] | HM217546 | HM217417 |
| R3459 | Loei (Thailand) | Niviventer sp. | Niviventer fulvescens | [21] | HM217548 | HM217419 |
| R4525 | Loei (Thailand) | Niviventer sp. | Niviventer fulvescens | [21] | HM217589 | HM217464 |
| R4723 | Loei (Thailand) | Niviventer fulvescens | Niviventer fulvescens | [21] | HM217591 | HM217465 |
| R4497 | Phrae (Thailand) | Niviventer sp. | Niviventer fulvescens. | [21] | HM217587 | HM217461 |
| R3795 | Khammouane (LPDR) | – | Niviventer langbianis | [21] | HM217561 | HM217433 |
| R3796 | Khammouane (LPDR) | – | Niviventer langbianis | [21] | HM217562 | HM217434 |
| R3212 | Kanchanaburi (Thailand) | Niviventer langbianis | Niviventer sp1. | [21] | HM217539 | HM217409 |
| LC104 | Lincang, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793910 | JQ793904 |
| LC135 | Lincang, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793914 | JQ793902 |
| LC136 | Lincang, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793912 | JQ793906 |
| LC140 | Lincang, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793913 | JQ793903 |
| RL038 | Ruili, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793916 | JQ793908 |
| RL039 | Ruili, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793917 | JQ793909 |
| RL055 | Ruili, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793911 | JQ793905 |
| DQ366 | Deqin, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793920 | JQ793901 |
| DQ372 | Deqin, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793918 | JQ793899 |
| DQ373 | Deqin, Yunnan (China) | Rattus rattus sladeni | Rattus andamanensis | This paper | JQ793919 | JQ793900 |
Phylogenetic analyses
All sequences were aligned using CLUSTALW [36] and manually confirmed. The COI and Cyt-b gene sequences of specimens of Rattus and Niviventer were aligned separately, and trimmed to a common length before concatenation. Neighbour Joining (NJ) trees based on COI sequences were generated using K2P distances, calculated in Paup*4b [37]. Missing data were ignored for distance calculation, and ties were broken at random. Phylogenies were generated from the complete dataset using Maximum likelihood (ML) and Bayesian inference (BI) approaches. Micromys minutus (HM217360, HM217482) was selected as the outgroup in all analyses. For use in model based tree inferences, the best fit substitution models were determined for the two partitions (COI and Cyt-b) using Likelihood ratio tests [38], [39] implemented in Jmodeltest0.1 [40]. The TPM1uf+G model was selected for COI of Rattus and Niviventer species, and the TIM2+I+G model was selected for Cyt-b sequences. ML trees were inferred using Garli v2.0 [41], a software allowing the implementation of partitioned evolutionary models. The best fit model for each gene was input via the starting model option (the ‘streefname’ option given in the configuration file), and these values fixed. Then a partitioned search was performed with otherwise default settings. Node support was obtained via bootstrapping, with the topology termination threshold (parameter: genthreshfortopoterm) reduced to 1000 to increase search speed. Bayesian trees were inferred using MrBayes v3.1.2 [42], again with a partitioned model. The Bayesian search was run for 2 million generations, sampling every 500, with two independent runs performed, each consisting of three heated and one cold chain. Convergence was assessed using the standard deviation of split frequencies, and the estimated sample sizes (ESS) of the sampled parameters, as calculated using Tracer [43].
Molecular delineation was carried out on a dataset from which identical haplotypes were removed (according to the algorithm given by [44]). The dereplicated dataset consisted of 52 sequences, with a total length of 1789 bp. A NJ tree was first generated, using Paup*4b [37]. Genetic distances were calculated under the K2P model, where missing data were ignored in distance calculation, and ties broken at random. ML and BI trees were also inferred for the dereplicated dataset, using the same method as used for the analysis of the complete dataset. The phylogenies from the three different methods were clock constrained using r8s 1.71 [45]. The root node was fixed at an arbitrary value of 1.0, then ultrametric trees formed by penalized likelihood (PL) and non-parametric rate smoothing (NPRS). For PL, smoothing parameters were compared by cross calibration (r8s command: divtime method = pl crossv = yes cvstart = −3 cvinc = 1 cvnum = 9), with the optimal value (10), used in further analyses. Finally, the putative species units on the ultrametric trees were determined using the general mixed Yule coalescent (GMYC) method [19]. This procedure detects the switch in the rate of lineage branching in a tree, from interspecific long branches to intraspecific short branching, and identifies clusters of specimens corresponding to putative species. A threshold (T) is optimized with the GMYC model so that nodes before the threshold are considered as species diversification events, therefore the number of species can be estimated. Significance was assessed by likelihood ratio test against a null model of a single coalescent population. This test was implemented using R code provided by T. G. Barraclough.
Results
A total of 205 small mammals were collected from three counties of Hainan. According to the morphological criteria we used, seven species belonged to three genera of Rodentia, two species belonged to two genera of Soricomorpha, and there was one species of Scandentia (Table 1).
