Abstract
RNA-seq data can be mined for sequence differences relative to the reference genome to identify both genomic SNPs and RNA editing events. We analyzed the long, polyA-selected, unstranded, deeply sequenced RNA-seq data from the ENCODE Project across 14 human cell lines for candidate RNA editing events. On average, 43% of the RNA sequencing variants that are not in dbSNP and are within gene boundaries are A-to-G(I) RNA editing candidates. The vast majority of A-to-G(I) edits are located in introns and 3′ UTRs, with only 123 located in protein-coding sequence. In contrast, the majority of non–A-to-G variants (60%–80%) map near exon boundaries and have the characteristics of splice-mapping artifacts. After filtering out all candidates with evidence of private genomic variation using genome resequencing or ChIP-seq data, we find that up to 85% of the high-confidence RNA variants are A-to-G(I) editing candidates. Genes with A-to-G(I) edits are enriched in Gene Ontology terms involving cell division, viral defense, and translation. The distribution and character of the remaining non–A-to-G variants closely resemble known SNPs. We find no reproducible A-to-G(I) edits that result in nonsynonymous substitutions in all three lymphoblastoid cell lines in our study, unlike RNA editing in the brain. Given that only a fraction of sites are reproducibly edited in multiple cell lines and that we find a stronger association of editing and specific genes suggests that the editing of the transcript is more important than the editing of any individual site.
RNA editing is a post-transcriptional process that modifies the primary RNA and microRNA transcripts. This process can result in nonsynonymous protein coding substitutions, alternative splicing, nuclear retention of mRNA, or alterations of microRNA seed regions (for a review, see Nishikura 2010). The most common form of RNA editing in mammals is A-to-I editing, in which adenosine is deaminated to produce inosine by members of the ADAR (adenosine deaminases acting on RNA) family of enzymes (Wagner et al. 1989; Kim et al. 1994; Kumar and Carmichael et al. 1997). C-to-U editing in mammals by the APOBEC family of enzymes is thought to be much less frequent and much more specific (Gerber and Keller 2001). In mammals, ADAR is found within several tissues, while ADARB1 is known to be active in the brain. Abnormal RNA editing has been reported in epilepsy, amyotrophic lateral sclerosis (ALS), brain ischemia, depression, and brain tumors (Maas et al. 2006; Nishikura 2006; Peng et al. 2006; Paz et al. 2007; Cenci et al. 2008).
ADARs recognize double-stranded RNA as their major substrate, but editing at some sites is very selective for specific A residues, while other sites are edited promiscuously and mainly in clusters (Nishikura et al. 1991; Polson and Bass 1994). Because inosine pairs preferentially with cytidine, I is read as G during protein synthesis or during reverse transcription for RNA-seq. Known functional consequences of this post-transcriptional modification include changes in amino acids in the protein product such as in the glutamate and serotonin receptors, creation or deletion of entire exons by changes in splicing, retention of mRNA in the nucleus, changes in RNA stability, heterochromatin formation, protection against viral RNA, and microRNA modification (Zheng et al. 1992; Burns et al. 1997; Seeburg et al. 1998; Athanasiadis et al. 2004; Luciano et al. 2004; Prasanth et al. 2005; Wang et al. 2005; Agranat et al. 2008). Although the best-studied RNA editing sites are in coding sequences that qualitatively change the protein product, the majority of known RNA edits in human occur within Alu sequences embedded within introns and UTRs (Kim et al. 2004). ADAR mouse knockouts are embryonic lethal at day E11.5 (Wang et al. 2004), where it plays an important role in suppressing interferon signaling to block premature apoptosis in hematopoiesis (Iizasa and Nishikura 2009). While microRNAs are known to be important RNA editing targets, this present study focuses on RNA editing in messenger RNA as measured from polyA-selected RNA.
