Abstract
Human neural progenitor cells (hNPCs) form a new prospect for replacement therapies in the context of neurodegenerative diseases. The Wnt/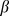 -catenin signaling pathway is known to be involved in the differentiation process of hNPCs. RVM cells form a common cell model of hNPCs for in vitro investigation. Previous observations in RVM cells raise the question of whether observed kinetics of the Wnt/
-catenin signaling pathway is known to be involved in the differentiation process of hNPCs. RVM cells form a common cell model of hNPCs for in vitro investigation. Previous observations in RVM cells raise the question of whether observed kinetics of the Wnt/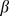 -catenin pathway in later differentiation phases are subject to self-induced signaling. However, a concern when investigating RVM cells is that experimental results are possibly biased by the asynchrony of cells w.r.t. the cell cycle.
-catenin pathway in later differentiation phases are subject to self-induced signaling. However, a concern when investigating RVM cells is that experimental results are possibly biased by the asynchrony of cells w.r.t. the cell cycle.
In this paper, we present, based on experimental data, a computational modeling study on the Wnt/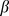 -catenin signaling pathway in RVM cell populations asynchronously distributed w.r.t. to their cell cycle phases. Therefore, we derive a stochastic model of the pathway in single cells from the reference model in literature and extend it by means of cell populations and cell cycle asynchrony. Based on this, we show that the impact of the cell cycle asynchrony on wet-lab results that average over cell populations is negligible. We then further extend our model and the thus-obtained simulation results provide additional evidence that self-induced Wnt signaling occurs in RVM cells. We further report on significant stochastic effects that directly result from model parameters provided in literature and contradict experimental observations.
-catenin signaling pathway in RVM cell populations asynchronously distributed w.r.t. to their cell cycle phases. Therefore, we derive a stochastic model of the pathway in single cells from the reference model in literature and extend it by means of cell populations and cell cycle asynchrony. Based on this, we show that the impact of the cell cycle asynchrony on wet-lab results that average over cell populations is negligible. We then further extend our model and the thus-obtained simulation results provide additional evidence that self-induced Wnt signaling occurs in RVM cells. We further report on significant stochastic effects that directly result from model parameters provided in literature and contradict experimental observations.
Introduction
Human neural progenitor cells (hNPCs) potentially form a new basis for the in vitro growing of neuron populations that can be used for replacement therapies in the context of neurodegenerative diseases, such as Parkinson's or Huntington's diseases [1], [2]. They undergo the processes of proliferation, i.e., successive cell division controlled by the cell cycle, and differentiation into neural cells, i.e., neurons and glial cells. In order to deploy hNPCs for replacement therapies, a clear understanding of their proliferation and differentiation processes is essential.
ReNcell VM cells (RVM cells) derived from the ventral midbrain of a ten week old fetus [3] form an appropriate cell model for the in vitro study of hNPCs differentiation: they stay in proliferation as long as growth factors are present and differentiate into neurons and glial cells after growth factors removal [4]–[6].
The Wnt/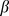 -catenin signaling pathway is known to be involved in the proliferation and differentiation processes of neural cells [7]–[9], particularly in those of RVM cells [10]–[12]. It denotes a cascade of reactions that is induced by extracellular Wnt molecules at the cell membrane and that leads to an accumulation of
-catenin signaling pathway is known to be involved in the proliferation and differentiation processes of neural cells [7]–[9], particularly in those of RVM cells [10]–[12]. It denotes a cascade of reactions that is induced by extracellular Wnt molecules at the cell membrane and that leads to an accumulation of 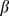 -catenin in the cytosol. Consecutively,
-catenin in the cytosol. Consecutively, 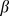 -catenin is relocated to the nucleus where it activates the transcription of genes including the gene encoding for Axin protein. In this way, a negative feedback loop is established, since Axin forms the major component of the
-catenin is relocated to the nucleus where it activates the transcription of genes including the gene encoding for Axin protein. In this way, a negative feedback loop is established, since Axin forms the major component of the 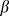 -catenin destruction complex assembling in the cytosol [13].
-catenin destruction complex assembling in the cytosol [13].
Our previous in vitro analyses [12] show that Wnt signaling pathway is active during the early differentiation (first 6 hours) of RVM cells and suggest that Wnt molecules are expressed by RVM cells themselves, i.e., self-induced Wnt signaling. That is, cells secrete Wnt molecules without any exogenous stimulus but only due to the growth factor removal that induces the differentiation process. Self-induced Wnt signaling occurs in embryonic stem cells [14], both in an autocrine (cells signal to themselves) and paracrine (signaling to neighbor cells) fashion. Autocrine Wnt/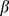 -catenin signaling has been shown to occur in neural stem cells [15] and in brain development [16], [17] but not in hNPCs, in particular. Evidence for self-induced signaling in RVM cells are: endogenous expression of Wnt ligands and signaling proteins, as well as spatio-temporal traffic of the pathway signaling proteins, in both cases without addition of external Wnt signal [12]. Mainly, the two hallmarks of the pathway activation: expression of Axin gene and cytosolic accumulation of
-catenin signaling has been shown to occur in neural stem cells [15] and in brain development [16], [17] but not in hNPCs, in particular. Evidence for self-induced signaling in RVM cells are: endogenous expression of Wnt ligands and signaling proteins, as well as spatio-temporal traffic of the pathway signaling proteins, in both cases without addition of external Wnt signal [12]. Mainly, the two hallmarks of the pathway activation: expression of Axin gene and cytosolic accumulation of 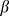 -catenin, have also been observed.
-catenin, have also been observed.
Investigations of RVM cells in vitro are hampered by the heterogeneity of cell populations w.r.t. cell cycle states. That is, cells that are in phases S, G2, or M, rather than G1, cannot adapt to growth factor withdrawal right away. Thus, only a fraction of a RVM cell population starts differentiation immediately [12]. This asynchrony may bias the results of experimental work. For the time being, techniques to synchronize RVM cell populations during proliferation could not be successfully applied.
Computational modeling provides a way to circumvent the limitations of wet-lab experiments. The basic idea is to create an abstract representation of the system under study, a formal model, which is then analyzed with the help of computers. Models to describe a system's dynamics are in need of kinetic parameters, such as rate constants. The closer kinetic parameters relate to experimental data the more reliable the results of a modeling study are.
Stochastic modeling, as described in [18], considers models in terms of chemical reactions and multisets of molecules, which represent chemical solutions. Molecular interactions are regarded as discrete events randomly distributed in time. Analysis of stochastic models in terms of stochastic simulation provides distinct sequences of molecular interactions, with each simulation run being a different sequence. Stochastic effects have been shown to have significant impact on the dynamics of biochemical systems, especially in signaling pathways where key players appear in relatively low abundance [19]. Spatial aspects, such as molecular location or crowding, may add to this [20].
Deterministic modeling studies usually transform chemical reactions into ordinary differential equations (ODEs) and regard concentrations instead of multisets of molecules. The equivalent to simulation in the context of ODEs is numerical integration. Studies based on ODEs form an approximation of the stochastic approach that neglects the stochastic effects and thus may miss significant variations in the dynamics of systems [21], [22]. Furthermore, models expressed in ODEs often largely abstract from chemical reactions by aggregating several chemical species and reactions, e.g., in order to deal with a lack of kinetic parameters. This may largely hamper the switch back to the stochastic realm.
In this paper, we present, based on experimental data, a computational modeling study on cell cycle asynchrony and self-induced signaling in the context of the Wnt/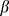 -catenin pathway in RVM cells. Therefore, we derive a model of the core components of the Wnt/
-catenin pathway in RVM cells. Therefore, we derive a model of the core components of the Wnt/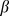 -catenin pathway from the reference model of this pathway in Xenopus oocyte (referred to as the Lee model subsequently) [23] and validate it with experimental data for RVM cells, as obtained in our prior work [12]. Our model extends on the Lee model as it is fully specified in terms of chemical reactions, which allows us to easily switch between the stochastic and deterministic domain. Furthermore, it covers spatial aspects w.r.t. molecule location in compartments. For this, we provide additional experimental data on compartment volumes and molecule distribution in space.
-catenin pathway from the reference model of this pathway in Xenopus oocyte (referred to as the Lee model subsequently) [23] and validate it with experimental data for RVM cells, as obtained in our prior work [12]. Our model extends on the Lee model as it is fully specified in terms of chemical reactions, which allows us to easily switch between the stochastic and deterministic domain. Furthermore, it covers spatial aspects w.r.t. molecule location in compartments. For this, we provide additional experimental data on compartment volumes and molecule distribution in space.
We extend this core model with means of cell populations and cell cycle asynchrony based partly on our own experimental data and partly on data from the literature for the distribution of RVM cell populations over cell cycle states. We show, based on the comparison of experimental in vitro and in silico simulation data, that the impact of the cell cycle asynchrony on the average 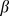 -catenin dynamics in cell populations as observed in wet-lab experiments is negligible. We additionally extend our model with mechanisms for self-induced signaling. Comparing further results from simulation studies to experimental data allows us to provide additional evidence that self-induced Wnt signaling may occur in RVM cells. Moreover, we show that in our model of RVM cells, low Axin amounts, such as suggested by [23] for Xenopus oocyte, lead to significant stochastic effects that contradict experimental observations and that are not observable in deterministic investigations. To the best of our knowledge, so far, no results on stochastic investigations have been presented in the context of the Wnt/
-catenin dynamics in cell populations as observed in wet-lab experiments is negligible. We additionally extend our model with mechanisms for self-induced signaling. Comparing further results from simulation studies to experimental data allows us to provide additional evidence that self-induced Wnt signaling may occur in RVM cells. Moreover, we show that in our model of RVM cells, low Axin amounts, such as suggested by [23] for Xenopus oocyte, lead to significant stochastic effects that contradict experimental observations and that are not observable in deterministic investigations. To the best of our knowledge, so far, no results on stochastic investigations have been presented in the context of the Wnt/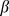 -catenin pathway.
-catenin pathway.
Related work
The reference model of the Wnt/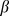 -catenin pathway, the Lee model, is based on ordinary differential equations (ODEs) [23]. It represents the pathway in the Xenopus oocyte and is based on experimental data. Model analysis revealed that Axin is the limiting protein of the system due to its low abundance. Furthermore, Axin turnover strongly regulates
-catenin pathway, the Lee model, is based on ordinary differential equations (ODEs) [23]. It represents the pathway in the Xenopus oocyte and is based on experimental data. Model analysis revealed that Axin is the limiting protein of the system due to its low abundance. Furthermore, Axin turnover strongly regulates 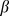 -catenin dynamics.
-catenin dynamics.
Three follow-ups of the Lee model exist. The work presented in [24] and [25] extend the Lee model with one and two negative feedback loops, respectively. The latter shows that the addition of the negative feedbacks leads to an adaptation of the parameters given in the Lee model, especially an increase of 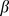 -catenin and Axin turnover. The work presented in [26] aims to reduce the Lee model.
