Life on earth has an unbelievable adaptive capacity. Except for centers of volcanic activity, the entire surface of our planet is a biosphere. In this context, the most surprising discovery in our lifetime was the expansion from the anthropocentrically defined “normal temperature” of mesophiles (<40°C) to the optimum temperature range of hyperthermophiles around and above the boiling point of water. That in this class of microorganisms high temperature is required for growth rather than tolerated implies that the whole repertoire of their biomolecules must be sufficiently stable to allow the cellular microcosm to work. The strategies nature has used to stabilize the inventory of the cell, especially proteins, under extreme conditions are still enigmatic, despite 25 years of active research. What has become clear is that proteins, independent of their mesophilic or extremophilic origin, consist exclusively of the canonical 20 natural amino acids; if other protein constituents are found, they originate from covalent chemical modifications (1). Thus, enhanced stability can come from only improved attractive forces: within the core, between domains and subunits, or from extrinsic protectants such as compatible solutes, conjugating components, and specific metabolites. The second alternative has been explained in terms of preferential solvation or specific ligand binding (2). For the first, a variety of “rules” has been proposed without providing an unambiguous solution to the problem. The reason for this failure is simple: considering the thermodynamic characteristics of proteins from mesophiles and extremophiles, the free energies of stabilization differ only marginally. The adaptive changes in terms of free energy differences (ΔΔGstab) correspond to the equivalent of just a few weak intermolecular interactions (3). Given the large number of small increments from hydrogen bonds, as well as hydrophobic, coulombic, and van der Walls interactions in molecules with hundreds or even thousands of atoms, there may be an astronomical number of combinations yielding ΔΔGstab as a small difference between large numbers (4). Based on the complete genome sequences of hyperthermophiles and systematic comparisons of their protein inventories with those of suitable mesophilic counterparts, a wealth of data has been accumulated that indicated that stabilization involves all levels of the hierarchy of protein structure, i.e., secondary, supersecondary, tertiary, and quaternary interactions. The common conclusion from model studies was that the stability of proteins from extremophiles is optimized to maintain corresponding functional states under a given set of environmental conditions. For the standard state at 25°C, enhanced thermal stability of hyperthermophile proteins would then be the result of enhanced conformational rigidity in their folded native state (5).
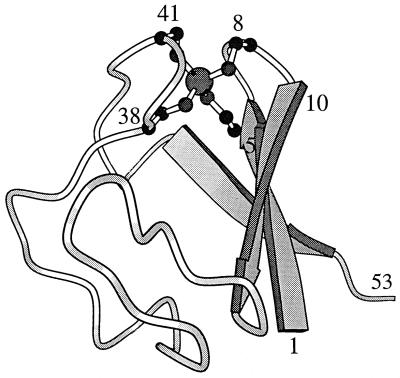
Evidence from recent amide hydrogen exchange experiments reported in this issue of PNAS seems to indicate that this hypothesis has no general validity. Using rubredoxin from the archaeon Pyrococcus furiosus, Hernández et al. (6) report that conformational opening processes for solvent access occurs within milliseconds for all amide positions along the 53-residue polypeptide chain. Considering first the object the authors have chosen, Pyrococcus rubredoxin seems to be ideally suited for the project: the protein contains three antiparallel β-strands joined by two loops, each containing two cysteine residues liganded to a metal ion to form C-X-X-C-X-X “knuckles” (Fig. 1). In the present study, for technical reasons, the normally occurring paramagnetic Fe2+ was replaced by Zn2+, essentially without any effect on the native three-dimensional structure. With its conformation unaltered over a wide temperature and pH range, together with an extrapolated denaturation temperature close to 200°C and an estimated global unfolding rate of unfolding of 10−6 s−1 at 100°C, the protein represents the most thermostable system presently known (9). Regarding the experimental approach, the protein allows all advantages of amide hydrogen exchange measurements to be exploited. Using the 15N-labeled protein to monitor local conformational fluctuations at 3–53°C, the authors succeeded in observing the open–closed conformational transitions for all amide hydrogens of the protein.
Figure 1.
Three-dimensional structure of rubredoxin from P. furiosus. Numbered residues mark the most slowly exchanging hydrogens, close to the two cysteine knuckles (7–9).
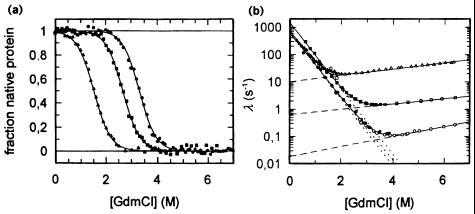
Because of the extremophilic characteristics and the presence of a metal-ion cluster, it is not surprising that there is no way to obtain quantitative thermodynamic data the easy (calorimetric) way. Therefore, denaturation kinetics were used to compare the hyperthermophilic protein with its thermophilic and mesophilic homologs. Indeed, the unfolding rates of the hyperthermophilic protein and its mesophilic counterpart differ by two to three orders of magnitude. Related studies with hyperthermophilic dihydrofolate reductase (10) and the 7-kDa cold-shock protein (Csp) from a mesophile, a thermophile, and a hyperthermophile (11) gave similar results. In the latter case, thermodynamic data confirmed the kinetic analysis; the differences in the equilibrium stabilities (ΔGstab) were found to increase with increasing optimal growth temperature (Topt; Fig. 2a). The corresponding activation energies for the unfolding reaction differed by ≈7 and 16 kJ·mol−1 (Fig. 2b, open symbols). Interestingly, the refolding kinetics were shown to differ only slightly, suggesting that the activated states of folding for all three proteins are similarly native-like in their interactions with the solvent (Fig. 2b, closed symbols).
