Abstract
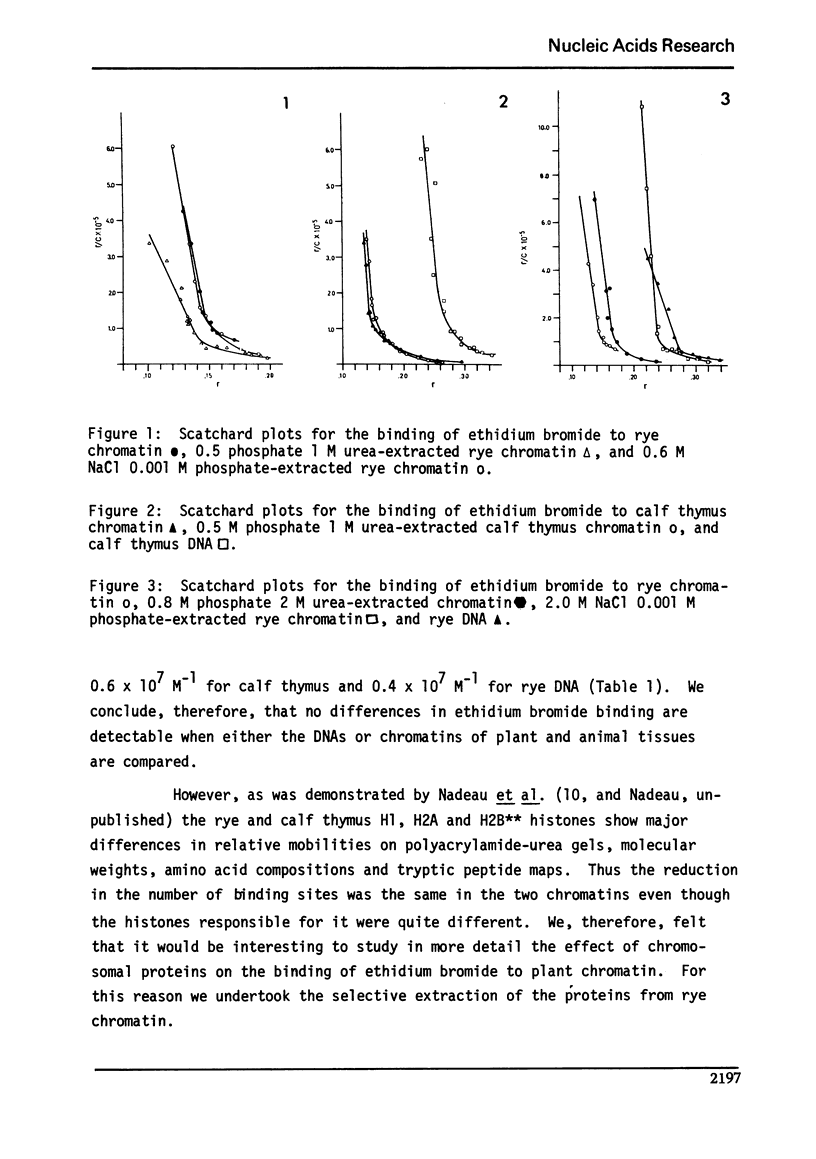
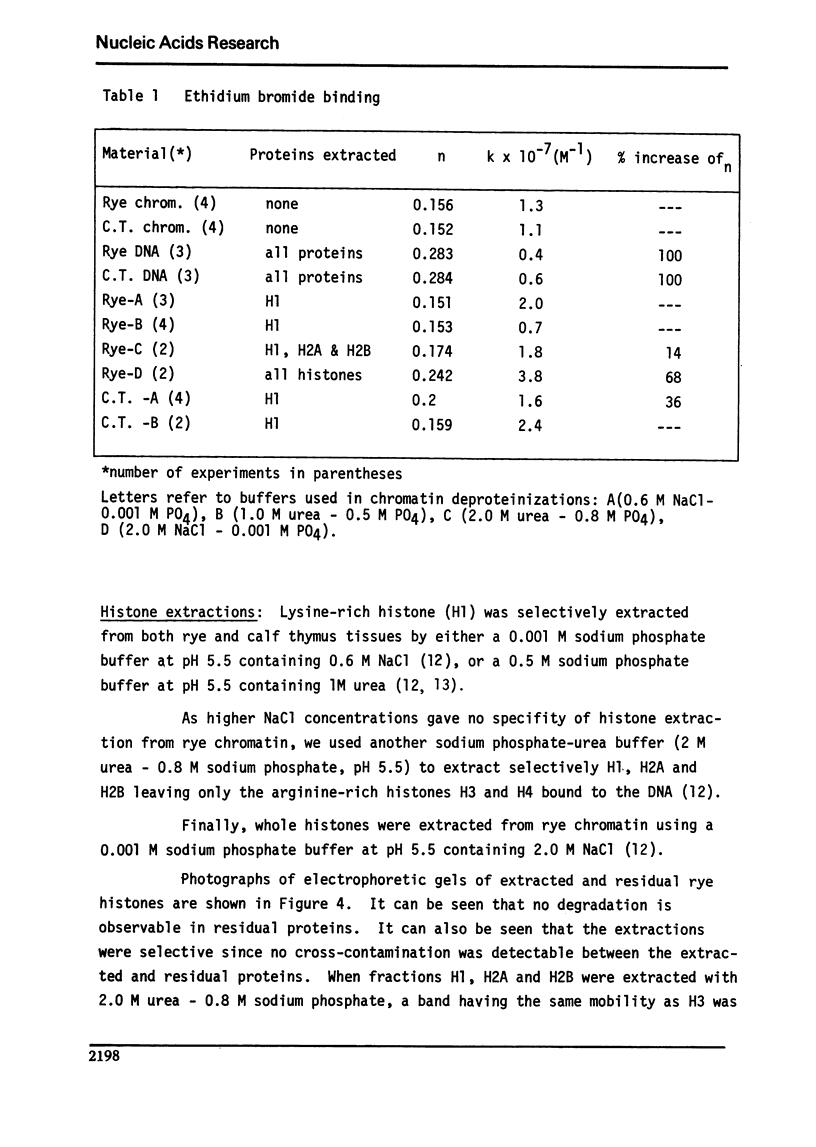
We studied the interaction of ethidium bromide with rye and calf thymus chromatin. Both types of chromatin have the same dye accessibility, which is about 50% of that of DNA. From this result we conclude that the molecular structure of these two chromatins is similar. For rye, the extraction of H1 produces no change in the binding of ethidium bromide. The subsequent extraction of H2A and H2B produces a 14% increase in the binding, and the removal of H3 and H4, another 54% increase. At this stage, the number of binding sites is still less than that of DNA. This is presumably due to the presence of some tightly bound non-histones. Thus, the arginine-rich histones and the tightly bound non-histones are most responsible for limiting the binding of ethidium bromide to rye chromatin.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angerer L. M., Georghiou S., Moudrianakis E. N. Studies on the structure of deoxyribonucleoproteins. Spectroscopic characterization of the ethidium bromide binding sites. Biochemistry. 1974 Mar 12;13(6):1075–1082. doi: 10.1021/bi00703a003. [DOI] [PubMed] [Google Scholar]
- Angerer L. M., Moudrianakis E. N. Interaction of ethidium bromide with whole and selectively deproteinized deoxynucleoproteins from calf thymus. J Mol Biol. 1972 Feb 14;63(3):505–521. doi: 10.1016/0022-2836(72)90444-5. [DOI] [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Bartley J. A., Chalkley R. The binding of deoxyribonucleic acid and histone in native nucleohistone. J Biol Chem. 1972 Jun 10;247(11):3647–3655. [PubMed] [Google Scholar]
- Bradbury E. M., Carpenter B. G., Rattle H. W. Magnetic resonance studies of deoxyribonucleoprotein. Nature. 1973 Jan 12;241(5385):123–126. doi: 10.1038/241123a0. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Molgaard H. V., Stephens R. M., Bolund L. A., Johns E. W. X-ray studies of nucleoproteins depleted of lysine-rich histone. Eur J Biochem. 