-catenin and Axin turnover. The work presented in [26] aims to reduce the Lee model.
A first study of 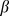 -catenin's roles in correlation with its cellular location is presented in [27]. Based on the Lee model, they analyze, in the context of colorectal cancer, the balance between the two roles of
-catenin's roles in correlation with its cellular location is presented in [27]. Based on the Lee model, they analyze, in the context of colorectal cancer, the balance between the two roles of 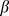 -catenin: its participation in cell adhesion at the membrane and in the Wnt pathway in the cytosol. However, their computational model only considers single cells and does not take into account compartments such as the cytosol or the membrane.
-catenin: its participation in cell adhesion at the membrane and in the Wnt pathway in the cytosol. However, their computational model only considers single cells and does not take into account compartments such as the cytosol or the membrane.
An exhaustive review of models related to the Wnt pathway is available in [28]. It covers models of the Wnt pathways and their implications in animal development and in cross-talks.
Results and Discussion
In the following, we first recapitulate the experimental data resulting from our prior work [12]. They form the crucial basis of any conclusion drawn in this study as they are the reference to which we compare our simulation results of 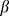 -catenin dynamics in RVM cells. Then, we describe our core model of the Wnt/
-catenin dynamics in RVM cells. Then, we describe our core model of the Wnt/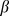 -catenin pathway and its evaluation to experimental and literature data. This is followed by a report on the stochastic effects on our model's behavior. Subsequently, we discuss our investigations on cell cycle asynchrony in the context of the Wnt/
-catenin pathway and its evaluation to experimental and literature data. This is followed by a report on the stochastic effects on our model's behavior. Subsequently, we discuss our investigations on cell cycle asynchrony in the context of the Wnt/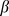 -catenin pathway and on self-induced Wnt signaling in RVM cells.
-catenin pathway and on self-induced Wnt signaling in RVM cells.
Experimental data on  -catenin dynamics in RVM cells
-catenin dynamics in RVM cells
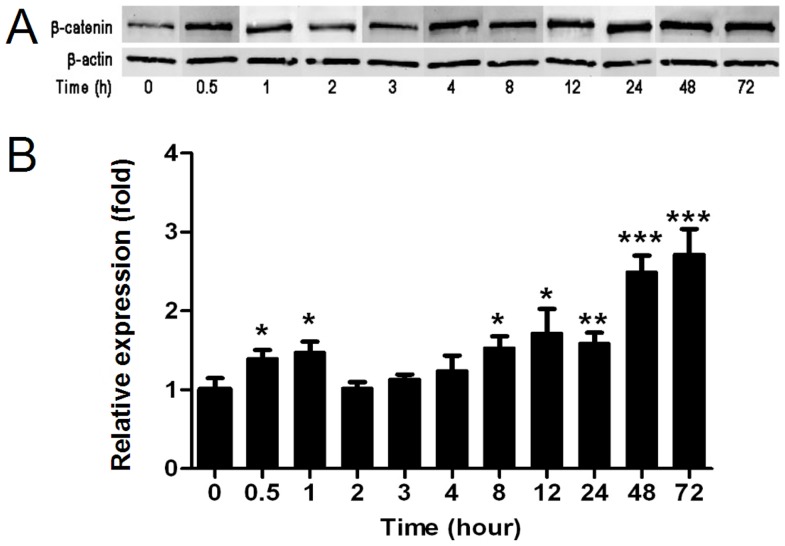
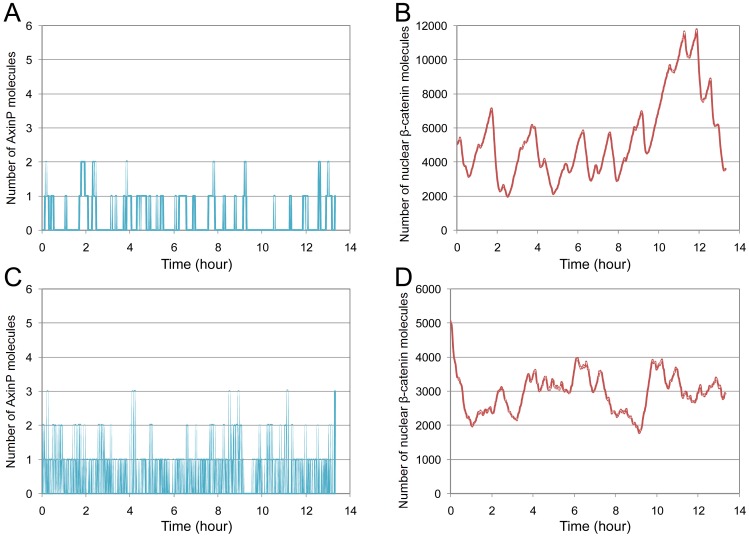
Our study is based on wet-lab data obtained from Western blot experiments [29] presented in Figure 1, and reproduced from the Figure 8 in [12]. These data present the dynamics of nuclear  -catenin in RVM cell populations from the starting point of differentiation, i.e., growth factor removal (time point 0 hour) until 72 hours after.
-catenin in RVM cell populations from the starting point of differentiation, i.e., growth factor removal (time point 0 hour) until 72 hours after.
Figure 1. Nuclear  -catenin during RVM cell differentiation.
-catenin during RVM cell differentiation.
(A) Representative Western-blot from which the  -catenin protein amount was quantified (B). Time point 0 stands for control using proliferating cells and
-catenin protein amount was quantified (B). Time point 0 stands for control using proliferating cells and 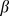 -actin was used as a loading control. The signal intensities at 0 hour are normalized to 1.0 and the values are presented as mean
-actin was used as a loading control. The signal intensities at 0 hour are normalized to 1.0 and the values are presented as mean  standard error on the mean from at least 3 independent experiments. The figure is reproduced from [12] (Figures 8C and 8D) where details about experimentation can be found, as well as in the section Materials & Methods.
standard error on the mean from at least 3 independent experiments. The figure is reproduced from [12] (Figures 8C and 8D) where details about experimentation can be found, as well as in the section Materials & Methods.
The data show two significant increases of 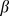 -catenin during the cell differentiation. The first with a peak at ca. 1 hour and the second continuously growing from 8 hours on (Figure 1B). Such biphasic activity of Wnt is known during embryonic stem cell development [30], [31]. We expect the first increase to be a direct result of the Wnt/
-catenin during the cell differentiation. The first with a peak at ca. 1 hour and the second continuously growing from 8 hours on (Figure 1B). Such biphasic activity of Wnt is known during embryonic stem cell development [30], [31]. We expect the first increase to be a direct result of the Wnt/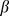 -catenin pathway activity, e.g., through crosstalk with growth factor pathways [32]. The source of the second increase, however, remains controversial. On one hand it may result from cell cycle asynchrony. On the other hand, it may be caused by a subsequent self-induced signaling from time point 8 hours on, as the Wnt signal is also developmental-stage specific [30]. Whereas we show in the 3rd section of this chapter that the first hypothesis can be rejected, we provide evidence for the second in the last section of this chapter. After 24 hours, another accumulation of
-catenin pathway activity, e.g., through crosstalk with growth factor pathways [32]. The source of the second increase, however, remains controversial. On one hand it may result from cell cycle asynchrony. On the other hand, it may be caused by a subsequent self-induced signaling from time point 8 hours on, as the Wnt signal is also developmental-stage specific [30]. Whereas we show in the 3rd section of this chapter that the first hypothesis can be rejected, we provide evidence for the second in the last section of this chapter. After 24 hours, another accumulation of 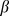 -catenin can be observed. However, at that time, the cell population is already heterogeneous due to differentiation, such that the accumulation may originate from multiple sources. Therefore, our study presented focuses only on the first 12 hours.
-catenin can be observed. However, at that time, the cell population is already heterogeneous due to differentiation, such that the accumulation may originate from multiple sources. Therefore, our study presented focuses only on the first 12 hours.
The measurements presented in Figure 1 are in terms of relative fold changes compared to time point 0 hour, whose value is set to 1.0 (see Materials & Methods). Throughout this paper, when comparing wet-lab experiments to simulation results, we therefore consider relative amounts of nuclear 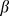 -catenin (
-catenin (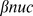 in our model), where value 1.0 represents the initial concentration or initial amount in deterministic or stochastic simulations, respectively.
in our model), where value 1.0 represents the initial concentration or initial amount in deterministic or stochastic simulations, respectively.
Model of the Wnt/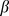 -catenin pathway in RVM cells
-catenin pathway in RVM cells
In this section, we describe our core model and detail its underlying assumptions. We evaluate the model based on two deterministic simulation experiments, whereby the first compares to the Lee model to show that we cover the basic machinery of the Wnt/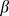 -catenin pathway, as it is currently known, and the second compares to our own experimental data. Parameter sets are partly derived from literature [23], [33] and from wet-lab experiments [10], [12]. Furthermore, due to unknown parameters, each evaluation procedure involves some in silico experiments fitting the behavior of the model to the respective reference behaviors (parameter fitting experiments). Therefore each evaluation study comes with its individual sets of parameters. The parameter set resulting from the comparison to our own experimental data serves as a basis for the investigations as reported in the subsequent sections.
-catenin pathway, as it is currently known, and the second compares to our own experimental data. Parameter sets are partly derived from literature [23], [33] and from wet-lab experiments [10], [12]. Furthermore, due to unknown parameters, each evaluation procedure involves some in silico experiments fitting the behavior of the model to the respective reference behaviors (parameter fitting experiments). Therefore each evaluation study comes with its individual sets of parameters. The parameter set resulting from the comparison to our own experimental data serves as a basis for the investigations as reported in the subsequent sections.
Model definition
Our core model of the Wnt/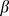 -catenin pathway is derived from the Lee model and covers the basic components and processes in two compartments. In the cytosol,
-catenin pathway is derived from the Lee model and covers the basic components and processes in two compartments. In the cytosol, 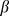 -catenin is destructed by the degradation complex, which forms around Axin and is deactivated by Wnt. In the nucleus the negative feedback loop is established that consists in
-catenin is destructed by the degradation complex, which forms around Axin and is deactivated by Wnt. In the nucleus the negative feedback loop is established that consists in 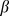 -catenin triggering Axin production, leading to a higher amount of degradation complex in the cytosol. The nucleus is assumed to be spherical and the cytosol, as it is surrounding the nucleus, to form a spherical shell, see Figure 2.
-catenin triggering Axin production, leading to a higher amount of degradation complex in the cytosol. The nucleus is assumed to be spherical and the cytosol, as it is surrounding the nucleus, to form a spherical shell, see Figure 2.