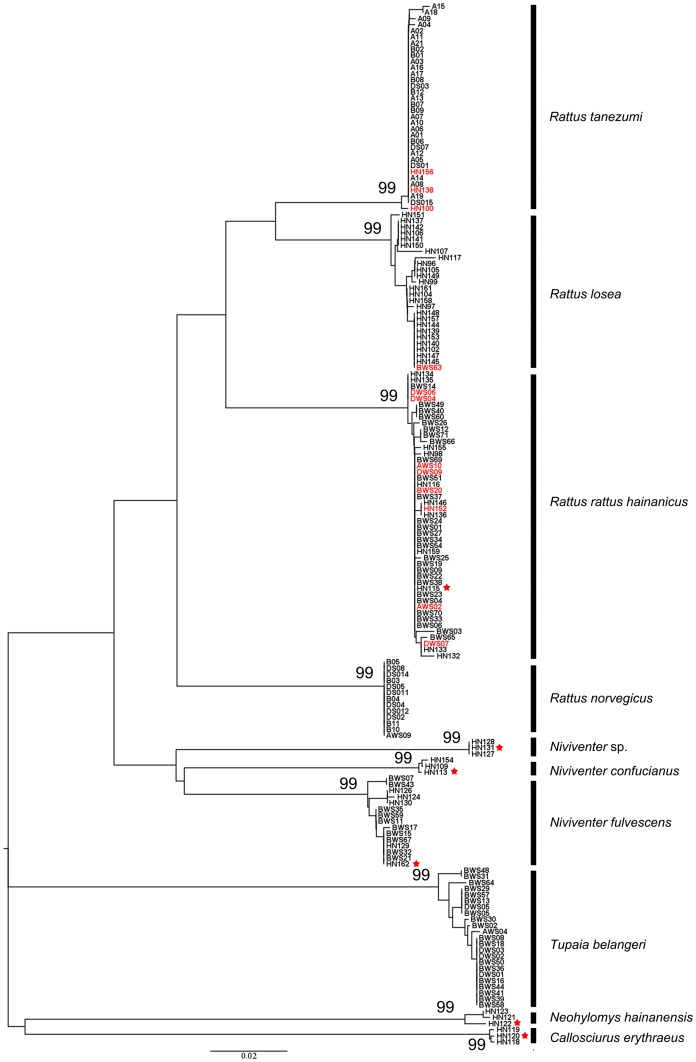
COI amplicons, each approximately 650 bp in length, were recovered from 172 individual animals (83%). Of note, amplification failed in all specimens of Suncus murinus, even with the cocktail primer set. The NJ tree of COI sequence from Hainan showed that there were ten well supported lineages (Figure 2). Nine of these lineages corresponded to field identified species, but three specimens identified (via morphology) as N. confucianus were not clustered with other members of this species in the tree. These three specimens (one cluster with two members, and a singleton) are labeled as Niviventer sp in Figure 2. According to the NJ tree, 12 specimens were clustered in a lineages different to that given by the field identification, indicating field misidentification (Table 3). These specimens were identified as species of Rattus, of which juveniles were particularly difficult to be distinguished.
Figure 2. Neighbor-joining tree of COI sequence for small mammals collected from Hainan.
Samples separated by small genetic distance (<2%) were labeled with one vertical black bar and regarded as one species. The red sample names meant they were misidentified in the field. The samples labeled with red star indicated the availability of voucher specimens.
Table 3. Information of samples misidentified in the field of Hainan.
| Sample number | Locality | Field identification | Barcoding identification | GenBank accession numbers of COI |
| HN100 | Qiongzhong | Rattus norvegicus | Rattus tanezumi | HM031870 |
| HN138 | Qiongzhong | Rattus losea | Rattus tanezumi | HM031864 |
| HN152 | Qiongzhong | Rattus losea | Rattus rattus hainanicus | HM031818 |
| HN156 | Qiongzhong | Rattus losea | Rattus tanezumi | HM031865 |
| AWS02 | Chengmai | Niviventer fulvescens | Rattus rattus hainanicus | HM031797 |
| AWS10 | Chengmai | Niviventer fulvescens | Rattus rattus hainanicus | HM031796 |
| BWS20 | Chengmai | Rattus losea | Rattus rattus hainanicus | HM031823 |
| BWS63 | Chengmai | Rattus rattus hainanicus | Rattus losea | HM031872 |
| DWS04 | Chengmai | Rattus losea | Rattus rattus hainanicus | HM031806 |
| DWS06 | Chengmai | Rattus losea | Rattus rattus hainanicus | HM031807 |
| DWS07 | Chengmai | Rattus losea | Rattus rattus hainanicus | HM031790 |
| DWS09 | Chengmai | Rattus losea | Rattus rattus hainanicus | HM031813 |
The average K2P distances between individuals of R. norvegicus was 0, the distance between individuals of R. tanezumi 0.08%, and that of R. rattus hainanicus and R. losea was 0.21% and 0.41% respectively. The average intraspecific distance among different Niviventer species ranged from 0 to 0.40%, similar with that of Rattus species. We determined that the divergences between Rattus species ranged from 7% to 13%, and 11%–14% for Niviventer species.