We surveyed our human polyA+ ENCODE RNA-seq data from 14 cell types for RNA editing events using a rigorous computational pipeline designed to filter out sequencing and read mapping artifacts. We further filtered private genomic single nucleotide variants (SNVs) for 12 of the 14 cell types using either 1000 Genomes resequencing data (The 1000 Genomes Project Consortium 2010) or ENCODE ChIP-seq data sets. We identified between 500 and 3000 reproducible A-to-I RNA editing events per cell type in biological duplicate RNA-seq samples. We then focused on genes that are frequently edited across multiple cell types for further analysis, and found enrichment for genes involved in basic housekeeping processes such as cell division, viral defense, and translation.
Results
Development and refinement of an RNA-editing pipeline tuned based on data from the GM12878 lymphoblastoid cell line
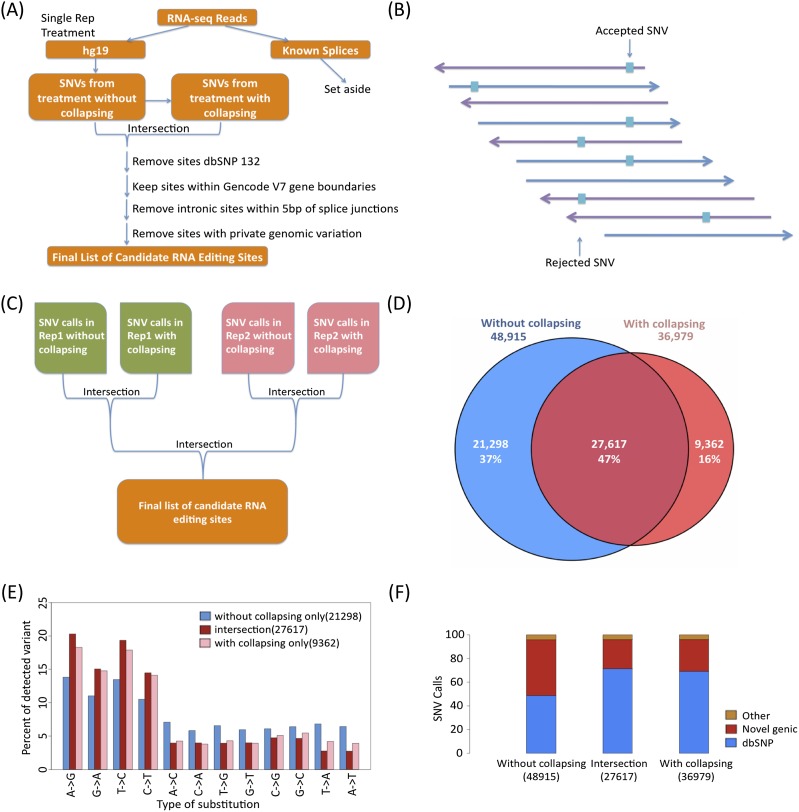
RNA-seq data has been mined for known SNPs in expressed genes to study allele-specific expression (Montgomery et al. 2010). While sequence variants in RNA-seq that are not in the genome are RNA editing candidates, we expect some level of mapping artifact and sequencing error in the data, and these could be mistaken for RNA editing. We reasoned that a pipeline that maximizes the fraction of called known SNPs would then produce the most conservative set of RNA editing candidates. One ENCODE cell line, GM12878, was particularly well-suited for tuning our pipeline, as it was deeply sequenced as part of the 1000 Genomes Project with the results incorporated into dbSNP (Sherry et al. 2001). GM12878 2 × 75 bp RNA-seq reads were mapped onto an expanded genome that includes known splice junctions (Mortazavi et al. 2008) using Bowtie as described in the Methods (Figs. 1A,B; Supplemental Table 1). Reads that mapped onto splice junctions were set aside because they are more prone to mismapping artifacts, and the remainder were used for SNV calling. An additional source of false-positive SNVs are reads with errors that are amplified during the library construction process. We can avoid PCR artifacts by collapsing reads, i.e., counting reads with identical starts only once. We further restricted ourselves to sites that had a minimum 10% frequency in both the entire data set (“uncollapsed set”) as well as in the smaller data set of nonredundant reads (“collapsed set”). We kept only candidate RNA editing sites called from two independent RNA-seq replicates (Fig. 1C). Of the SNVs present in the union of collapsed and uncollapsed sets, 47% were in the intersection, while 37% were only in the uncollapsed set and 16% were only in the collapsed set (Fig. 1D). We found that using the intersection strategy delivered a higher fraction of A-to-G calls (20%), while the sets that were only in the uncollapsed or only in the collapsed sets had A-to-G calls of 14% and 18%, respectively (Fig. 1E). We then scored our SNVs (Supplemental Table 2) for occurrence within dbSNP and found that the intersection had the highest percentage of known SNPs (71%) (Fig. 1F).