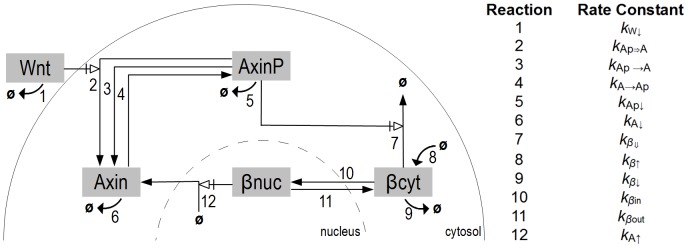
Figure 2. Schematic representation of the intracellular Wnt/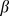 -catenin pathway model.
-catenin pathway model.
A cell is composed of two compartments, the cytosol and the nucleus, separated by the dashed lane. The five species are framed in gray. Only the species Wnt is extra-cellular. For a given reaction, an arrow-less lane shows the reactant(s), and an arrow lane points to the product. Bended arrows represent protein production for reaction  or protein decay for reactions
or protein decay for reactions  ,
,  ,
,  , and
, and 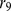 . An open arrow coming after a large vertical bar represents a trigger effect (according to the Systems Biology Graphical Notation [60]) where the reactant is not consumed in the reaction, but necessary for the process to take place. The reactions' numbers correspond to the ones in Table 1.
. An open arrow coming after a large vertical bar represents a trigger effect (according to the Systems Biology Graphical Notation [60]) where the reactant is not consumed in the reaction, but necessary for the process to take place. The reactions' numbers correspond to the ones in Table 1.
We only consider the three main proteins: Wnt, 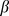 -catenin, and Axin. They are represented by five species. We introduce two species for
-catenin, and Axin. They are represented by five species. We introduce two species for 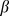 -catenin,
-catenin, 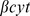 and
and 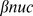 , to represent its cellular location in the cytosol and in the nucleus, respectively, and two species for Axin reflecting the phosphorylation state of Axin (
, to represent its cellular location in the cytosol and in the nucleus, respectively, and two species for Axin reflecting the phosphorylation state of Axin (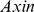 and
and 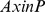 ). Following the ideas in [34], Axin entirely abstracts the degradation complex:
). Following the ideas in [34], Axin entirely abstracts the degradation complex: 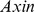 and
and 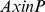 representing the inactive and active degradation complex, respectively. Axin only occurs in the cytosol. Furthermore also Wnt is located in the cytosol, since we do not consider the processes of transferring the Wnt signal from the outside to the inside of the cell.
representing the inactive and active degradation complex, respectively. Axin only occurs in the cytosol. Furthermore also Wnt is located in the cytosol, since we do not consider the processes of transferring the Wnt signal from the outside to the inside of the cell.
Our model is entirely expressed in terms of chemical reactions (listed in Table 1). can either decay (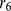 ,
, 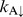 ) or phosphorylate to become
) or phosphorylate to become 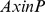 (
(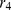 ,
, 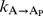 ). The latter can again dephosphorylate (
). The latter can again dephosphorylate (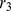 ,
, 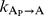 ), decay (
), decay (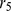 ,
, 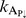 ), or degrade
), or degrade 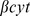 (
(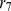 ,
, 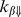 ). We represent the Wnt-dependent deactivation of the degradation complex by
). We represent the Wnt-dependent deactivation of the degradation complex by 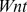 promoting
promoting 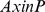 dephosphorylation (
dephosphorylation (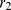 ,
, 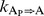 ). Notice that by this the amount of
). Notice that by this the amount of 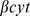 is indirectly increased. Moreover, this forms also the only way in which the Wnt signal influences
is indirectly increased. Moreover, this forms also the only way in which the Wnt signal influences 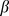 -catenin dynamics.
-catenin dynamics.
Table 1. Reactions of the intracellular Wnt/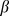 -catenin pathway model.
-catenin pathway model.
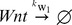
|
(1) |
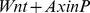
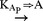
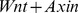
|
(2) |
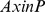
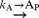
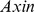
|
(3) |
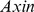
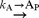
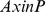
|
(4) |
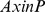
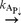
|
(5) |
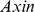
|
(6) |
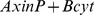
|
(7) |
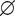
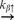
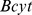
|
(8) |
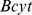
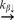
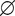
|
(9) |
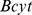
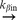
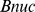
|
(10) |
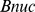
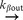
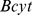
|
(11) |
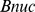
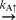
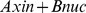
|
(12) |
All reactions are following Mass action kinetics, but 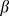 -catenin production (1) that is a constant flux. The reaction numbers correspond to the ones in the model schema (Figure 1).
-catenin production (1) that is a constant flux. The reaction numbers correspond to the ones in the model schema (Figure 1).
Previous wet-lab experiments suggest that the passage from cell proliferation to differentiation after removal of growth factors is accompanied with an extra-cellular presence of active Wnt molecules that is self-induced by the cell population [12]. Our model reflects this fact by considering a given initial amount of 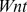 . The effect of the signal decreases over time, since
. The effect of the signal decreases over time, since 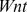 decays (
decays (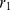 ,
, 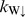 ) and is not further produced.
) and is not further produced.
For 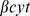 , there exists a constant flux of production (
, there exists a constant flux of production (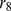 ,
, 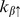 ) and decay (
) and decay (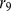 ,
, 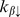 ). Furthermore,
). Furthermore, 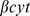 travels between the cytosol and the nucleus in both directions (
travels between the cytosol and the nucleus in both directions (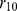 ,
, 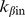 , and
, and 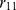 ,
, 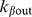 ). In the nucleus,
). In the nucleus, 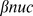 induces
induces 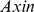 production (
production (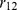 ,
, 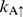 ), representing the pathway's negative feedback loop. Except for
), representing the pathway's negative feedback loop. Except for 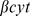 production (
production (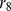 ,
, 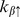 ) that is modeled as a constant flux, all the reactions follow Mass action kinetics.
) that is modeled as a constant flux, all the reactions follow Mass action kinetics.
It is worth noting that the Lee model contains reactions for both basal Axin and basal  -catenin production. By contrast, our model only contains basal
-catenin production. By contrast, our model only contains basal  -catenin production. The reason is that basal Axin production is a constant flux, i.e., its reaction speed is independent of species concentration levels. As the reaction rate constant of this flux is, according to the Lee model, five orders of magnitudes lower than the one of basal
-catenin production. The reason is that basal Axin production is a constant flux, i.e., its reaction speed is independent of species concentration levels. As the reaction rate constant of this flux is, according to the Lee model, five orders of magnitudes lower than the one of basal  -catenin production, its impact is negligible and can therefore be omitted.
-catenin production, its impact is negligible and can therefore be omitted.
Model assumptions
We adopt the usual assumptions made when modeling cell-biological systems, i.e., constant compartment volumes, molecules without volumes, etc. [18]. Besides focussing only on the major components of the Wnt/ -catenin pathway, we also neglect possible crosstalks with other pathways, such as Ryk [35] or Notch [36]. Following the ideas in [34], we abstract the degradation complex by only one of its components, Axin. This is possible since Axin is the limiting factor of the degradation complex due to its low amount in comparison to the other three components, GSK3
-catenin pathway, we also neglect possible crosstalks with other pathways, such as Ryk [35] or Notch [36]. Following the ideas in [34], we abstract the degradation complex by only one of its components, Axin. This is possible since Axin is the limiting factor of the degradation complex due to its low amount in comparison to the other three components, GSK3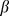 , APC, and CKI
, APC, and CKI [23]. The binding of Wnt molecules to the membrane receptors is not represented as it still remains, biologically, poorly understood. The reaction of Wnt decay (
[23]. The binding of Wnt molecules to the membrane receptors is not represented as it still remains, biologically, poorly understood. The reaction of Wnt decay (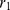 ) represents both its consumption and deactivation after signaling. The nucleo-cytoplasmic shuttling of
) represents both its consumption and deactivation after signaling. The nucleo-cytoplasmic shuttling of 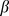 -catenin remains yet unclear [37], thus we introduce the motion of
-catenin remains yet unclear [37], thus we introduce the motion of 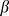 -catenin as a simple diffusion [38] with rate constants based on experimental data [33].
-catenin as a simple diffusion [38] with rate constants based on experimental data [33].
Model evaluation by comparison to the Lee model
In the following, we provide an evaluation study for the overall concept of our model, with its reactions and the assumptions made, based on a comparison to the Lee model. It focuses on comparing the concentration of 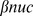 to the concentration of free
to the concentration of free 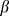 -catenin in the Lee model (that considers only the cytosolic compartment).
-catenin in the Lee model (that considers only the cytosolic compartment).
In our model, initial concentrations are taken from the Lee model, as they were retrieved from experiments in Xenopus oocyte. An exception forms the concentration of Wnt. In the Lee model, Wnt molecules are not considered but replaced by a rather abstract signal called  . The signal
. The signal  has a real value between 0 and 1 that decreases exponentially following:
has a real value between 0 and 1 that decreases exponentially following: 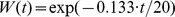 . In our model, Wnt molecules decay over time following a Mass action kinetics (
. In our model, Wnt molecules decay over time following a Mass action kinetics (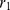 ). The kinetic rate value for this reaction is obtained from the previous function
). The kinetic rate value for this reaction is obtained from the previous function  :
: 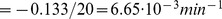 , and an initial Wnt concentration of 100 (unitless) is assumed. Reactions for decay and production and for Axin decay are conserved from the Lee model. Table 2 presents the parameter names and values as used in the Lee model and their respective names in our model.
, and an initial Wnt concentration of 100 (unitless) is assumed. Reactions for decay and production and for Axin decay are conserved from the Lee model. Table 2 presents the parameter names and values as used in the Lee model and their respective names in our model.
Table 2. Parameters from the Lee model [23] used in our model and their stochastic conversion.
| Lee model | our model | ||
| Parameters | Deterministic values | Parameters | Stochastic values |
| Species initial values | |||
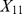
|
25.01 nM | nβcyt | 13141 molecules |
| nβnuc | 5347 molecules | ||
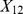
|
0.02 nM | nAxin | 11 molecules |
| nAxinP | 11 molecules | ||
| W | 0 W W 1 1 |
nWnt | 100 (dimensionless) |
| Rate constants | |||
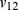
|
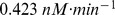
|
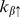
|
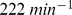
|
| W(t) |
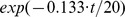
|
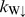
|
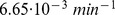
|
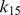
|
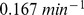
|
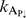
|
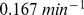
|
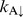
|
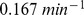
|
||
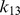
|
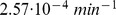
|
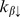
|
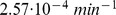
|
Parameters' names and values are given as in [23]. Their corresponding names in our model are given with their stochastic values.
Table 3, Set 1 provides the values of all model parameters: The kinetic rates of reactions 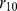 and
and 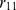 are taken from literature [33]. As several other rate constants are unknown, we performed parameter fitting experiments (parameter estimation details are given in Materials & Methods Section).
are taken from literature [33]. As several other rate constants are unknown, we performed parameter fitting experiments (parameter estimation details are given in Materials & Methods Section).