Figure 2.
Unfolding/folding mechanism of the Csp from Bacillus subtilis, Bacillus caldolyticus, and Thermotoga maritima with Topt= 52, 72, and 90°C, respectively. (a) Guanidinium chloride (GdmCl)-induced equilibrium unfolding transitions at 25°C, monitored by intrinsic fluorescence. Least-squares fit according to two-state model U ↔ N yields ΔGstab = 11.3, 20.1, and 26.2 kJ/mol for B. subtilis Csp, B. caldolyticus Csp, and T. maritima Csp, respectively. (b) Unfolding kinetics (open symbols) and refolding kinetics (closed symbols) of B. subtilis Csp (▵, ▴), B. caldolyticus Csp (□, ■), and T. maritima Csp (○, ●), respectively. The apparent rate constant λ is plotted against the denaturant concentration. Fits according to the two-state model (5, 11). [Reproduced with permission from ref. 11 (Copyright 1998, Nat. Struct. Biol.).]
The most remarkable findings for Pyrococcus rubredoxin are the following. (i) The native hydrogen bonding of the protein close to its temperature of maximal thermodynamic stability is disrupted for all amide hydrogens in less than a second. (ii) At 28°C, conformational fluctuations for solvent access occur in the millisecond time range or faster through the entire protein. (iii) At alkaline pH, the activation energy analysis of hydrogen bonded amides yields maximum enthalpy contributions of less than 5% of the activation enthalpies commonly observed for protein unfolding (12). (iv) The corresponding distribution of amide protection factors is indistinguishable from data reported for typical mesophilic homologs. (v) The slowest local unfolding process involves a tertiary hydrogen bonding interaction, in accordance with similar configurations in other thermostable proteins.
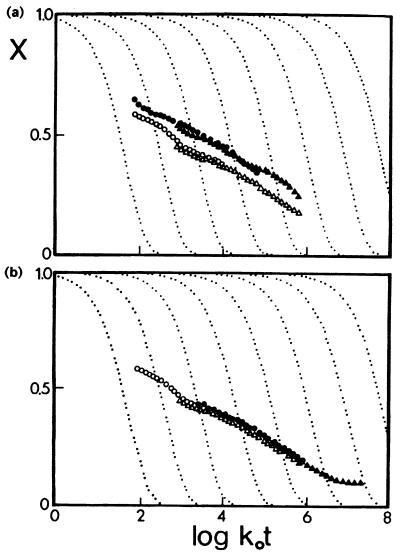
For those who have been working in the field for some time, these findings are unexpected, because in short, they seem to show that moderate-scale conformational dynamics in the millisecond time range are ubiquitous throughout the polypeptide chain of the most thermostable globular protein presently known. The critical comment of the authors questioning “the common hypothesis that enhanced conformational rigidity in the folded native state underlies the increased thermal stability of hyperthermophile proteins” might be viewed by some as going a bit too far, having in mind the catalog of stabilizing increments that has been worked out during the past decade (5). Based on a vast amount of structural and thermodynamic data, the view is now widely accepted that the anomalous stability of hyperthermophile proteins correlates with strong local interactions and/or improved packing of the polypeptide chain. For a mumber of ultrastable proteins, X-ray analysis and classical hydrogen–deuterium exchange kinetics clearly indicated anomalous conformational rigidity paralleled by decreased biological activity. As an example, the upper profile in Fig. 3a illustrates the slow hydrogen–deuterium exchange of an ultrastable protein from Thermotoga maritima compared with the lower curve for its mesophilic counterpart from rabbit, both measured at constant (low) temperature; coming closer to physiological conditions for both proteins (Fig. 3b), exchange rates become superimposable in agreement with Somero's “corresponding states” concept (13). Regarding the mechanisms of orthologous enzymes with different temperature optima, it is interesting to note that their active site residues are always conserved such that differences in the catalytic properties must be caused by substitutions elsewhere in the molecules (5, 14, 15). Analyzing the stabilities and activities of large numbers of random mutants, it turns out that the two properties are not necessarily inversely correlated. However, mutations that increase thermostability while maintaining low-temperature activity are extremely rare (16, 17).
Figure 3.
Hydrogen–deuterium exchange of glyceraldehyde phosphate dehydrogenase from T. maritima (open symbols) and rabbit (closed symbols), measured at pH 6.0 (○, ●) and pH 7.0 (▵, ▴), plotted as relaxation spectra. (a) Measurements at 25°C; increased X values reflect increased rigidity. (b) Measurements for the rabbit enzyme at 25°C and for the Thermotoga enzyme at 68°C. Coincidence of the curves indicated similar flexibility (5). [Reproduced with permission from ref. 5 (Copyright 1998, Curr. Opin. Struct. Biol.).]