Figure 1.
RNA SNV calling strategy. (A) Flowchart of analysis: 75-bp paired-end RNA-seq reads were mapped onto an extended genome (genome + known splice junctions + spikes) using Bowtie. Reads mapping onto splice sites and spikes were set aside, and reads mapping onto hg19 were used to call single nucleotide variants (SNVs). A parallel set of analyses was done using a collapsed set of reads with unique coordinates, and the intersections of SNVs from the uncollapsed and collapsed treatments were obtained. Known SNPs annotated in dbSNP132, sites outside gene boundaries, and intronic sites within 5 bp of splice junctions were removed. For the GM trio, any candidate with evidence of a private genomic variation was also removed. (B) Example of candidate editing site. Purple arrows pointing to the left represent reads on the (−) strand, while blue arrows pointing to the right represent reads on the (+) strand. The blocks represent variants between the reference DNA and the RNA-seq. A SNV is kept when at least three nonidentical reads support the SNV, with a minimum SNV frequency of 10%, and at least one edit per strand. (C) Intersection strategy for two replicates. For cell types with two replicates, the SNVs remaining after collapsing were intersected between the replicates. (D) The number of SNVs remaining after collapsing for the prefiltered sites. Number of SNVs that are only in the uncollapsed set are in blue; the intersection, purple; and collapsed set, red. (E) Collapsing increases the relative amount of A-to-G SNVs and also increases the relative number of transitions. Number of SNVs that are only in the uncollapsed set are in blue; the intersection, purple; and collapsed set, red. (F) The fraction of dbSNP is highest in the intersection of the full and collapsed sets. The relative amount of calls found in dbSNP132, novel genic SNVs, and other SNVs in the uncollapsed set are at the left; the collapsed set, right; and the intersection of the two, middle.
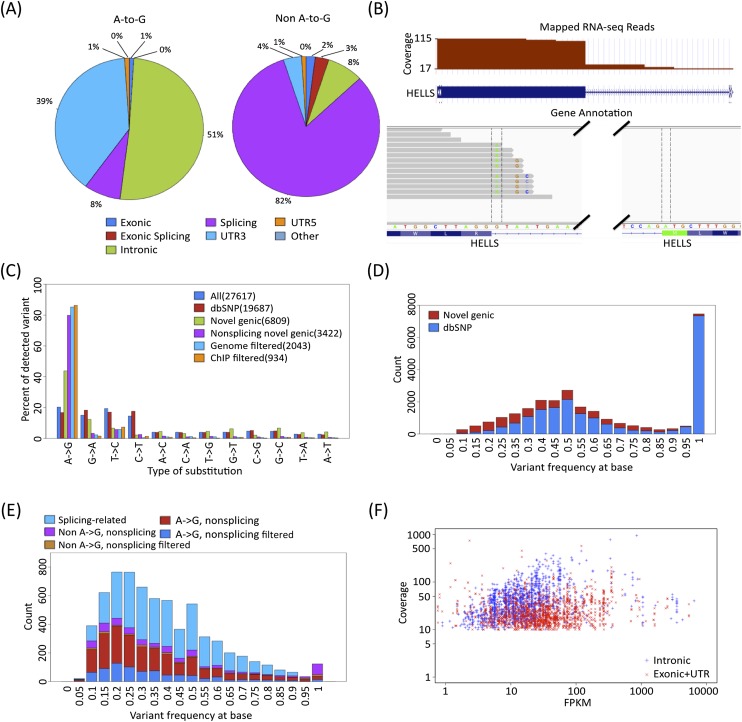
We further filtered the set of SNVs absent in dbSNP (candidates for editing because they are not known polymorphisms) for their overlap with known transcript boundaries from GENCODE v7 protein coding genes, which retained 86% of the candidates. SNV calls within known transcripts on the minus strand were reassigned to reflect the appropriate substitution in the sense of transcription. We then used ANNOVAR (Wang et al. 2010) to annotate the part of the transcript in which the variants occurred. We found that A-to-G variants were present primarily in introns and UTRs, whereas 82% of the non–A-to-G calls were annotated as “splicing,” which is defined here as intronic and within 5 bp of the splice junction (Fig. 2A). Inspection of these calls revealed that they were primarily due to mismapped reads that should have mapped across splice junctions (Fig. 2B). After removing all calls within 5 bp of splice sites, we found that >80% of our novel SNVs were A-to-G calls, which is 20-fold higher than the 