Table 3. List of model parameters.
| Parameters | Deterministic values | Stochastic values | ||
| Set 1 | Set 2 | Set 3 | Set4 | |
| Species initial values | ||||
| nβcyt | 24.9 nM | 23.6 nM | 11145 molecules | 12989 molecules |
| nβnuc | 24.9 nM | 23.6 nM | 4532 molecules | 5282 molecules |
| nAxin | 0.007 nM | 0.051 nM | 144 molecules | 252 molecules |
| nAxinP | 0.042 nM | 0 nM | 125 molecules | 219 molecules |
| nWnt | 100 | 1000 | 1000 | 1000 |
| Rate constants | ||||
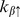
|
0.232* | 0.571* | 420 | 600 |
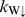
|
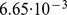
|
0.27 | 0.6 | 0.27 |
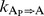
|
7 | 10 | 10 | 20 |
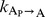
|
0.3 | 0.0182 | 0.03 | 0.03 |
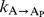
|
3 |
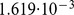
|
0.03 | 0.03 |
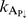
|
0.167 | 0.167 |
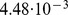
|
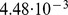
|
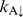
|
0.367 | 0.0167 |
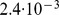
|
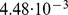
|
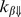
|
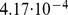 * * |
52.5* |
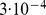
|
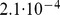
|
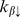
|
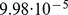
|
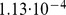
|
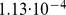
|
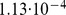
|
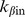
|
0.0549 | 0.0549 | 0.0549 | 0.0549 |
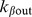
|
0.135 | 0.135 | 0.135 | 0.135 |
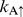
|
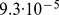
|
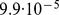
|
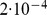
|
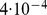
|
Parameters used in our model for the different in silico experiments. Sets 1 and 2 are the results of the parameter fitting experiments comparing to the Lee model and our experimental data, respectively. Sets 3 and 4 are obtained from our stochastic and our cell cycle investigation, respectively. All rate constants are in 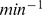 but the ones indicated by “*”: the deterministic values of
but the ones indicated by “*”: the deterministic values of 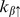 are given in
are given in 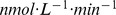 , and the deterministic values of
, and the deterministic values of 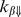 are given in
are given in 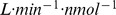 .
.
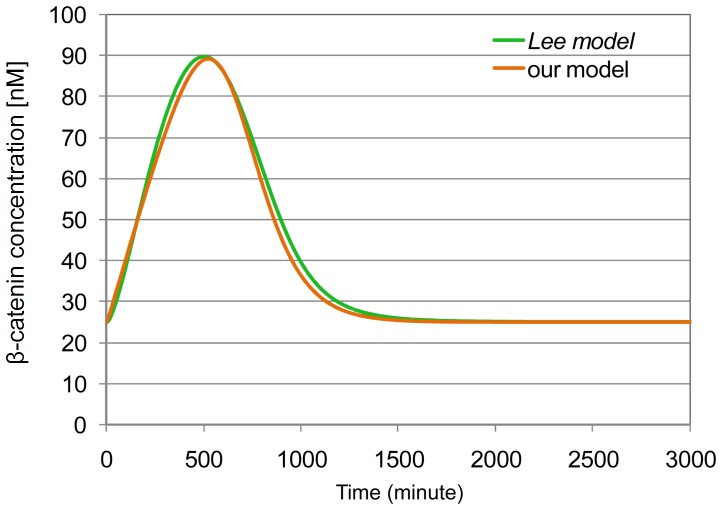
The results of simulation experiments with Wnt set to 100 (transient Wnt stimulation) in our model, and 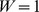 in the Lee model are provided in Figures 3. They show a good fit between the concentrations of
in the Lee model are provided in Figures 3. They show a good fit between the concentrations of 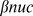 in our model and
in our model and 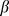 -catenin in the Lee model.
-catenin in the Lee model.
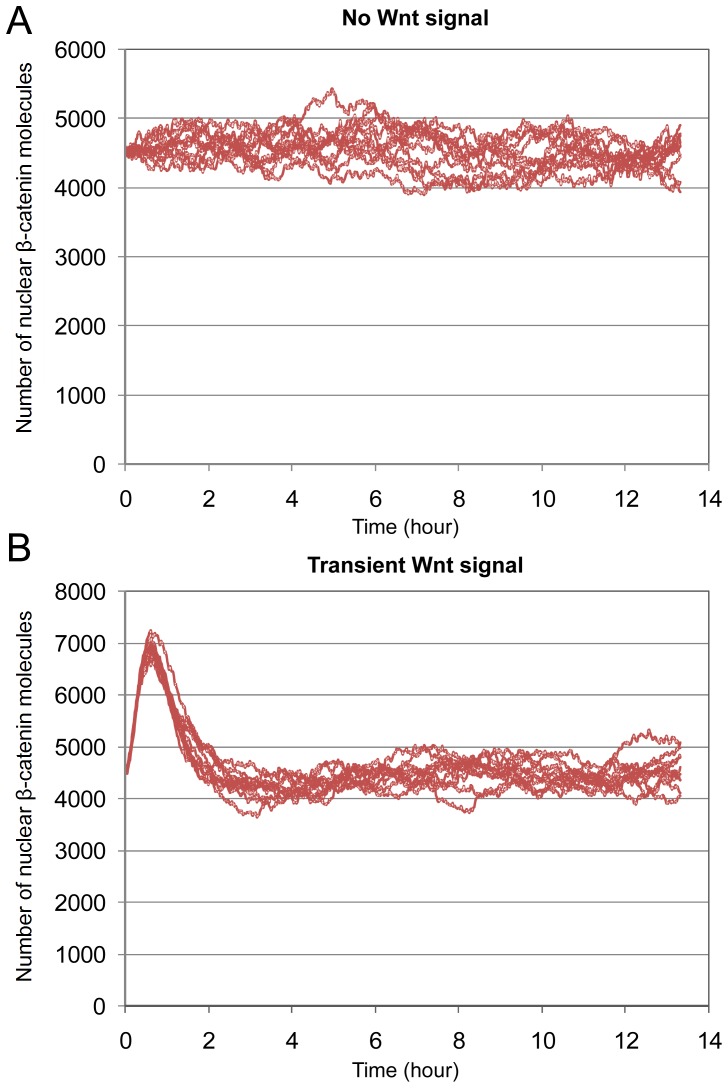
Figure 3. Comparison of 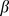 -catenin dynamics between our core model and the Lee model.
-catenin dynamics between our core model and the Lee model.
Time-dependent response of 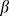 -catenin under transient Wnt signal (
-catenin under transient Wnt signal (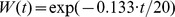 introduced into the system at the steady-state, at
introduced into the system at the steady-state, at 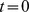 ). The dynamics of total in the and of nuclear
). The dynamics of total in the and of nuclear 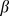 -catenin (
-catenin (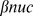 ) in our core model are corresponding. Simulations of the Lee model are performed through the JWS platform [55].
) in our core model are corresponding. Simulations of the Lee model are performed through the JWS platform [55].
Model evaluation by comparison to experimental data
Here, we present the results of a parameter fitting experiment that compares the dynamics of in 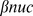 our model to those provided by our wet-lab experiments (see Figure 1). Only the first increase in the data is subject to the fitting, since it is the one considered to reflect the dynamics of the Wnt/
our model to those provided by our wet-lab experiments (see Figure 1). Only the first increase in the data is subject to the fitting, since it is the one considered to reflect the dynamics of the Wnt/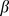 -catenin pathway as initially induced by growth factor removal. Notice that at this point we compare single in silico cells to the average behavior of entire populations as measured in our wet-lab experiments. This means in particular that we implicitly assume a synchronized RVM cell population w.r.t. the cell cycle.
-catenin pathway as initially induced by growth factor removal. Notice that at this point we compare single in silico cells to the average behavior of entire populations as measured in our wet-lab experiments. This means in particular that we implicitly assume a synchronized RVM cell population w.r.t. the cell cycle.
The results of our experiment are provided in Table 3, Set 2. Whereas initial species values remain as before, the values of most rate constants are adjusted. In particular, the value of the rate constant of Axin-dependent 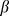 -catenin degradation (
-catenin degradation (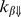 ) shows a significant increase (see the section on stochastic investigations for further details).
) shows a significant increase (see the section on stochastic investigations for further details).
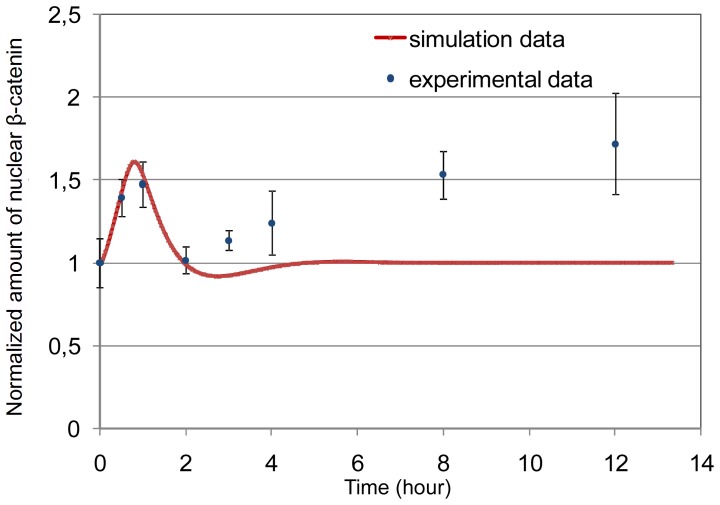
In Figure 4, results of simulation experiments with the obtained parameters are provided. We obtain a good fit to the first increase in our data. Moreover, after two hours of differentiation, 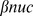 remains constant and no second increase can be observed.
remains constant and no second increase can be observed.
Figure 4. Model evaluation in comparison to the experimental data.
Comparison of the model time course to the experimental data for nuclear 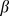 -catenin dynamics. The simulation data fit the experimental ones for the first 2 hours.
-catenin dynamics. The simulation data fit the experimental ones for the first 2 hours.
Stochastic effects due to low AxinP amounts
In this section, we report on stochastic fluctuations in the dynamics of our model that contradict our wet-lab observations. We show that these fluctuations directly result from the low Axin amounts as suggested by the Lee model [23] rather than from our specific choices of parameter values. This study requires a transformation of the concentrations and kinetic rate constants in Table 3 (Set 2) into molecule numbers and stochastic rate constants, following, e.g. [39], (Table 3, Set 3).