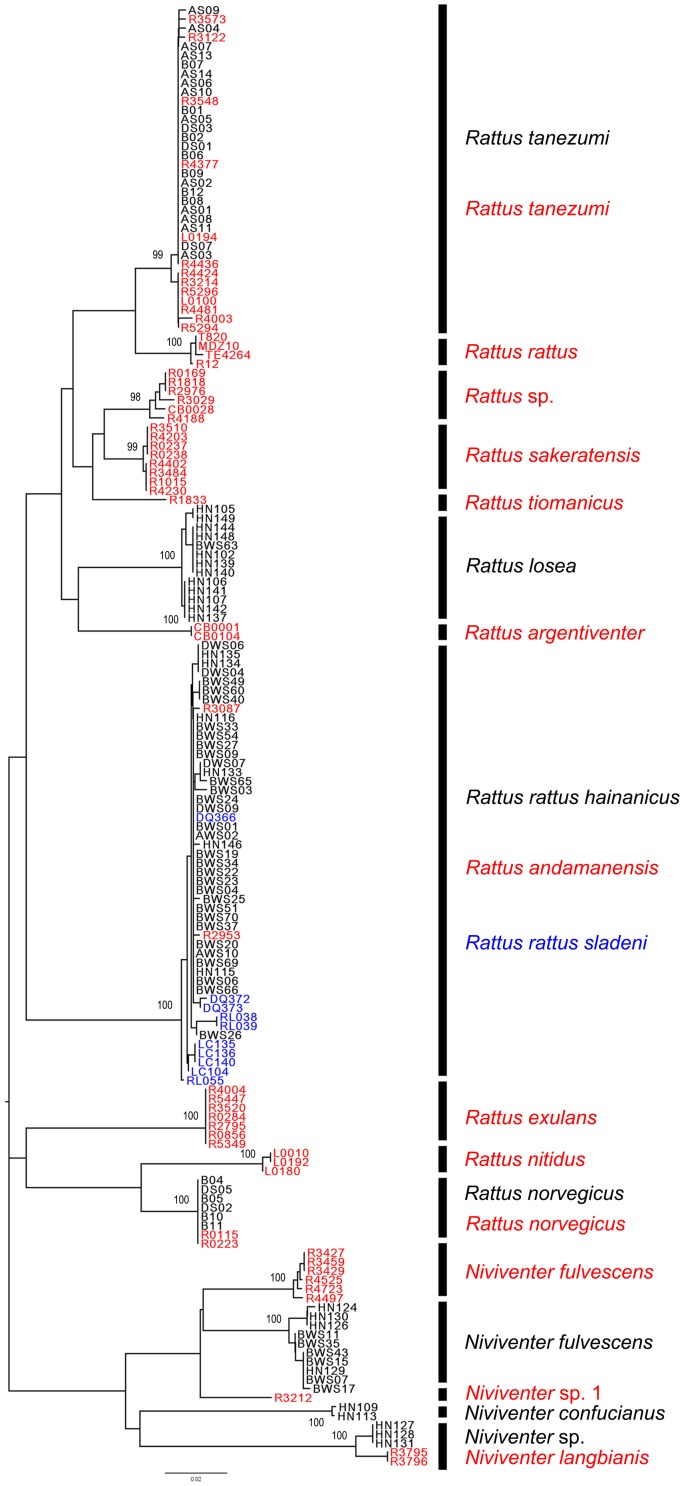
The NJ tree of the combined COI dataset from Hainan and Southeast Asia (Figure 3) showed that R. tanezumi and R. norvegicus of Hainan clustered with samples of the same species from Southeast Asia, R. losea of Hainan formed an independent cluster from the R. sakeratensis (R. losea-like in Pagès et al. [21]) collected from Southeast Asia, and R. rattus hainanicus, and R. rattus sladeni from Yunnan grouped with R. andamanensis from Southeast Asia. The average intraspecific distance of Rattus species was 0.23%, ranging from 0 (R. norvegicus) to 1.30% (R. andamanensis). The interspecific distance of Rattus species ranged from 5.5% to 15%. For Niviventer species, N. fulvescens of Hainan clustered independently from N. fulvescens of Southeast Asia, and two samples of Hainan grouped with two samples of N. langbianis from Laos. The average intraspecies distance of Niviventer species ranged from 0.20% (N. confucianus) to 1.60% (N. langbianis). The interspecies distance of Niviventer species ranged from 14.0%–18.9%.
Figure 3. Neighbor-joining tree of COI sequence of Rattus and Niviventer collected from Hainan, Yunnan and Southeast Asia.
Samples with black name and species name were collected from Hainan, those with blue name and species name were from Yunnan, and the red from Southeast Asia [21]. Samples separated by small genetic distance (<2%) were labeled with one vertical black bar and regarded as one species. Low bootstrap support value (<90%) on deep branches were not shown.