4% of SNV calls passing all the same filters that are G-to-A calls (Fig. 2C). This enrichment is 20% higher in the intersection of the collapsed and uncollapsed sets than it is in either of the outersects (Supplemental Fig. 1). However, there remains a chance that there are private genomic SNVs that are not yet represented in dbSNP because of low-coverage or read mapping issues. We therefore used the genomic reads for GM12878 (and GM12891 and GM12892 below) from the 1000 Genomes Project to remove transcriptome SNVs with one or more genomic reads also supporting that candidate variant (Supplemental Table 1). This further increased our A-to-G SNV to >85% of the remaining calls. If we assume that our G-to-A calls are false positives with respect to editing (whether they are true SNPs not found in dbSNP or are sequencing/mapping artifacts), then our false-discovery rate (FDR) for A-to-G RNA editing would be <2%. No other SNV class accounted for >6% of our most stringently filtered set. T-to-C calls were the second most frequent class of SNV. Thity-three percent of these T-to-C were found in regions with overlapping GENCODE gene models on opposite strands, and another 33% had an unannotated transcript in the opposite sense as the overlapping gene model.
Figure 2.
RNA editing calls in GM12878. (A) Most non–A-to-G SNVs are near splicing boundaries. The distribution relative to gene boundaries of A-to-G SNVs (left) versus non–A-to-G SNVs (right). (B) Example of reads mapped incorrectly across a known splice junction. Overhanging RNA-seq reads are mapped incorrectly into the intron when the correct position is in the adjacent exon, even though the splice junction was provided to the read mapper. (C) Distribution of SNVs at different steps in the pipeline. Prefiltered SNVs defined by having at least three nonidentical reads support the SNV, with a minimum SNV frequency of 10%, at least one edit per strand, and no more than one type of SNV for the same position in blue. SNVs annotated in dbSNP132 are red, SNVs that are not in dbSNP132 and within gene boundaries are green, SNVs that are not in dbSNP132 and within gene boundaries without splicing sites are purple, SNVs that had no matching 1000 Genome sequencing reads are in light blue, and SNVs passing ChIP filtering are in orange. (D) Frequency distribution of SNVs primarily reflects expression of homozygous and heterozygous SNPs. The SNVs that were found in dbSNP132 are in blue; the novel genic SNVs, red. (E) Most nonsplice adjoining SNPs are A-to-G. The nonsplicing novel genic A-to-G calls in filtered calls are in blue; nonsplicing novel genic A-to-G calls, red; nonsplicing novel genic non–A-to-G, brown; nonsplicing novel genic non–A-to-G in filtered calls, purple; and splicing-only novel genic, light blue. (F) Distribution of gene expression versus coverage of exonic sites are in red and intronic sites are in blue for genic SNVs. SNVs in more lowly expressed genes are primarily on exons, due to our minimum depth of coverage requirements.