The stochastic fluctuations in the 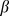 -catenin dynamics resulting from Parameter Set 3 are shown in Figures 5A, 5B. These figures present the amount of
-catenin dynamics resulting from Parameter Set 3 are shown in Figures 5A, 5B. These figures present the amount of 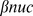 and
and 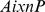 molecules over time in absence of Wnt signal, as the result of a single simulation run. Large fluctuations in the
molecules over time in absence of Wnt signal, as the result of a single simulation run. Large fluctuations in the 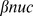 amounts can be observed that were not visible in the deterministic investigations of the previous section. Along with the fluctuations, we see only small differences in the number of
amounts can be observed that were not visible in the deterministic investigations of the previous section. Along with the fluctuations, we see only small differences in the number of 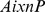 , which underlines the high impact of
, which underlines the high impact of 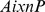 changes on the
changes on the 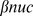 dynamics. This impact results from a comparatively high rate of
dynamics. This impact results from a comparatively high rate of 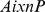 -dependent
-dependent 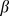 -catenin degradation (
-catenin degradation (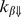 ) that is necessary to fit our experimental data. The stochastic fluctuations contradict our in vitro results, since they do not allow for a clear transient peak of
) that is necessary to fit our experimental data. The stochastic fluctuations contradict our in vitro results, since they do not allow for a clear transient peak of 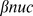 in response to the Wnt signal (not shown).
in response to the Wnt signal (not shown).
Figure 5. Effects of AxinP on 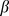 -catenin dynamics according to stochastic simulation.
-catenin dynamics according to stochastic simulation.
In absence of Wnt signal, small variations of 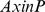 (A) influence
(A) influence 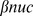 dynamics (B) leading to high variance. Increase of
dynamics (B) leading to high variance. Increase of 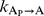 and
and 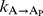 by 1000 accelerate
by 1000 accelerate 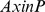 changes (C), but
changes (C), but 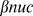 fluctuations still remain too high (D). Each plot represents a single simulation run.
fluctuations still remain too high (D). Each plot represents a single simulation run.
In order to exclude that these stochastic fluctuations are the result of our specific choice of parameter values, we explored different strategies to change parameter values in order to lower stochastic effects and at the same time to maintain the amounts of 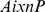 and the average behavior of the model w.r.t.
and the average behavior of the model w.r.t. 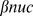 dynamics.
dynamics.
An obvious solution would be to decrease the rate constant of 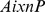 -dependent
-dependent 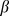 -catenin degradation (
-catenin degradation (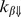 ) and maintaining the
) and maintaining the 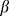 -catenin level by simultaneously decreasing the flux of
-catenin level by simultaneously decreasing the flux of 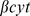 production (
production (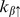 ). However, simulation experiments showed that already through small changes of this sort, the amount of
). However, simulation experiments showed that already through small changes of this sort, the amount of 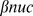 is prevented from increasing in response to Wnt signal (results not shown).
is prevented from increasing in response to Wnt signal (results not shown).
Only one other parameter modification strategy seems to be plausible to us: to deploy the inertia of 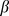 -catenin reacting on changes in
-catenin reacting on changes in 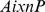 amounts. Since the amount of
amounts. Since the amount of 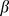 -catenin is relatively high, observable changes due to differences in
-catenin is relatively high, observable changes due to differences in 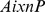 amounts can only take place with a certain delay. The amount of
amounts can only take place with a certain delay. The amount of 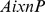 is impacted only by its decay (
is impacted only by its decay (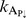 ) and its de-/phosphorylation (
) and its de-/phosphorylation (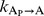 ,
, 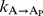 ). Instead of increasing one of the corresponding rate constants separately, which would result in a change in the overall amount of
). Instead of increasing one of the corresponding rate constants separately, which would result in a change in the overall amount of 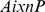 , one can increase the rate constants for de-/phosphorylation simultaneously. Figures 5C, 5D show the results when increasing the rate constants for dephosphorylation (
, one can increase the rate constants for de-/phosphorylation simultaneously. Figures 5C, 5D show the results when increasing the rate constants for dephosphorylation (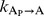 ) by about 1000 times and the one for phosphorylation (
) by about 1000 times and the one for phosphorylation (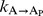 ) accordingly. Changes in
) accordingly. Changes in 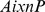 numbers happen much faster. However, although they are lower, the stochastic fluctuations in the
numbers happen much faster. However, although they are lower, the stochastic fluctuations in the 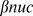 dynamics are still too high. Raising the dephosphorylation rate (
dynamics are still too high. Raising the dephosphorylation rate (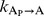 ) even more seems not plausible to us, since then changes on the few
) even more seems not plausible to us, since then changes on the few 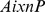 molecules happen in periods of milliseconds.
molecules happen in periods of milliseconds.
We are therefore convinced that in our model with low 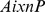 level, as derived from the Lee model, stochastic fluctuations in the
level, as derived from the Lee model, stochastic fluctuations in the 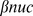 level cannot be limited to fit our experimental data. As additional support for this statement, we also provide an Sbml version of our model in the supplementary material (Model S1).
level cannot be limited to fit our experimental data. As additional support for this statement, we also provide an Sbml version of our model in the supplementary material (Model S1).
Our results may indicate on one hand that our RVM cells contain higher amounts of Axin than suggested for the Xenopus oocyte by [23]. This is supported by the recent work of Tan and colleagues [40] as they report that mammalian cells have a higher Axin concentration than the Xenopus extract and that yet 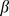 -catenin concentrations remain higher than Axin concentration. Notice, however, that in prior wet-lab experiments, we failed to detect Axin. On the other hand, our model and thus our understanding of the system may miss an important mechanism to reduce stochastic noise, such as dimerization [41], [42] or additional feedback loops [43].
-catenin concentrations remain higher than Axin concentration. Notice, however, that in prior wet-lab experiments, we failed to detect Axin. On the other hand, our model and thus our understanding of the system may miss an important mechanism to reduce stochastic noise, such as dimerization [41], [42] or additional feedback loops [43].
As a basis for our subsequent investigations, we performed additional parameter fitting experiments, thereby stepwise increasing the amount of 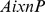 and
and 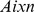 . As a result, we found a parameter set with acceptable stochastic fluctuations at an initial amount of
. As a result, we found a parameter set with acceptable stochastic fluctuations at an initial amount of 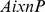 of 125, see Table 3, Set 3. In Figure 6A, the dynamics of
of 125, see Table 3, Set 3. In Figure 6A, the dynamics of 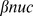 for 10 simulation runs are shown, where the Wnt signal is switched off (Wnt = 0). One can see that the fluctuations in
for 10 simulation runs are shown, where the Wnt signal is switched off (Wnt = 0). One can see that the fluctuations in 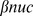 levels have been effectively decreased. The t-test provides that at the time point of maximum deviation (ca. 426 minutes), the mean value of
levels have been effectively decreased. The t-test provides that at the time point of maximum deviation (ca. 426 minutes), the mean value of 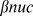 numbers in the 10 simulation runs lies in the interval of
numbers in the 10 simulation runs lies in the interval of 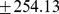 (ca. 5.5% of the simulated mean value) around the simulated mean value with a confidence of 95%. Figure 6B shows the dynamics of
(ca. 5.5% of the simulated mean value) around the simulated mean value with a confidence of 95%. Figure 6B shows the dynamics of 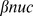 for 10 simulation runs when the Wnt signal is switched on. We observe in our simulation a transient peak within the standard deviation of the experimental data.
for 10 simulation runs when the Wnt signal is switched on. We observe in our simulation a transient peak within the standard deviation of the experimental data.
Figure 6. Nuclear  -catenin dynamics with higher number of AxinP.
-catenin dynamics with higher number of AxinP.
(A) In absence of Wnt signal. (B) Under transient Wnt signal. The plots represent 10 simulation runs.
Impact of the Cell Cycle on 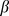 -catenin dynamics in RVM cells
-catenin dynamics in RVM cells
In this section, we show that in the results of wet-lab experiments, the impact of the cell cycle on the average dynamics of 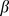 -catenin in RVM cell populations can be neglected.
-catenin in RVM cell populations can be neglected.
The idea of extending our model to represent asynchronous cell populations is to consider a number of copies of our core model, each representing an individual cell. Following [12], cells have to first complete the cell cycle before they can process the Wnt signal. As for our previous model Wnt is not produced and decays overtime. However, we assign an individual delay to the start time of Wnt degradation (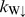 ) and Wnt-dependent Axin dephosphorylation (
) and Wnt-dependent Axin dephosphorylation (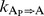 ) of each cell that represents the cell delay to exit the cell cycle. That is the corresponding reactions
) of each cell that represents the cell delay to exit the cell cycle. That is the corresponding reactions 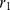 and
and 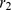 are ensured not to happen before a fixed time point has passed. Cells, which are already in the G1 phase at the beginning of a simulation experiment, i.e., at time point 0 hour, start to perform their reactions without any delay. Notice that, since each cell has its own pool of Wnt and also reaction
are ensured not to happen before a fixed time point has passed. Cells, which are already in the G1 phase at the beginning of a simulation experiment, i.e., at time point 0 hour, start to perform their reactions without any delay. Notice that, since each cell has its own pool of Wnt and also reaction 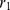 is delayed for each cell individually, we assume self-induced signaling to happen in an autocrine fashion. The integration of delays into our stochastic model is described in the Materials & Methods Section.
is delayed for each cell individually, we assume self-induced signaling to happen in an autocrine fashion. The integration of delays into our stochastic model is described in the Materials & Methods Section.
Cell cycle states are assigned to cells following the distribution of cells over the cycle phases as obtained from experiments (see Figure 2 in [12] and details in Materials & Methods). For example, in a population of  cells,
cells, 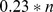 cells are assigned to state
cells are assigned to state 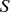 . Potential rounding problems are solved by randomly assigning states to remaining cells with probabilities to be in certain states following our experimental data. Notice that we do not separate G2 and M phases. The delay for each cell is computed in the following way: cells are assumed to be equally distributed over their respective states. That is, to a cell in state
. Potential rounding problems are solved by randomly assigning states to remaining cells with probabilities to be in certain states following our experimental data. Notice that we do not separate G2 and M phases. The delay for each cell is computed in the following way: cells are assumed to be equally distributed over their respective states. That is, to a cell in state 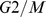 a delay
a delay 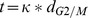 is assigned, where
is assigned, where  is equally distributed in
is equally distributed in 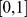 and
and 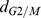 is the duration of phase
is the duration of phase 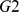 . Similarly, since each cell in state
. Similarly, since each cell in state 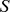 has to additionally pass state
has to additionally pass state 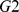 , the delay of a cell in state
, the delay of a cell in state 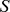 is given by
is given by 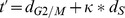 , with
, with 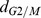 being the duration of phase
being the duration of phase 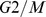 . The duration of each cell cycle phase is obtained from literature [10], [44] (details in Materials & Methods).
. The duration of each cell cycle phase is obtained from literature [10], [44] (details in Materials & Methods).