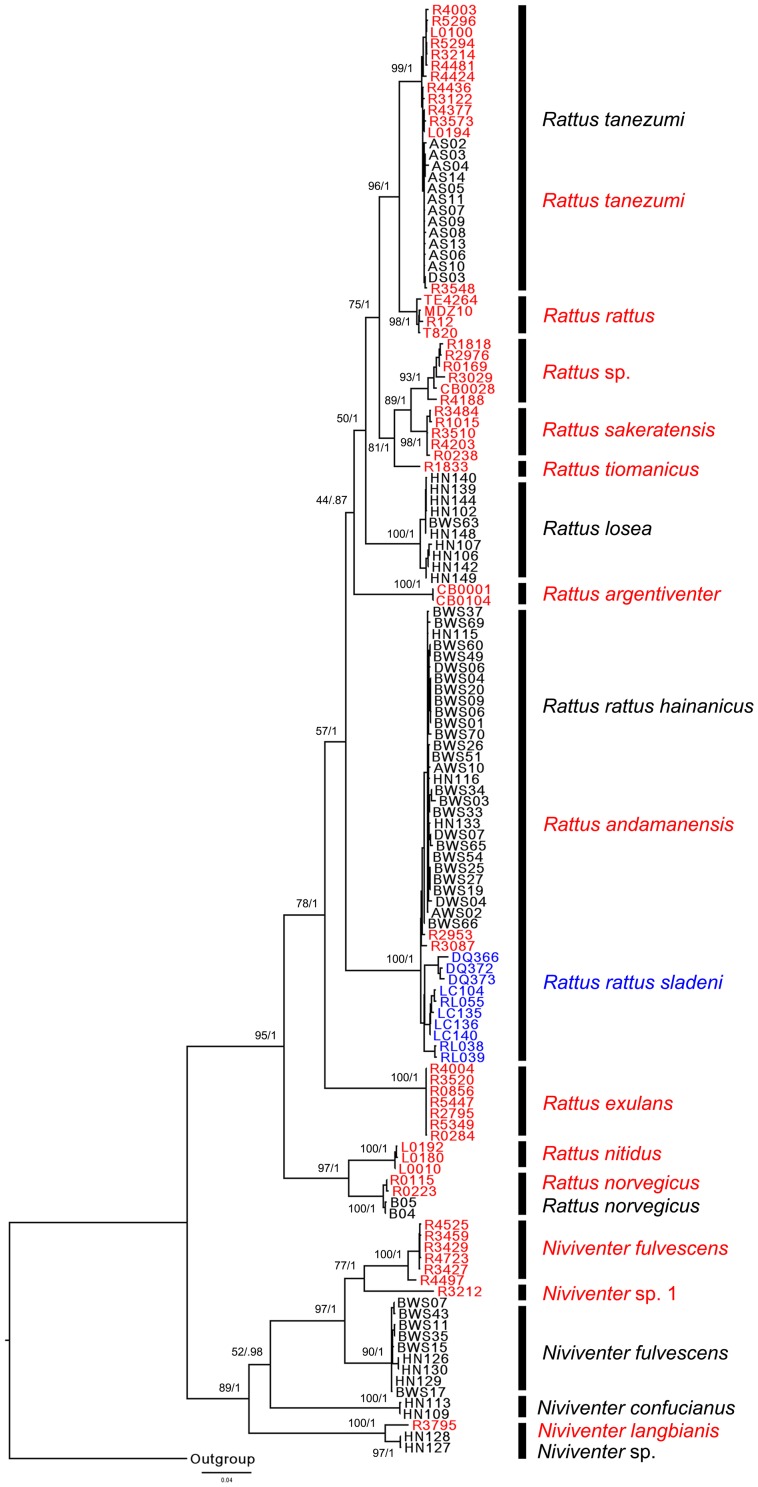
The ML tree and BI tree showed identical topology (Figure 4). According to the trees, samples of R. norvegicus, and R. tanezumi were clustered with samples of the same species collected from Southeast Asia, while samples of R. rattus hainanicus and R. rattus sladeni formed a branch with samples of R. andamanensis, and samples of R. losea formed a branch independent from R. sakeratensis from Southeast Asia. Samples of N. fulvescens and N. confucianus from Hainan did not cluster with any samples from Southeast Asia. Two samples of Niviventer sp. from Hainan showed close relationship with samples of N. langbianis from Southeast Asia.
Figure 4. Maximum likelihood tree of Rattus and Niviventer species from Hainan, Yunnan and Southeast Asia based on the combined COI and Cyt-b dataset.
ML and BI analyses of the dataset gave identical tree topology. Numbers beside the nodes reflect support obtained from the analysis of the dataset following two different reconstruction methods: ML/BI. The meaning of different colors of samples and lineages names is the same as in the Figure 3.
Phylogenetic based species delimitation was carried out on a series of ultrametric trees. For trees in which the NPRS method was applied, the GMYC model showed no significant fit (NJ, p = 0.1162, ML, p = 0.1756, BI, p = 0.1666). Whereas trees adjusted by PL showed significant GMYC structure in all cases, irrespective of the optimization method used (Powell, TN or Qnewt). 17 (likelihood ratio: 19.12398, p = 0.0002), 17 (likelihood ratio:13.21194, p = 0.00419) and 18 (likelihood ratio = 12.37016, p = 0.00621) species were inferred (not including the outgroup) on the NJ, ML and BI trees, respectively, including 11 and 12 species of Rattus and 6 species of Niviventer. The confidence interval for the number of species ranged from 17 to 24, which is demarcated in the blue shadow in Figure 5.
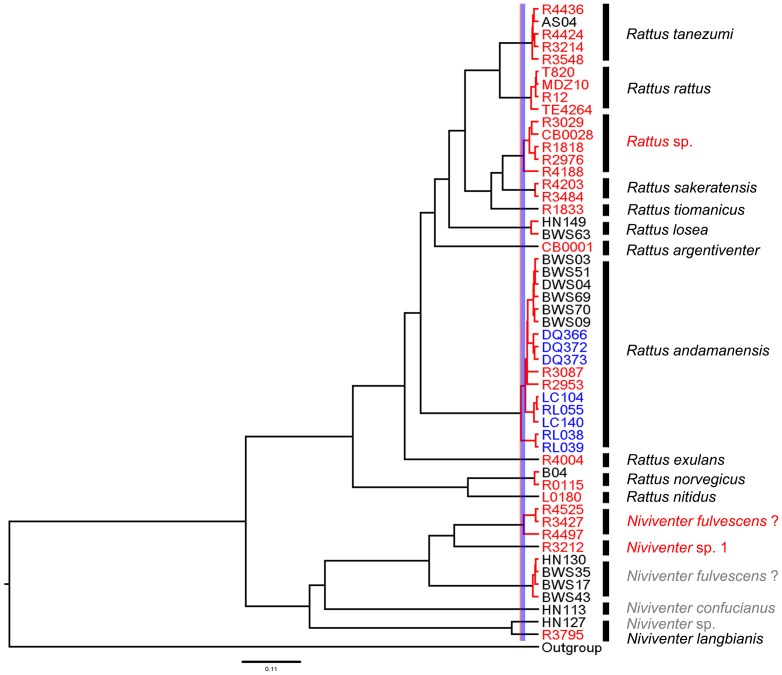
Figure 5. Ultrametric tree of Rattus and Niviventer species from Hainan, Yunnan and Southeast Asia based on the combined Cyt-b and COI dataset.