We then tested whether using ChIP-seq reads could be used to filter candidate SNVs in cases for which genomic reads are not available, as is the case for many ENCODE cell lines analyzed in the next section. Surprisingly, we found that we filtered more SNVs using ChIP-seq than using the 1000 Genomes data. In order to better understand filtering the SNV calls using ChIP-seq data versus 1000 Genomes data, we compared the coverage of SNVs in the two data-types. Although there were fewer total reads in the ENCODE ChIP-seq data than in the 1000 Genomes data, we found overall higher coverage in the ChIP data (mean coverage, 57.4) than in the 1000 Genomes data (mean coverage, 33.8). Our ChIP-seq mean coverage was even higher (60.6) over the 1457 SNVs that were filtered when using only the ChIP data, thus allowing us to detect rare variants (Supplemental Fig 2). However, there were other regions for which the 1000 Genomes data filter out SNVs that were missed using the ChIP data.
SNVs that matched dbSNP have an expected bimodal distribution with one mode at a frequency of 1.0 and the second mode at a frequency of 0.5, which are due to homozygous and heterozygous SNPs, respectively (Fig. 2D). In contrast, we find that the distribution of A-to-G RNA editing calls are skewed rightward with a mode at a frequency of 0.2. Nonsplicing, non–A-to-G SNVs show a distribution similar to dbSNP SNVs, with a mode at a frequency 1.0 (Fig. 2E). Relatively few candidate RNA editing events were in open reading frames (Supplemental Fig. 3). As expected, our power to call SNVs within exons exceeded that of calling them in introns for a given expression level (Fig. 2F), given the much greater depth of RNA-seq coverage in exons versus introns. We next asked whether genes with A-to-(G)I candidate edits had related functions, and found that they are enriched for Gene Ontology terms that are mainly broad anabolic functions such as translation, translational elongation, ribonucleoprotein complex, chromosome, centromeric region, ribosome, cytosolic ribosome, mitochondrial nucleoid, melanosome, and coated pit (Supplemental Table 3).
Survey of 14 ENCODE cell lines
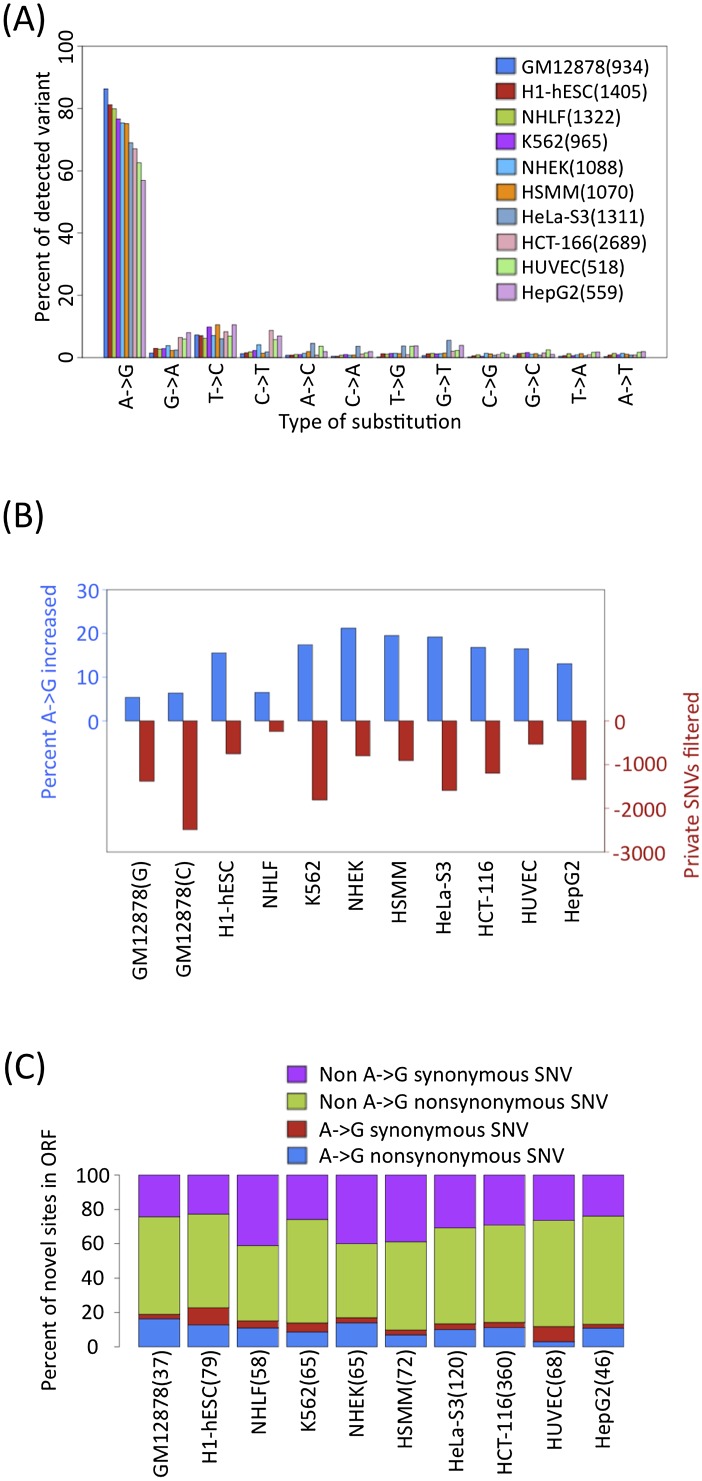
We then applied the above pipeline to polyA RNA-seq from 14 ENCODE cell types with 2 × 75 bp non-strand-specific protocol, all of which express only ADAR (Supplemental Fig. 4). Since sequenced genomes were not available for any cell line other than the three lymphoblastoid cell lines, we substituted ENCODE ChIP-seq data from HudsonAlpha and histone modifications data from the Broad Institute to filter out private genomic SNVs that were not represented in dbSNP (Supplemental Table 1). The fraction of editing candidates that were of the A-to-G class ranged from 50%–85% (Fig. 3A; Supplemental Fig. 5). HepG2 and HUVEC cells had the lowest number of candidate RNA editing sites with about 500 calls each. The filtering of private genomic SNVs using ChIP-seq data increased the percentage of A-to-G calls by 5%–20%, with the most SNVs filtered out for growth-transformed tumor cell types (Fig. 3B). The number of candidate sites called by our pipeline did not depend on the sequencing depth of the RNA-seq data set (Supplemental Fig. 6) over the range represented in our samples (28–135 million reads per replicate), but the amount of filtering did depend on the aggregate depth of coverage in the ChIP-seq data sets. The individual non–A-to-G classes ranged between 0%–10% of the total, with T-to-C again being the second most prevalent modification. Based on the results in GM12878, it is likely that the bulk of the T-to-C edits are A-to-G(I) edits from transcripts on the opposite strand relative to their original annotation. Although members of the APOBEC family are detectably expressed in all cell lines (Supplemental Fig. 7), we observed a very modest proportion of C-to-T(U) editing events in HepG2, HUVEC, and HCT116. While some of these C-to-T(U) sites might prove to be true RNA edits, only a handful of these C-to-T(U) sites are located in AU-rich regions known to be associated with APOBEC editing. We therefore provisionally conclude that most C-to-T(U) candidates are false positives.
Figure 3.
Survey of SNV calls across ENCODE cell lines. (A) Distribution of nonsplicing novel genic SNVs for all data sets. (B) In every cell type, the percentage of A-to-G SNVs increase and the number of candidate sites decrease (red) after filtering for private SNVs using ChIP-seq. GM12878 calls were filtered with 1000 Genomes or ChIP-seq reads are labeled with G or C, respectively. (C) Relatively few non–A-to-G synonymous SNVs (purple), non–A-to-G nonsynonymous SNVs (green), A-to-G synonymous SNVs (red), A-to-G nonsynonymous SNVs (blue) are found in ORFs.
The number of candidate SNVs within coding domains summed over all cell types are below 1000, and these are primarily nonsynonymous for the non–A-to-G SNVs (Fig. 3C). We find that 94% (5349 of 5695) of the candidate A-to-G(I) calls are within known repeat families, and 98% (5247 of 5349) of these are in Alu's. Alu families with the most members also had the most edits (Supplemental Table 4). This is certainly an underestimate, as our conservative mapping strategy would underreport hyper-edited regions with more than three simultaneous edits within our 75-bp reads.