We performed simulation experiments with the parameters in Table 3, Set 3, as obtained from our stochastic investigation. In Figure 7A, the sum of the number of 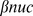 (nuclear
(nuclear 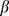 -catenin) for a single simulation run with 100 cells is presented. Similar to the results of the previous section (Figure 6B), we observe a single transient increase. This, however, shows only a value about 1.28 instead of 1.48, i.e., we obtain only 86% of the expected amount. The reason is that, following our experimental data on the cell cycle in RVM cells, the
-catenin) for a single simulation run with 100 cells is presented. Similar to the results of the previous section (Figure 6B), we observe a single transient increase. This, however, shows only a value about 1.28 instead of 1.48, i.e., we obtain only 86% of the expected amount. The reason is that, following our experimental data on the cell cycle in RVM cells, the 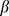 -catenin amounts in most of the cells initially dedicated to the cell cycle, does not start to increase before 30 minutes, i.e., after the time of the first peak is over. Thus, only about 60% of the cells contribute to the first peak, resulting in the limited increase.
-catenin amounts in most of the cells initially dedicated to the cell cycle, does not start to increase before 30 minutes, i.e., after the time of the first peak is over. Thus, only about 60% of the cells contribute to the first peak, resulting in the limited increase.
Figure 7. Dynamics of nuclear 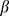 -catenin in heterogeneous cell population.
-catenin in heterogeneous cell population.
A population of 100 cells is simulated. (A, B, and D) represent the sum of nuclear in 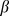 -catenin 100 cells as compared to the experimental data. (C) represents the number of nuclear
-catenin 100 cells as compared to the experimental data. (C) represents the number of nuclear 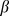 -catenin in each cell individually for a population of 100 cells. (A) The total cell population is analyzed. The simulation data do not correlate with the experimental ones. (B) The cell population asynchrony toward the cell cycle does not give rise to a second increase of nuclear
-catenin in each cell individually for a population of 100 cells. (A) The total cell population is analyzed. The simulation data do not correlate with the experimental ones. (B) The cell population asynchrony toward the cell cycle does not give rise to a second increase of nuclear 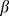 -catenin after 2 hours. (C) Single cell analysis shows the delay in
-catenin after 2 hours. (C) Single cell analysis shows the delay in 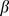 -catenin dynamics due to the cell cycle asynchrony. (D) The addition of a continuous and autocrine Wnt signal to the previous experimental setting produces simulation data in perfect agreement to the experimental ones. (A, C) Parameters used are given in Table 3 (Set 3). (B, D) Parameters used for both experiments are given in Table 3 (Set 4).
-catenin dynamics due to the cell cycle asynchrony. (D) The addition of a continuous and autocrine Wnt signal to the previous experimental setting produces simulation data in perfect agreement to the experimental ones. (A, C) Parameters used are given in Table 3 (Set 3). (B, D) Parameters used for both experiments are given in Table 3 (Set 4).
As mentioned earlier fitting experiments to obtain the parameters (Table 3, Set 3) implicitly assume that cells are homogeneously distributed over the cell cycle as we compared the behavior of single cells, i.e., one instance of our core model, with the average behavior of entire cell populations as observed in experiments. Thus, the results of our simulation experiments imply that this assumption is not feasible and that when modeling the Wnt/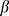 -catenin pathway in RVM cells, the cells' commitment to the cell cycle may need to be considered.
-catenin pathway in RVM cells, the cells' commitment to the cell cycle may need to be considered.
We present an additional parameter set to fit the first increase in our experimental data with the simulation data in asynchronous cell populations (see Table 3, set 4). Only a few parameters are adjusted, as compared to the previous one. Corresponding simulation results are presented in Figure 7B. It can be seen that with the new parameter set, the first increase of nuclear 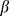 -catenin is well-fitted. Furthermore, no other increase than the first one can be observed in our simulation results.
-catenin is well-fitted. Furthermore, no other increase than the first one can be observed in our simulation results.
Our simulation results indicate that when studying the Wnt pathway in RVM cell populations by wet-lab experiments the impact of the cell cycle on the average 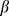 -catenin dynamics can be neglected. No significant changes occur other than the increase caused by Wnt pathway activation. In particular, the second increase in the experimental data shows not to be a result of the cells' asynchrony. The reason for this becomes obvious when looking at the dynamics of
-catenin dynamics can be neglected. No significant changes occur other than the increase caused by Wnt pathway activation. In particular, the second increase in the experimental data shows not to be a result of the cells' asynchrony. The reason for this becomes obvious when looking at the dynamics of 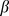 -catenin for each cell separately, as presented in Figure 7C. Although, all the cells, which are initially dedicated to the cell cycle, start their
-catenin for each cell separately, as presented in Figure 7C. Although, all the cells, which are initially dedicated to the cell cycle, start their 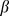 -catenin increase comparatively late, they do not perform it simultaneously but rather widely distributed over time (between 0.5 and 10 hours). Thus, summing up over all cells, the individual peaks only lead to small deviations. It is important to notice here that this observation is rather independent from the particular design decisions of our model but results mainly from the distribution of RVM cells over the cell cycle as it is obtained experimentally and from the basic knowledge about the Wnt/
-catenin increase comparatively late, they do not perform it simultaneously but rather widely distributed over time (between 0.5 and 10 hours). Thus, summing up over all cells, the individual peaks only lead to small deviations. It is important to notice here that this observation is rather independent from the particular design decisions of our model but results mainly from the distribution of RVM cells over the cell cycle as it is obtained experimentally and from the basic knowledge about the Wnt/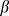 -catenin pathway as represented in our model.
-catenin pathway as represented in our model.
Self-induced Wnt signaling
In this section, we provide new evidence for the hypothesis that the 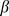 -catenin dynamics from 8 hours on after the start of differentiation arises from a second wave of Wnt signal that could be self-induced. This hypothesis is in the lines with our previous study [12] listing various indicators from wet lab experiments for late self-induced Wnt signaling. Among these are an increase of Axin, Wnt ligands, and receptor gene expression, as well as an accumulation of the pathway's intracellular proteins during cell differentiation without addition of external Wnt signal (see also this article's introduction). Notice that the goal here is not to explain the detailed underlying mechanisms of self-induced Wnt signaling, but rather to explore whether our experimental data actually suggest that such a process may occur in RVM cell populations. In particular, we leave possible spatial extensions to our model to distinct autocrine from paracrine signaling as subject to future work and consider only autocrine signaling here.
-catenin dynamics from 8 hours on after the start of differentiation arises from a second wave of Wnt signal that could be self-induced. This hypothesis is in the lines with our previous study [12] listing various indicators from wet lab experiments for late self-induced Wnt signaling. Among these are an increase of Axin, Wnt ligands, and receptor gene expression, as well as an accumulation of the pathway's intracellular proteins during cell differentiation without addition of external Wnt signal (see also this article's introduction). Notice that the goal here is not to explain the detailed underlying mechanisms of self-induced Wnt signaling, but rather to explore whether our experimental data actually suggest that such a process may occur in RVM cell populations. In particular, we leave possible spatial extensions to our model to distinct autocrine from paracrine signaling as subject to future work and consider only autocrine signaling here.
To model self-induced Wnt signaling, we extend our model with a single reaction of Wnt production representing the overall process in a very abstract way. This reaction occurs with a given delay after cells exit the cell cycle. The delay is to reflect the time necessary to induce the signal. It is implemented in the same way as the one for the cell cycle reaction (details in Materials & Methods section). Once the delay is over, cells continuously produce Wnt molecules with rate constant 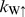
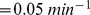 , each for themselves (autocrine signaling).
, each for themselves (autocrine signaling).
We performed a simulation experiment with a cell population of 100 cells and a Wnt induction delay of 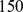 minutes (2.5 hours). Such value for the delay is plausible, since our Wnt-producing reaction encompasses various biological processes, i.e., gene transcription of Wnt molecules, their subsequent intracellular trafficking, post-translational modifications [45], secretion of Wnt in the extracellular environment, and its binding to the cell receptors.
minutes (2.5 hours). Such value for the delay is plausible, since our Wnt-producing reaction encompasses various biological processes, i.e., gene transcription of Wnt molecules, their subsequent intracellular trafficking, post-translational modifications [45], secretion of Wnt in the extracellular environment, and its binding to the cell receptors.
In Figure 7D, the results of a single simulation run are presented. The first peak of  is still fitting the experimental data. Furthermore, the simulation data at the other time points, i.e., 3, 4, 8, and 12 hours, are also fitting the experimental ones. Since the previous variants of the model failed to reproduce the biphasic kinetics of nuclear
is still fitting the experimental data. Furthermore, the simulation data at the other time points, i.e., 3, 4, 8, and 12 hours, are also fitting the experimental ones. Since the previous variants of the model failed to reproduce the biphasic kinetics of nuclear  -catenin, our results suggest that self-induced Wnt signaling is responsible of such dynamics and occurs in RVM cells during differentiation from 2.5 to 12 hours.
-catenin, our results suggest that self-induced Wnt signaling is responsible of such dynamics and occurs in RVM cells during differentiation from 2.5 to 12 hours.
It should be noticed here that the investigations of Wnt production in RVM cells at 3, 6, and 24 hours of differentiation shows only for the latter time point an increase of Wnt mRNA level (mRNA of Wnt5a and Wnt7a) [11], [12]. These observations are, however, not necessarily in contradiction to the hypothesis of self-induced signaling whitin the first hours of cell differentiation. On one hand, without measurements of intermediate time points, Wnt production could start as soon as 8 hours. On the other hand, even without an increase in the Wnt mRNA level, self-induced signaling could occur. It is known that Wnt molecules are produced in the cell and are subsequently stored in cytosolic vesicles that can then fuse with the membrane in order to release Wnt molecules outside of the cell, inducing signaling [46]. Thus, the self-induced Wnt signaling can result not only from a constant production and secretion during RVM cell differentiation but also from an anticipated production of Wnt molecules followed by a vesicle storage and a delayed continuous secretion during differentiation. Clarification on this point could be achieved by further in vitro experiments, based, e.g., on a constant inhibition of the Wnt/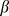 -catenin pathway, in particular with Dickkopf 1, Mesd, or Porcupin [46]–[48], followed by kinetic analysis of nuclear
-catenin pathway, in particular with Dickkopf 1, Mesd, or Porcupin [46]–[48], followed by kinetic analysis of nuclear 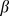 -catenin between 2 and 12 hours of differentiation.
-catenin between 2 and 12 hours of differentiation.
Furthermore, the occurrence of crosstalk remains to be a valid alternative to the hypothesis of self-induced Wnt signaling. Previous works show that during nervous system development [49], [50], although not during neural differentiation in particular, crosstalks occur between the Wnt/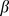 -catenin pathway and other pathways, among which are the Ryk [35], Notch [36], and extracellular signal regulated-kinase (ERK) pathways [51]. Therefore, to finally answer the question of self-induced Wnt signaling in human neural progenitor cells, further in vitro or in silico investigations are required.
-catenin pathway and other pathways, among which are the Ryk [35], Notch [36], and extracellular signal regulated-kinase (ERK) pathways [51]. Therefore, to finally answer the question of self-induced Wnt signaling in human neural progenitor cells, further in vitro or in silico investigations are required.