Red clusters of specimens were recognized as putative species by the method of Pons et al. [19]. The blue shadow on the tree indicated the confidence interval of the threshold, and the red vertical line was the threshold point obtained from the GMYC model. The meaning of different colors of samples names is the same as in the Figure 3. The black species names were confirmed species names in published papers and this paper; the red and gray species names means they were not confirmed in published papers and this paper.
According to the species delimitation results, samples of R. rattus hainanicus and R. rattus sladeni grouped with samples of R. andamanensis from Southeast Asia [21] as one putative species, partly supporting the downgrading of hainanicus to a synonym of R. andamanensis by Musser and Carleton [24]. Niviventer specimens from Hainan were split into three putative species, and did not group with three species from Southeast Asia, even though two putative species were morphologically identified as N. fulvescens (Figure 5). Niviventer specimens collected from Kanchanaburi of Thailand, labeled as Niviventer sp1 [21], were related to N. fulvenscens, although with a molecular distance (∼0.1) outside that expected for a single species. The specimens labeled as Niviventer sp were placed close to N. langbianis, although it can not be concluded that they belong to this species, due to the position of the threshold confidence interval.
Discussion
The distinct gap between intraspecific and interspecific variation is the cornerstone of the DNA barcoding tool for species identification [14], [15], [46]. In the present study, the range of intraspecific variation of COI for Rattus and Niviventer was low (<1.60%) while interspecific variation was high (>5.50%), almost 5–30 times higher than the average differences within species. Based on this gap, morphospecies generally formed well-supported clusters on the NJ tree. The COI data from Hainan gives 10 well-supported branches (Figure 2), meaning 10 putative species in the included samples, whereas the combined COI dataset from Hainan and Southeast Asia gave 16 well-supported groups (Figure 3). Among these branches, two Niviventer samples from Hainan were clustered with samples of N. langbianis from Laos with an average genetic distance of 1.60%, which was very small compared with the range of interspecific variation of Niviventer species.
In this paper, only the mitochondrial COI and Cyt-b genes (and no genes of the nuclear genome) were used in phylogenetic analysis and species delimitation. According to Figure 4, the phylogeny of Rattus species is concordant with the result in Figure 2 of Pagès et al. [21], the latter of which was based on the analysis of combined Cyt-b, COI and IRBP genes. Figure 4 of Pagès et al. [21], a ML tree based on Cyt-b and COI only, also show an identical topological relationship of Rattus species with Figure 2 of their paper. IRBP is a nuclear gene frequently used in phylogenetic analysis of mammals [47], [48], [49], [50]. While for the species of Rattus, the IRBP gene reported by Pagès et al. [21] could give only limited phylogenetic information (data not shown), thus we did not use this gene for this paper.
Species delimitation is an issue fundamental to taxonomic, evolutionary and ecological research. Use of morphological data alone in traditional species delimitation may underestimate the number of species and, in particular, may fail to identify cryptic species. Phylogenetic-based species delimitation using molecular information gives an opportunity to overcome the above weakness, hence the development of a series of analysis methods [19], [51], [52], [53]. In this paper, the GMYC method delimited 17 (NJ, ML) or 18 (BI) putative species, including 11 or 12 species of Rattus and 6 species of Niviventer from combined COI and Cyt-b data from Hainan and Southeast Asia (Figure 5 for the NJ tree). While the NJ tree of combined COI data (Figure 3) supported 11 species of Rattus and 5 species branches of Niviventer, which was also supported by a ML tree of combined COI and Cyt-b (Figure 4). According to the position of the threshold (Figure 5), N. langbianis (Figure 3) comprises two distinct lineages. Additionally, R. andamanensis (Figures 3 and 5) was split into two clusters when the BI tree was used for GMYC analysis (Data not shown). The samples of N. langbianis and R. andamanensis showed a greater range of intraspecific distance in COI than other species, with 0–1.30% for R. andamanensis and 0–1.60% for N. langbianis. According to the confidence interval of the threshold, R4188, R4497, RL038 and RL039 could be delimited as independent species (Figure 5). These samples all show a relatively large distance (0.7%–1.6%). These results suggest that where the range of intraspecific distance is great, GMYC analysis tends towards an increase in the number of species units.
The DNA barcoding results were mostly congruent with that of the species delimitation (Figure 3 and Figure 5), with 15 equivalent species assignments. The remaining clade was considered one species with the barcoding method (Figure 3), including N. langbianis from Southeast Asia and two samples from Hainan, but was split into two species units by the GMYC model (Figure 5), because of the relative high genetic distance between them. The congruent results add weight to the 15 molecular species assignments, and suggest the presence of a strong signal in the molecular data of the groups researched here. On the other hand, the diversity of Rattus and Niviventer species of China and Southeast Asia revealed by molecular data indicate that further taxonomic study is required for these two genera.