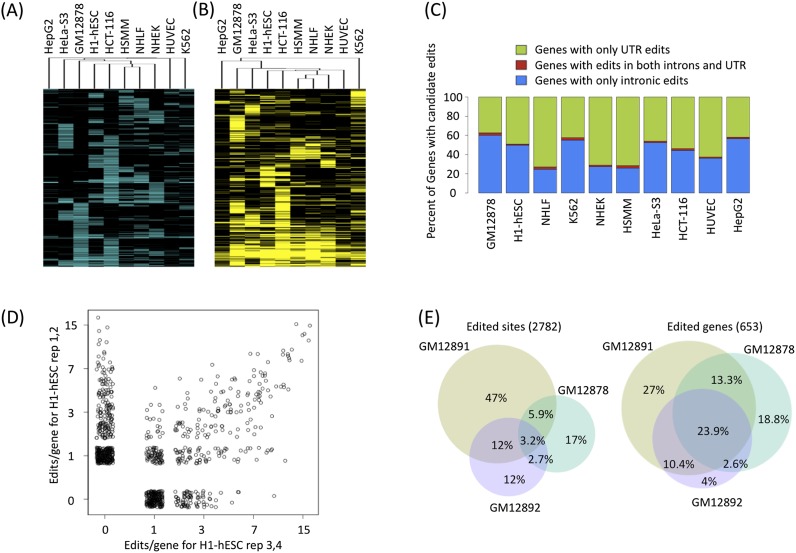
We next focused on individual sites with evidence of editing in multiple cell types. Overall 33.5% (1905 out of 5695 possible) of individual A-to-G(I) candidate editing sites occur independently in two or more different cell types (Fig. 4A; for non–A-to-G classes, see Supplemental Fig 8). We also found that 24% (1386/5695) of our candidate A-to-G(I) calls intersect with the DARNED database of RNA editing in human (Kiran and Baranov 2010), which was generated from mining human ESTs. This low overlap is expected, since the ENCODE RNA-seq data analyzed here did not include neuronal tissues or cell lines. We found 28 genes that were edited in all of our cell types, and 20 genes of these (71%) are also in the DARNED set. We found that 47.4% (662 out of 1396 possible) of genes edited are called as edited in at least two or more different cell types (Fig. 4B). We therefore conclude that gene-level association with editing is more robust than the identity of an individual site edited.
Figure 4.
Gene level analysis of RNA editing after private SNV filtering. (A) Hierarchical clustering of the editing frequency of the 33.5% (1905 out of 5695 possible) individual A-to-G candidate editing sites occurring in at least two distinct cell types. (B) Hierarchical clustering of the number of edits in the 47.4% (662 out of 1395 possible) of genes edited in at least two distinct cell types. (C) RNA editing in genes cluster in the UTR or in the introns with few genes having edits in both UTR and introns. Percentage of genes with only UTR edits are in green; intronic edits, blue; and edits in both introns and UTR, red. (D) Reproducibility of calling RNA edits for human H1 ES cells. Scatter plot of RNA edit calls for rep 1,2 versus rep 3,4 is on a log2-log2 scale with a pseudocount of 1. A Gaussian noise was added to points to visualize density. (E) Venn diagrams of A-to-G candidate edits in lymphoblastoid cells from a hapmap trio. The Venn diagram of the individual sites (left) and edited genes (right); 35.8% of the union of edited sites are found in two or more cell types, while 54.2% of the union of edited genes are found in two or more cell types.
We then focused on the distribution of calls within gene models. For most genes, A-to-G(I) RNA editing candidates were either all in intronic regions or all in UTRs, although 1%–4% of edited genes had both intronic and UTR sites, depending on the cell type (Fig. 4C). Because editing events are—overall—rare in the transcriptome and because editing often covers only a fraction of transcripts from a given gene, we wanted to probe our sensitivity for calling events in replicate samples. While most ENCODE data is in duplicates, we had an instance of data from four replicate determinations for Human H1 ES. We compared calls from Human H1 ES cell reps 1 and 2 with calls from reps 3 and 4 (Fig. 4D) and found that the number of edits per gene that we had called was quite noisy when there were only a few candidate sites that were called per gene. This is possibly due to the stochastic and promiscuous nature of ADAR's ability to hyper-edit dsRNA (Polson and Bass 1994) or due to the sensitivity of pipeline to coverage at the low end of the RNA expression spectrum.