Materials and Methods
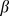 -catenin Western-blotting
-catenin Western-blotting
Western-blots were performed according to the protocol described previously in [10]. Visualization and quantification of 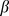 -catenin were performed using the Odyssey Infrared Imaging System (LI-COR Biosciences GmbH, Bad Homburg, Germany). For the purpose of quantification, time-series samples were loaded onto the gel in a randomized and non-chronological order to reduce systematic errors [52]. Expression of
-catenin were performed using the Odyssey Infrared Imaging System (LI-COR Biosciences GmbH, Bad Homburg, Germany). For the purpose of quantification, time-series samples were loaded onto the gel in a randomized and non-chronological order to reduce systematic errors [52]. Expression of 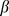 -actin protein was used as a loading control to normalize the expression of
-actin protein was used as a loading control to normalize the expression of 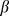 -catenin. Thereby relative expression levels of
-catenin. Thereby relative expression levels of 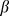 -catenin proteins were determined. For each quantified Western-blot, the relative expression at a given time point is normalized to the mean of the time series. Normalization is performed for each experiment separately, as the raw data obtained for each Western-blot are not absolute values. For each time point, the mean of all experiments is computed. Thus, the values observed in Figure 1 show the mean of 26 experiments (9 cell cultures with 2 to 3 Western-blots performed for each cell culture) for the time points 0, 0.5, 1, 3, 8, 24, and 48 hours, and the mean of 11 experiments (4 cell cultures with 2 to 3 Western-blots performed for each cell culture) for the other time points 2, 4, 12, and 72 hours. Statistical evaluation was carried out using the Mann-Whitney test. An increase was considered to be statistically significant when the p-value
-catenin proteins were determined. For each quantified Western-blot, the relative expression at a given time point is normalized to the mean of the time series. Normalization is performed for each experiment separately, as the raw data obtained for each Western-blot are not absolute values. For each time point, the mean of all experiments is computed. Thus, the values observed in Figure 1 show the mean of 26 experiments (9 cell cultures with 2 to 3 Western-blots performed for each cell culture) for the time points 0, 0.5, 1, 3, 8, 24, and 48 hours, and the mean of 11 experiments (4 cell cultures with 2 to 3 Western-blots performed for each cell culture) for the other time points 2, 4, 12, and 72 hours. Statistical evaluation was carried out using the Mann-Whitney test. An increase was considered to be statistically significant when the p-value  (
( ,
,  , and
, and  ) as compared to control (time point 0 hour). The values of the mean and the standard error of the mean were rescaled to the mean at time point 0 hour which is set to 1.0.
) as compared to control (time point 0 hour). The values of the mean and the standard error of the mean were rescaled to the mean at time point 0 hour which is set to 1.0.
Cell volume determination
Data on compartment volumes is a prerequisite for calculating stochastic parameters as the reaction speed is dependent on the probability of molecules to collide. As the cell volume and thus the compartment volumes might change along cell differentiation, the volume measurement is restricted to proliferating RVM cells, as at this stage their general shape is regular, following a cobble-stone pattern [3]. The global volume of proliferating RVM cells was determined using the electric cell counter CASY® (Innovatis AG, Germany) which uses impedance technique. The RVM cells were cultivated according to the protocol described previously in [10]. The cells were detached from the coated cell culture flask T75 with addition of Trypsin-Benzonase solution (2.5 ml/flask) followed by trypsin-inhibition solution (5 ml/flask). As the solutions are isotonic they do not cause cell volume changes. In presence of Trypsin, RVM cells are in nearly spherical shape within the solution suspension. This condition is necessary and sufficient for the device CASY® to retrieve the mean volume of the cells analyzed in the sample. Cell suspension (25 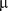 l) was diluted in 10 ml of CasyTon buffer and analyzed by CASY®. The analysis results contain the mean volume of the spherical cells passing through the capillary. Thus, given 79 cell samples, from cell passage 9 to 27, the average volume of RVM cells is
l) was diluted in 10 ml of CasyTon buffer and analyzed by CASY®. The analysis results contain the mean volume of the spherical cells passing through the capillary. Thus, given 79 cell samples, from cell passage 9 to 27, the average volume of RVM cells is 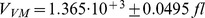 . To obtain the compartment volumes, the average area of the cytosol and nucleus in proliferating cells, was measured via microscopy. The cytosol represents an average of 64% of the cell surface, whereas the nucleus represents 26% of the cell surface. These values were considered to hold true for the volume proportions, leading to:
. To obtain the compartment volumes, the average area of the cytosol and nucleus in proliferating cells, was measured via microscopy. The cytosol represents an average of 64% of the cell surface, whereas the nucleus represents 26% of the cell surface. These values were considered to hold true for the volume proportions, leading to:
Duration and distribution of cell cycle phases
The duration of the RVM cell cycle is given in [10] with a doubling time of 19.8 0.6 hours (ca. 1188 minutes). In order to estimate the duration of each phase, we took reference values from [44] that describes a relative duration of 50% for G1, 33.3% for S, and 16.7% for G2/M of the cell cycle entire duration in mammalian cells. Thus, the delay for the G2/M phase amounts to
0.6 hours (ca. 1188 minutes). In order to estimate the duration of each phase, we took reference values from [44] that describes a relative duration of 50% for G1, 33.3% for S, and 16.7% for G2/M of the cell cycle entire duration in mammalian cells. Thus, the delay for the G2/M phase amounts to 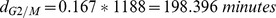 and for the S phase to
and for the S phase to 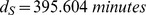 . The amount of RVM cells committed in the cell cycle was retrieved experimentally as described in [12]. Around 20% of RVM cells are in G2 phase and around 23% in S phase when differentiation is induced (time point 0 hour).
. The amount of RVM cells committed in the cell cycle was retrieved experimentally as described in [12]. Around 20% of RVM cells are in G2 phase and around 23% in S phase when differentiation is induced (time point 0 hour).
Parameter estimation & simulation
Parameter estimation for the unknown values of rate constants and species amount was done using COPASI [53]. The method chosen is Hooke & Jeeves [54]. Estimation experiments were supported by sensitivity analysis available in the supplementary material (Text S1). The Lee model simulations were performed using the online tool JWS Online [55]. Deterministic simulations were also done with the tool COPASI [53] using the deterministic LSODA method. Stochastic simulations were performed using a small extension of the stochastic simulation algorithm, the version presented in [56], that allows to directly reflect normally distributed delays (see paragraph below). We implemented this extension and performed stochastic simulation experiments based on the modeling and simulation framework JamesII [57].
Modeling delays
We used the following approach to model delays: for each delay 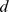 a species
a species 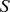 and a reaction
and a reaction 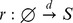 is introduced. Initially, the amount of
is introduced. Initially, the amount of 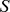 is set to
is set to  . Reaction
. Reaction  produces a single instance of
produces a single instance of  to denote the end of the delay. Reactions, which are supposed not to happen before the delay has ended, are equipped with an additional reactant and product of species
to denote the end of the delay. Reactions, which are supposed not to happen before the delay has ended, are equipped with an additional reactant and product of species  , e.g., reaction
, e.g., reaction  is transformed to
is transformed to  . Notice that when ensuring that the amount of
. Notice that when ensuring that the amount of 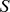 is always 1, the rate of reactions with Mass action kinetics is not affected by this modification. Reaction
is always 1, the rate of reactions with Mass action kinetics is not affected by this modification. Reaction  cannot be directly mapped to a stochastic reaction since it is required to happen only once and at a time point not exponentially distributed, but normally distributed in time that denotes the end of delay
cannot be directly mapped to a stochastic reaction since it is required to happen only once and at a time point not exponentially distributed, but normally distributed in time that denotes the end of delay 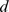 . This can be achieved by using the next-reaction method [56] version of the stochastic simulation algorithm [18], which schedules the time point of each reaction in an event queue. The firing of a reaction at a fixed time point that models the end of a delay can thus be just scheduled as an additional event. We assumed that the end time point of a delay is drawn from a normal distribution on the length of the delay with a standard deviation of 5%.
. This can be achieved by using the next-reaction method [56] version of the stochastic simulation algorithm [18], which schedules the time point of each reaction in an event queue. The firing of a reaction at a fixed time point that models the end of a delay can thus be just scheduled as an additional event. We assumed that the end time point of a delay is drawn from a normal distribution on the length of the delay with a standard deviation of 5%.
Similar ideas to integrate delays into stochastic models have already been presented in [58]. Alternatively, one can also approximate events at normally distributed time points with sets of fast intermediate reactions [59].
Supporting Information
We provide the details of the model in Sbml format. The parameter values correspond to the ones found in Table 3 , Set 2.
(XML)
We provide details regarding the parameter sensitivity analysis.
(PDF)
Acknowledgments
We would like to thank Tareck Rharass for discussions regarding FRAP experiments and cell volumes, as well as the reviewers, Fiete Haack, and Hans-Jörg Schulz for their valuable comments on the paper.