The combined results of morphological identification, DNA barcoding and molecular species delimitation showed that there are four Rattus species (R. tanezumi, R. norvegicus, R. losea, and R. andamanensis), three Niviventer species, and Callosciurus erythraeus, Neohylomys hainanensis, Tupaia belangeri and Suncus murinus, in the investigated area of Hainan (especially in and nearby residential environments). According to the collection records, almost all specimens of R. tanezumi and R. norvegicus were collected indoors, while R. losea and R. andamanensis were all trapped in farmlands and forests around residential sites. In the laboratory, collected blood samples of small mammals were checked to determine whether they were infected by Richettsiae bacteria. Only N. fulvescens from Chengmai county were found to harbor the bacteria, with over half of the specimens PCR positive (seven), and seven isolates obtained [22]. The high infection rate of N. fulvescens in Hainan and another report [54] indicate that this species is an important host animal of Rickettsia bacteria in Southern China. The accurate identification of small mammals can give information on, not only the host animal of specific pathogens, but also the possible distribution of related diseases according to the distribution of host animals, which is very important in zoonotic disease control and prevention.
There are four subspecies of R. rattus recorded in China: R. rattus rattus, R. rattus alexandrinus, R. rattus sladeni and R. rattus hainanicus. The last two subspecies were only recorded in China, and all are wild species as opposed to commensal rodents [23]. In contrast, Musser and Carleton [24] regarded sladeni as a synonym of R. tanezumi, and hainanicus as a synonym of R. andamanensis. The results from this study confirmed that samples of R. rattus hainanicus from Hainan Island, R. rattus sladeni of Yunnan and R. andamanensis from Southeast Asia belong to one species. The genetic distance in COI of these samples ranged from 0 to 1.3%, with an average value of 0.34%. The preferred habitation of R. rattus hainanicus recorded in the investigation was also similar to that of R. andamanensis (R. sikkimensis in Aplin et al. [25]).
Rattus losea was described from Taiwan, with morphologically identical or similar specimens recorded from East and Southeast Asia. However, Aplin et al. [25] reported that R. losea was discontinuously distributed across mainland Southeast Asia and East Asia, and that morphological variation existed between two geographic populations. Accordingly, Pagès et al. [21] did not name samples collected only from Southeast Asia as R. losea although these samples possessed the morphological characters of this species and formed a single independent group. Aplin et al. [27] confirmed that the losea-like rats of Southeast Asia should be named R. sakeratensis. Our research further confirmed the caution of Pagès et al. [21] and the result of Aplin et al. [27] with the samples from Hainan.
The Niviventer genus is a diverse group distributed throughout East and Southeast Asia, with nine individual species recorded in China to date [31], [55]. Among them, Wang [55] reported that N. confucianus lotipes and N. fulvescens had been recorded in Hainan. In the list of Musser and Carleton [24], N. tenaster and N. fulvescens occurred on Hainan Island, since these authors regarded lotipes as a synonyms of N. tenaster.
The NJ tree of COI in our study demonstrates that there are three independent lineages of Niviventer collected in Hainan (Figure 2). After checking the morphological characteristics of the three voucher specimens of each putative species, the two clades could be named as N. confucianus (HN113) and N. fulvescens (HN162). Whereas the voucher specimen (HN131) for the third clade was a white-bellied rat with a mono-colored dark brown tail. There are only two known Niviventer species with mono-colored dark tails, N. cremoriventer and N. langbianis [28]. N. cremoriventer is recorded in Yunnan of China [31], [55], while N. langbianis has no record in China, but likely to be found in Southern China according to its distribution in Southeast Asia [28]. The voucher specimen has a relatively large bullae and long anterior incisive foramina, and could be identified as N. langbianis. The DNA-based species delimitation gave more complex results than that of the COI NJ tree. The N. fulvescens from Hainan and Southeast Asia formed two independent clades. We could not confirm which one was the true fulvescens although it was also discussed by Pagès et al. [21]. As with the N. langbianis-like individual from Hainan and Southeast Asia, the langbianis-like specimens from Hainan should not be named until further studies of morphology, ecology and genetics are carried out. For the third N. confucianus-like species found in Hainan, sufficient molecular characters of N. tenaster were necessary to confirm whether it was N. confucianus or N. tenaster. However, a series of works were carried out using only the Cyt-b gene, in order to explore the species level phylogenetics and phylogeography of members from China and Vietnam [31], [56], [57], [58]. Using these sequences there was insufficient information (data not shown) supporting whether the confucianus-like specimens found in Hainan were N. tenaster. Accordingly, more molecular data and increased sampling are necessary to confirm the systematic position of these individuals from Hainan.
Acknowledgments
We would like to thank anonymous reviewers for comments that helped us to improve the manuscript. We are particularly grateful to the people who gave us help in the field work: Zhao Wei of Hainan Provincial Center for Disease Control and Prevention, and Gong Zhengda and Cai Wenfeng of the Institute of Epidemiology of Yunnan Province. We thank Professor Yong Ma of the Institute of Zoology, Chinese Academy of Sciences, for identifying all voucher specimens included in this research.