What can be done to resolve the different views? Experienced protein chemists know that proteins show individual features, like different species; thus, more examples need to be studied to draw general conclusions. Apart from this one swallow does not a summer make argument, the chemist wonders whether even rubredoxin under the extreme experimental conditions might be damaged, e.g., by oxidation of cysteine residues or perturbation of the metal-ion cluster; similarly, the physicochemist might argue that, close to pH 12, the protein has 14 negative charges that may cause drastic alterations of its dynamics because of local structural changes (18).
On the other hand, also the “rigidity hypothesis” must not be taken dogmatically. For instance, in the case of phage T4 lysozyme, a topological separation of functional and stabilizing regions was observed—functional amino acids being optimized for flexibility rather than stability; however, mutating residues involved in catalysis or substrate binding may enhance stability at the cost of reduced activity, supporting a relationship between stability and function (19). In other cases, part of the polypeptide backbone (predominantly secondary structure) was found to form a stable scaffold while more flexible regions are involved in catalysis (15). A drastic exception has been reported for a number of thermophile enzymes with unstable metabolic intermediates. Here, nature suspended the corresponding-states concept for good reasons, because only extremely high catalytic activity (even at low temperature) can salvage short-lived substrates or intermediates (20).
Thus, as usual in nature, we are left with a whole spectrum of different solutions to our problem. In connection with the present rubredoxin study, it would be most desirable to see more such data under conditions closer to physiological ones to come closer at last to the solution of one of the most challenging problems in physical biochemistry today.
Footnotes
See companion article on page 3166.
References
- 1.Jaenicke R. Biochemistry (Moscow) 1998;63:312–321. [PubMed] [Google Scholar]
- 2.Carpenter J F, Clegg J S, Crowe J H, Somero G N. Cryobiology. 1993;30:201–241. [Google Scholar]
- 3.Dill K A. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 4.Jaenicke R. Eur J Biochem. 1991;202:715–728. doi: 10.1111/j.1432-1033.1991.tb16426.x. [DOI] [PubMed] [Google Scholar]
- 5.Jaenicke R, Böhm G. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/s0959-440x(98)80094-8. [DOI] [PubMed] [Google Scholar]
- 6.Hernández G, Jenney F E, Jr, Adams M W W, LeMaster D M. Proc Natl Acad Sci USA. 2000;97:3166–3170. doi: 10.1073/pnas.040569697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day M W, Hsu B T, Joshua-Tor L, Park J B, Zhou Z H, Adams M W W, Rees D C. Protein Sci. 1992;1:1494–1507. doi: 10.1002/pro.5560011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blake P R, Park J B, Zhou Z H, Hare D R, Adams M W W, Summers M F. Protein Sci. 1992;1:1508–1521. doi: 10.1002/pro.5560011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiller R, Zhou Z H, Adams M W W, Englander S W. Proc Natl Acad Sci USA. 1997;94:11329–11332. doi: 10.1073/pnas.94.21.11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dams T, Jaenicke R. Biochemistry. 1999;38:9169–9178. doi: 10.1021/bi990635e. [DOI] [PubMed] [Google Scholar]
- 11.Perl D, Welker C, Schindler T, Schröder K, Marahiel M A, Jaenicke R, Schmid F X. Nat Struct Biol. 1998;5:229–235. doi: 10.1038/nsb0398-229. [DOI] [PubMed] [Google Scholar]
- 12.Pfeil W. Protein Stability and Folding: A Collection of Thermodynamic Data. Berlin: Springer; 1998. [Google Scholar]
- 13.Somero G N. Annu Rev Ecol Syst. 1978;9:1–29. [Google Scholar]
- 14.Field P A, Somero G N. Proc Natl Acad Sci USA. 1998;95:11476–11481. doi: 10.1073/pnas.95.19.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auerbach G, Ostendorp R, Prade L, Korndörfer I, Dams T, Huber R, Jaenicke R. Structure (London) 1998;6:769–781. doi: 10.1016/s0969-2126(98)00078-1. [DOI] [PubMed] [Google Scholar]
- 16.van den Burg B, Vriend G, Veltman O R, Venema G, Eijsink V G H. Proc Natl Acad Sci USA. 1998;95:2056–2060. doi: 10.1073/pnas.95.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giver L, Gershenson A, Freskgard P-O, Arnold F H. Proc Natl Acad Sci USA. 1998;95:12809–12813. doi: 10.1073/pnas.95.22.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavagnero S, Debe D A, Zhou Z H, Adams M W W, Chan S I. Biochemistry. 1998;37:3369–3376. doi: 10.1021/bi9721795. [DOI] [PubMed] [Google Scholar]
- 19.Shoichet B K, Baase W A, Kuroki R, Matthews B W. Proc Natl Acad Sci USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sterner R, Kleemann G R, Szadkowski H, Lustig L, Hennig M, Kirschner K. Protein Sci. 1996;5:2000–2008. doi: 10.1002/pro.5560051006. [DOI] [PMC free article] [PubMed] [Google Scholar]