1972 Dec 18;31(3):474–482. doi: 10.1111/j.1432-1033.1972.tb02555.x. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Keevert J. B. Subunit structure of a naturally occurring chromatin lacking histones F1 and F3. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3536–3540. doi: 10.1073/pnas.72.9.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon S., Johnson R. S., Wolf B., Chan A. Mixed conformations of deoxyribonucleic acid in chromatin: a preliminary report. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3263–3267. doi: 10.1073/pnas.69.11.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelm R. P., Jr, Huang R. C. The role of histones in the conformation of DNA in chromatin as studied by circular dichroism. Biochemistry. 1974 Dec 17;13(26):5275–5283. doi: 10.1021/bi00723a004. [DOI] [PubMed] [Google Scholar]
- Honda B. M., Baillie D. L., Candido E. P. Properties of chromatin subunits from developing trout testis. J Biol Chem. 1975 Jun 25;250(12):4643–4647. [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Littau V. C., Burdick C. J., Allfrey V. G., Mirsky S. A. The role of histones in the maintenance of chromatin structure. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1204–1212. doi: 10.1073/pnas.54.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Yeast chromatin subunit structure. Science. 1975 Apr 11;188(4184):165–166. doi: 10.1126/science.1090006. [DOI] [PubMed] [Google Scholar]
- Lurquin P. F., Seligy V. L. Binding of ethidium bromide to avian erythrocyte chromatin. Biochem Biophys Res Commun. 1972 Feb 16;46(3):1399–1404. doi: 10.1016/s0006-291x(72)80131-1. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Engel J. D. Subunit structure of chromatin is the same in plants and animals. Nature. 1975 Apr 3;254(5499):449–450. doi: 10.1038/254449a0. [DOI] [PubMed] [Google Scholar]