Funding Statement
This work was supported by the Special Infectious Disease Program (Grant No. 2008ZX10004-010) and the National Science and Technology Pillar Program (2008BAI56B02) of the Ministry of Science and Technology, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mills JN, Childs JE (1998) Ecologic studies of rodent reservoirs: their relevance for human health. Emerging Infectious Diseases 4: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meerburg BG, Singleton GR, Kijlstra A (2009) Rodent-borne diseases and their risks for public health. Critical Reviews in Microbiology 35: 221–270. [DOI] [PubMed] [Google Scholar]
- 3. Monroe MC, Morzunov SP, Johnson AM, Bowen MD, Artsob H, et al. (1999) Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerging Infectious Diseases 5: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson DE, Reeder DM (2005) Mammal species of the world: a taxonomic and geographic reference. Baltimore, Maryland: Johns Hopkins University Press. 2142 p. [Google Scholar]
- 5. Baker RJ, Bradley RD (2006) Speciation in mammals and the genetic species concept. Journal of Mammalogy 87: 643–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hebert PDN, Cywinska A, Ball SL, Dewaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London, Series B: Biological Sciences 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hebert PDN, Ratnasingham S, Jeremy RW (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society B: Biological Sciences 270: S96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lorenz JG, Jackson WE, Beck JC, Hanner RH (2005) The problems and promise of DNA barcodes for species diagnosis of primate biomaterials. Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1869–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clare EL, Lim BK, Engstrom MD, Eger JL, Hebert PDN (2007) DNA barcoding of Neotropical bats: species identification and discovery within Guyana. Molecular Ecology Notes 7: 184–190. [Google Scholar]
- 10. Borisenko AV, Lim BK, Ivanova NV, Hanner RH, Hebert PDN (2008) DNA barcoding in surveys of small mammal communities: a field study in Suriname. Molecular Ecology Resources 8: 471–479. [DOI] [PubMed] [Google Scholar]
- 11. Janzen DH, Hajibabaei M, Burns JM, Hallwachs W, Remigio E, et al. (2005) Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1835–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith MA, Fisher BL, Hebert PDN (2005) DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar. Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1825–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA (2007) DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends in Genetics 23: 167–172. [DOI] [PubMed] [Google Scholar]
- 14. Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America 101: 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN (2006) DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences 103: 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ross KG, Gotzek D, Ascunce MS, Shoemaker DDW (2010) Species delimitation: a case study in a problematic ant taxon. Systematic Biology 59: 162–184. [DOI] [PubMed] [Google Scholar]
- 17. Siler CD, Brown RM (2010) Phylogeny-based Species Delimitation in Philippine Slender Skinks (Reptilia: Squamata: Scincidae: Brachymeles): Taxonomic Revision of Pentadactyl Species Groups and Description of Three New Species. Herpetological Monographs 1–54. [Google Scholar]
- 18. Rivera PC, Di Cola V, Martínez JJ, Gardenal CN, Chiaraviglio M (2011) Species Delimitation in the Continental Forms of the Genus Epicrates (Serpentes, Boidae) Integrating Phylogenetics and Environmental Niche Models. PLoS One 6: e22199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, et al. (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology 55: 595–609. [DOI] [PubMed] [Google Scholar]
- 20. Fontaneto D, Boschetti C, Ricci C (2008) Cryptic diversification in ancient asexuals: evidence from the bdelloid rotifer Philodina flaviceps. Journal of Evolutionary Biology 21: 580–587. [DOI] [PubMed] [Google Scholar]
- 21. Pages M, Chaval Y, Herbreteau V, Waengsothorn S, Cosson JF, et al. (2010) Revisiting the taxonomy of the Rattini tribe: a phylogeny-based delimitation of species boundaries. BMC Evolutionary Biology 10: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin Y, Zhang L, Sun L, Lao S, Han J, et al. (2011) Epidemiological investigation of emerging spotted fever in Chengmai county, Hainan province. Disease Surveillance 26: 18–23. [Google Scholar]
- 23.Huang WJ, Chen YX, Wen YX (1995) Glires of China. Shanghai, China.(In Chinese): Publishing House of Fudan University. 309 p. [Google Scholar]
- 24.Musser GG, Carleton MD (2005) Superfamily Muroidea. [In: Mammal species of the World a taxonomic and geographic reference. DEWilson and DMReeder, eds]. Johns Hopkins University Press, Baltimore, Maryland. [Google Scholar]
- 25. Aplin KP, Brown PR, Jacob J, Krebs CJ, Singleton GR, et al. (2003) Field methods for rodent studies in Asia and the Indo-Pacific: Australian Centre for International Agricultural Research. 223. [Google Scholar]
- 26. Yu HT, Fang YP, Chou CW, Huang SW, Yew FH (1996) Chromosomal evolution in three species of murid rodents of Taiwan. Zoological Studies 35: 195–199. [Google Scholar]
- 27. Aplin KP, Suzuki H, Chinen AA, Chesser RT, Have J, et al. (2011) Multiple geographic origins of commensalism and complex dispersal history of black rats. PLoS One 6: e26357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Musser GG (1973) Species-limits of Rattus cremoriventer and Rattus langbianis, murid rodents of Southeast Asia and the Greater Sunda Islands. American Museum Novitates 2525: 1–65. [Google Scholar]