Funding Statement
This work has been supported as part of the research training group dIEM oSiRiS (The integrative development of modeling and simulation methods for regenerative systems) and the research project dIER MoSiS (Discrete event multi-level modeling and simulation for systems biology), both financed by the German foundation for research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lindvall O, Kokaia Z, Martinez-Serrano A (2004) Stem cell therapy for human neurodegenerative disorders-how to make it work. Nature Medicine 10: S42–S50. [DOI] [PubMed] [Google Scholar]
- 2. Clelland CD, Barker RA, Watts C (2008) Cell therapy in huntington disease. Neurosurgical Focus 24 3–4:E8. [DOI] [PubMed] [Google Scholar]
- 3. Donato R, Miljan EA, Hines SJ, Aouabdi S, Pollock K, et al. (2007) Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neuroscience 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wood-Kaczmar A, Gandhi S, Yao Z, Abramov ASY, Miljan EA, et al. (2008) Pink1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS ONE 3 6:e2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Le MT, Xie H, Zhou B, Chia PH, Rizk P, et al. (2009) Microrna-125b promotes neuronal differentiation in human cells by repressing multiple targets. Molecular and Cellular Biology 29 19:5290–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morgan PJ, Ortinau S, Frahm J, Krueger N, Rolfs A, et al. (2009) Protection of neurons derived from human neural progenitor cells by veratridine. Neuroreport 20: 1225–1229. [DOI] [PubMed] [Google Scholar]
- 7. Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, et al. (2003) Differential regulation of midbrain dopaminergic neuron development by wnt-1, wnt-3a, and wnt-5a. PNAS 100 22:12747–12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Michaelidis TM, Lie DC (2008) Wnt signaling and neural stem cells: caught in the Wnt web. Cell Tissue Research 331: 193–210. [DOI] [PubMed] [Google Scholar]
- 9. Israsena N, Hu M, Fu W, Kan L, Kessler JA (2004) The presence of FGF2 signaling determines whether beta-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Developmental Biology 268: 220–231. [DOI] [PubMed] [Google Scholar]
- 10. Schmöle AC, Brennführer A, Karapetyan G, Jaster R, Pews-Davtyan A, et al. (2010) Novel indolylmaleimide acts as GSK-3β inhibitor in human neural progenitor cells. Bioorganic & Medicinal Chemistry 18: 6785–6795. [DOI] [PubMed] [Google Scholar]
- 11. Hübner R, Schmöle AC, Liedmann A, Frech MJ, Rolfs A, et al. (2010) Differentiation of human neural progenitor cells regulated by Wnt-3a. Biochemical and Biophysical Research Communications 400: 358–362. [DOI] [PubMed] [Google Scholar]
- 12. Mazemondet O, Hubner R, Frahm J, Koczan D, Bader B, et al. (2011) Quantitative and kinetic profile of Wnt/β-catenin signaling components during human neural progenitor cell differentiation. Cellular & Molecular Biology Letters 16: 515–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, et al. (2002) Negative feedback loop of wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Molecular and Cellular Biology 22 4:1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, et al. (2011) Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nature Cell Biology 13: 1070–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wexler EM, Paucer A, Kornblum HI, Palmer TD, Geschwind DH (2009) Endogenous Wnt signaling maintains neural progenitor cell potency. Stem Cells 27: 1130–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chenn A, Walsh CA (2002) Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297: 365. [DOI] [PubMed] [Google Scholar]
- 17. Canning CA, Lee L, Irving C, Mason I, Jones CM (2007) Sustauned interactive Wnt and FGF signaling is required to maintain isthmic identity. Developmental Biology 305 1:276–286. [DOI] [PubMed] [Google Scholar]
- 18. Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. Journal of Physical Chemistry 81: 2340–2361. [Google Scholar]
- 19. McAdams HH, Arkin A (1999) It's a noisy business! Genetic regulation at the nanomolar scale. Trends in Genetics 15: 65–69. [DOI] [PubMed] [Google Scholar]
- 20. Kholodenko BN (2006) Cell-signalling dynamics in time and space. Nature Reviews Molecular Cell Biology 7: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilkinson DJ (2009) Stochastic modelling for quantitative description of heterogeneous biological systems. Nature Reviews Genetics 10: 122–133. [DOI] [PubMed] [Google Scholar]
- 22. Wolkenhauer O, Ullah M, Kolch W, Cho KH (2004) Modelling and simulation of intracellular dynamics: Choosing an appropriate framework. IEEE T Nanobiosci 3: 200–207. [DOI] [PubMed] [Google Scholar]
- 23. Lee E, Salic A, Krüger R, Heinrich R, Kirschner MW (2003) The roles of apc and axin derived from experimental and theoretical analysis of the wnt pathway. PLoS Biol 1: 116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cho KH, Baek S, Sung MH (2006) Wnt pathway mutations selected by optimal beta-catenin signaling for tumorigenesis. FEBS Letters 580: 3665–3670. [DOI] [PubMed] [Google Scholar]
- 25. Wawra C, Kühl M, Kestler HA (2007) Extended analyses of the wnt/beta-catenin pathway: Robustness and oscillatory behaviour. FEBS Letters 581: 4043–4048. [DOI] [PubMed] [Google Scholar]
- 26. Mirams GR, Byrne HM, King JR (2010) A multiple timescale analysis of a mathematical modelof the wnt/beta-catenin signalling pathway. Journal Mathematical Biology 60: 131–160. [DOI] [PubMed] [Google Scholar]
- 27. van Leeuwen IM, Byrne HM, Jensen OE, King JR (2007) Elucidating the interactions between the adhesive and transcriptional functions of β-catenin in normal and cancerous cells. Journal of Theoretical Biology 247: 77–102. [DOI] [PubMed] [Google Scholar]
- 28. Kofahl B, Wolf J (2010) Mathematical modelling of wnt/β-catenin signalling. Biochemical Society Transactions 38 5:1281–1285. [DOI] [PubMed] [Google Scholar]
- 29. Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. PNAS 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, et al. (2006) Developmental stagespecific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. PNAS 103: 19812–19817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, et al. (2007) Biphasic role for Wnt/β-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. PNAS 104: 9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Israsena N, Hu M, Fu W, Kan L, Kessler JA (2004) The presence of FGF2 signaling determines whether β-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Developmental Biology 268: 220–231. [DOI] [PubMed] [Google Scholar]
- 33. Krieghoff E, Behrens J, Mayr B (2006) Nucleo-cytoplasmic distribution of beta-catenin is regulated by retention. Journal of Cell Science 119: 1453–1463. [DOI] [PubMed] [Google Scholar]
- 34.Tymchyshyn O, Kwiatkowska M (2008) Combining intra- and inter-cellular dynamics to investigate intestinal homeostasis. In: Formal Methods in Systems Biology: First International Workshop, FMSB 2008, Cambridge, UK, June 4–5, 2008, Proceedings. Springer, pp. 63–76.
- 35. Fradkin LG, Dura JM, Noordermeer JN (2010) Ryks: new partners for Wnts in the developing and regenerating nervous system. Trends in Neurosciences 33: 84–92. [DOI] [PubMed] [Google Scholar]
- 36. Sethi JK, Vidal-Puig A (2010) Wnt signalling and the control of cellular metabolism. Biochemical Journal 427: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Städeli R, Hoffmans R, Basler K (2006) Transcription under the control of nuclear Arm/β-catenin. Current Biology 16: R378–R385. [DOI] [PubMed] [Google Scholar]
- 38. Schmitz Y, Wolkenhauer O, Rateitschak K (2011) Nucleo-cytoplasmic shuttling of apc can maximize β-catenin/TCF concentration. Journal of Theoretical Biology 279 1:132–142. [DOI] [PubMed] [Google Scholar]
- 39.Mazemondet O, John M, Maus C, Uhrmacher AM, Rolfs A (2009) Intergrating diverse reaction types into stochastic models - a signaling pathway case study in the imperative pi-calculus. In: Rossetti MD, Hill RR, Johansson B, Dunkin A, Ingalls RG, editors, Proceedings of the 2009 Winter Simulation Conference. pp. 932–943.
- 40. Tan CW, Gardiner BS, Hirokawa Y, Layton MJ, Smith DW, et al. (2012) Wnt signalling pathway parameters for mammalian cells. PLoS One 7: e31882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bundschuh R (2003) The role of dimerization in noise reduction of simple genetic networks. Journal of Theoretical Biology 220: 261–269. [DOI] [PubMed] [Google Scholar]
- 42. Morishita Y (2004) Noise-reduction through interaction in gene expression and biochemical reaction processes. Journal of Theoretical Biology 228: 315–325. [DOI] [PubMed] [Google Scholar]
- 43. Orrell D, Ramsey S, Marelli M, Smith J, Petersen T, et al. (2006) Feedback control of stochastic noise in the yeast galactose utilization pathway. Physica D: Nonlinear Phenomena 217: 64–76. [Google Scholar]
- 44. Alam S, Sen A, Behie LA, Kallos MS (2004) Cell cycle kinetics of expanding populations of neural stem and progenitor cells in vitro. Biotechnology and bioengineering 88: 332–347. [DOI] [PubMed] [Google Scholar]
- 45. Lorenowicz MJ, Korswagen HC (2009) Sailing with the wnt: Charting the wnt processing and secretion route. Experimental Cell Research 315: 2683–2689. [DOI] [PubMed] [Google Scholar]
- 46. Coudreuse D, Korswagen HC (2007) The making of Wnt: new insights into Wnt maturation, sorting and secretion. Development 134: 3–12. [DOI] [PubMed] [Google Scholar]
- 47. Semënov MV, Tamai H, Brott BK, Kühl M, Sokol S, et al. (2001) Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Current Biology 11: 951–961. [DOI] [PubMed] [Google Scholar]
- 48. Lu W, Liu CC, Thottassery JV, Bu G, Liang Y (2010) Mesd is a universal inhibitor of Wnt coreceptors LRP5 and LRP6 and block Wnt/beta-catenin signaling in cancer cells. Biochemistry 49: 4635–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McNeill H, Woodgett JR (2010) When pathways collide: collaboration and connivance among signalling proteins in development. Nature Reviews Molecular Cell Biology 11: 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cotter D, Honavar M, Lovestone S, Raymond L, Kerwin R, et al. (1999) Disturbance of Notch-1 and Wnt signalling proteins in neuroglial balloon cells and adbnormal large neurons in focal cortical dysplasia in human cortex. Acta Neuropathologica 98: 465–472. [DOI] [PubMed] [Google Scholar]
- 51. Kim D, Rath O, Kolch W, Cho KH (2007) A hidden oncogenic positive feedback loop caused by crosstalk between wnt and erk pathways. Oncogene 26: 4571–4579. [DOI] [PubMed] [Google Scholar]
- 52. Schilling M, Maiwalk T, Bohl S, Kollmann M, Kreutz C, et al. (2005) Computational processing and error reduction strategies for standardized quantitative data in biological networks. FEBS Journal 272: 6400–6411. [DOI] [PubMed] [Google Scholar]
- 53. Hoops S, Sahle S, Gauges R, Lee C, Pahle J, et al. (2006) Copasi-a complex pathway simulator. Bioinformatics 22 24:3067–3074. [DOI] [PubMed] [Google Scholar]
- 54. Hooke R, Jeeves TA (1961) “direct search” solution of numerical and statistical problems. Journal of the Association for Computing Machinery 8: 212–229. [Google Scholar]
- 55. Olivier BG, Snoep JJ (2004) Web-based kinetic modelling using jws online. Bioinformatics 20: 2143–2144. [DOI] [PubMed] [Google Scholar]
- 56. Gibson MA, Bruck J (2000) Efficient exact stochastic simulation of chemical systems with many species and many channels. Journal of Physical Chemistry 104: 1876–1889. [Google Scholar]
- 57.Himmelspach J, Uhrmacher A (2007) Plug'n simulate. In: Proc. of the Annual Simulation Symposium. IEEE Computer Society, pp. 137–143. URL http://dx.doi.org/10.1109/ANSS.2007.34.
- 58. Ciocchetta F (2009) Bio-pepa with events. Transactions on Computational Systems Biology XI: 45–68. [Google Scholar]
- 59. Schlicht R, Winkler G (2008) A delay stochastic process with applications in molecular biology. Journal of mathematical biology 57: 613–648. [DOI] [PubMed] [Google Scholar]
- 60. Novere NL, Hucka M, Mi H, Moodie S, Schreiber F, et al. (2009) The systems biology graphical notation. Nat Biotech 27: 735–741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We provide the details of the model in Sbml format. The parameter values correspond to the ones found in Table 3 , Set 2.
(XML)
We provide details regarding the parameter sensitivity analysis.
(PDF)