- More I. A., Paul J. Template activity and electron microscopic appearance of salt-extracted chromatin. Exp Cell Res. 1973 Jan;76(1):79–86. doi: 10.1016/0014-4827(73)90421-7. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Pederson T., Bhorjee J. S. A special class of non-histone protein tightly complexed with template-inactive DNA in chromatin. Biochemistry. 1975 Jul 15;14(14):3238–3242. doi: 10.1021/bi00685a033. [DOI] [PubMed] [Google Scholar]
- Remington J. A., Klevecz R. R. Poly-L-lysine binding to small quantities of different chromatin preparations. Biochim Biophys Acta. 1973 May 18;308(3):422–425. doi: 10.1016/0005-2787(73)90335-3. [DOI] [PubMed] [Google Scholar]
- Skidmore C., Walker I. O., Pardon J. F., Richards B. M. The structure of partially histone depleted nucleohistone. FEBS Lett. 1973 May 15;32(1):175–178. doi: 10.1016/0014-5793(73)80765-3. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Sommer K. R., Chalkley R. A new method for fractionating histones for physical and chemical studies. Biochemistry. 1974 Feb 26;13(5):1022–1027. doi: 10.1021/bi00702a029. [DOI] [PubMed] [Google Scholar]
- Tuan D. Y., Bonner J. Optical absorbance and optical rotatory dispersion studies on calf thymus nucleohistone. J Mol Biol. 1969 Oct 14;45(1):59–76. doi: 10.1016/0022-2836(69)90209-5. [DOI] [PubMed] [Google Scholar]
- Vandegrift V., Serra M., Moore D. S., Wagner T. E. The role of the arginine-rich histones in the maintenance of DNA conformation in chromatin. Biochemistry. 1974 Dec 3;13(25):5087–5092. doi: 10.1021/bi00722a005. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Complex formation between ethidium bromide and nucleic acids. J Mol Biol. 1965 Aug;13(1):269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- Williams R. E., Lurquin P. F., Seligy V. L. Circular dichroism of avian-erythrocyte chromatin and ethidium bromide bound to chromatin. Eur J Biochem. 1972 Sep 25;29(3):426–432. doi: 10.1111/j.1432-1033.1972.tb02005.x. [DOI] [PubMed] [Google Scholar]