- 29. Musser GG (1970) Species-limits of Rattus brahma, a murid rodent of northeastern India and northern Burma. American Museum Novitates 2406: 1–27. [Google Scholar]
- 30. Musser GG (1981) Notes on systematics of Indo-Malayan murid rodents, and descriptions of new genera and species from Ceylon, Sulawesi, and the Philippines. Bull Am Mus Nat Hist 168: 225–334. [Google Scholar]
- 31. Jing M, Yu HT, Wu SH, Wang W, Zheng X (2007) Phylogenetic relationships in genus Niviventer (Rodentia: Muridae) in China inferred from complete mitochondrial cytochrome b gene. Molecular Phylogenetics and Evolution 44: 521–529. [DOI] [PubMed] [Google Scholar]
- 32. Li YC, Wu Y, Harada M, Lin LK, Motokawa M (2008) Karyotypes of Three Rat Species (Mammalia: Rodentia: Muridae) from Hainan Island, China, and the Valid Specific Status of Niviventer lotipes. Zoological science 25: 686–692. [DOI] [PubMed] [Google Scholar]
- 33. Robins JH, Hingston M, Matisoo-Smith E, Ross HA (2007) Identifying Rattus species using mitochondrial DNA. Molecular Ecology Notes 7: 717–729. [Google Scholar]
- 34. Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN (2007) Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes 7: 544–548. [Google Scholar]
- 35. Irwin DM, Kocher TD, Wilson AC (1991) Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution 32: 128–144. [DOI] [PubMed] [Google Scholar]
- 36. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford DL (2003) PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4. [Google Scholar]
- 38. Goldman N (1993) Statistical tests of models of DNA substitution. Journal of Molecular Evolution 36: 182–198. [DOI] [PubMed] [Google Scholar]
- 39. Huelsenbeck JP, Crandall KA (1997) Phylogeny estimation and hypothesis testing using maximum likelihood. Annual Review of Ecology and Systematics 28: 437–466. [Google Scholar]
- 40. Posada D (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 41. Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion [PhD thesis]: The university of Texas at Austin. [Google Scholar]
- 42. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572. [DOI] [PubMed] [Google Scholar]
- 43. Rambaut A, Drummond AJ (2007) Tracer v1. 4. [Google Scholar]
- 44. Quicke DLJ, Smith MA, Janzen DH, Hallwachs W, Fernandez-Triana J, et al. (2012) Utility of the DNA barcoding gene fragment for parasitic wasp phylogeny (Hymenoptera: Ichneumonoidea): data release and new measure of taxonomic congruence. Molecular Ecology Resources [DOI] [PubMed] [Google Scholar]
- 45. Sanderson MJ (2003) r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19: 301–302. [DOI] [PubMed] [Google Scholar]
- 46. Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia's fish species. Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Montgelard C, Forty E, Arnal V, Matthee C (2008) Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evolutionary Biology 8: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lecompte E, Aplin KP, Denys C, Catzeflis F, Chades M, et al. (2008) Phylogeny and biogeography of African Murinae based on mitochondrial and nuclear gene sequences, with a new tribal classification of the subfamily. BMC Evolutionary Biology 8: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meredith RW, Westerman M, Springer MS (2009) A phylogeny of Diprotodontia (Marsupialia) based on sequences for five nuclear genes. Molecular Phylogenetics and Evolution 51: 554–571. [DOI] [PubMed] [Google Scholar]
- 50. Meredith RW, Mendoza MA, Roberts KK, Westerman M, Springer MS (2010) A phylogeny and timescale for the evolution of Pseudocheiridae (Marsupialia: Diprotodontia) in Australia and New Guinea. Journal of Mammalian Evolution 17: 75–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roe AD, Sperling FAH (2007) Population structure and species boundary delimitation of cryptic Dioryctria moths: an integrative approach. Molecular Ecology 16: 3617–3633. [DOI] [PubMed] [Google Scholar]
- 52. Bond JE, Stockman AK (2008) An integrative method for delimiting cohesion species: finding the population-species interface in a group of Californian trapdoor spiders with extreme genetic divergence and geographic structuring. Systematic Biology 57: 628–646. [DOI] [PubMed] [Google Scholar]
- 53. Yang Z, Rannala B (2010) Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences of the United States of America 107: 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang L, Han J, Xu J, Turchetto J, Mediannikov O, et al. (2009) Identification of a new serotype of Rickettsia heilongjiangensis in wild rats from Guangdong Province, China. Clinical Microbiology and Infection 15: 338–339. [DOI] [PubMed] [Google Scholar]
- 55.Wang YX (2003) A complete checklist of mammal species and subspecies in China: a taxonomic and geographic reference. Beijing, China: China Forestry Publishing House. 394 p. [Google Scholar]
- 56. Chen W, Liu S, Liu Y, Hao H, Zeng B, et al. (2010) Phylogeography of the large white-bellied rat Niviventer excelsior suggests the influence of Pleistocene Glaciations in the Hengduan Mountains. Zoological Science 27: 487–493. [DOI] [PubMed] [Google Scholar]
- 57. Balakirev AE, Rozhnov VV (2010) Phylogenic relationships and species composition in the genus Niviventer (Rodentia, Muridae) based on studies of the cytochrome b gene of mtDNA. Moscow University Biological Sciences Bulletin 65: 170–173. [Google Scholar]
- 58. Chen W, Li Y, Liu Y, Liu S, Yue B (2011) Complex topographic configuration in the Hengduan Mountains shaped the phylogeographic structure of Chinese white-bellied rats. Journal of Zoology 284: 215–223. [Google Scholar]