We compared the filtered A-to-G(I) SNV calls in GM12878 to those in its parents, GM12891 and GM12892, to help assess the stability of RNA editing within a single cell-type, i.e., EBV-transformed lymphoblastoid cells. GM12891 and GM12892 had 1885 (86%) and 843 (87.7%) candidate A-to-G(I) sites located in 479 and 265 genes, respectively. GM12878 shared 490 sites (337 genes) with GM12891 and 292 sites (218 genes) with GM12892. While 26.2% of the individual editing SNVs were found in at least two individuals of the trio, >49.6% of the genes were in common (Fig. 4E). Thus the gene-level association with editing is also more reproducible than the identity of individual sites edited within different cell lines of the same type.
Overall, there were 248 out of 1396 genes that were edited in at least five out of the 10 distinct cell types after we applied the ChIP-seq filter (Fig. 4A). Our GO analysis of these genes showed enrichments that included interspecies interaction between organisms, cell division, DNA metabolic process, positive regulation of defense response to virus by host, protein folding, ER-Golgi intermediate compartment, ribosome, and ribonucleoprotein complex (Supplemental Table 5). Two genes that are especially highly edited within multiple cell types are the inhibitor of apoptosis XIAP and the caspase-3 target DFFA, suggesting an explicit and direct link to the apoptosis phenotype of the mouse ADAR knockout.
The overwhelming prevalence of A-to-G(I) RNA editing candidates in our lymphoblastoid trio agrees with a recent publication (Peng et al. 2012) but differs qualitatively from a previously reported analysis of RNA editing in a nonoverlapping set of HapMap lymphoblastoid lines (Li et al. 2011). To make a more informed comparison, we applied our pipeline to the data sets from Li et al. (2011) though we note that their data were single replicate RNA-seq measurements with shorter 50-bp reads that were more shallowly sequenced (40 million reads vs. our average of 100 million reads). We found that some of their individual samples produced similar genic, nonsplicing A-to-G enrichments as in our lymphoblastoid lines, while others had an especially high percentage of A-to-C and T-to-G classes (Supplemental Figs. 9, 10). To check whether these could be DNA sequencing artifacts from earlier and different (2008–2009) Illumina chemistry, we checked the output of our pipeline on an independent data set (ENCODE GM12878 DNase-seq) from that same earlier timeframe. We found relatively low numbers of non-dbSNP SNVs in the DNase-seq data and found that A-to-C and T-to-G classes were again comparatively enriched (Supplemental Fig. 11), suggesting that a similar systematic bias exists in some of the Li et al. (2011) samples as in genomic DNA sequenced with that technology in that timeframe; these are not therefore attributable to RNA editing. Upon further inspection, we found that the particular sites most affected are embedded in G-rich regions (Supplemental Fig. 12). In addition, recent reports (Schrider et al. 2011; Pickrell et al. 2012) attribute the majority of the non–A-to-G calls in Li et al. (2011) to sequences that are paralogs of the gene that were reported as edited, i.e., a mismapping error. Therefore we only report our ChIP-filtered A-to-G(I) RNA editing candidates for each cell line in Supplemental Tables 6 through 19.
Discussion
We developed an intentionally conservative strategy to identify candidate RNA edits from RNA-seq data for the human ENCODE cell lines. We analyzed the transcriptomes of 14 cell types and compared the RNA sequences with the reference genome, filtered out known SNPs from dbSNP, and filtered out private SNPs detected in ChIP-seq data from each cell line. We found that SNVs in the intersection of our “collapsed” and “uncollapsed” mapping sets yielded the highest fraction of known SNPs; this observation functions as a positive control that our SNV calls are not significantly biased by mapping artifacts. We also found that incorrect read mapping across splice junctions was the source of the majority of non–A-to-G calls. We further developed a strategy to filter out nonediting transcriptome SNVs by using up to 2.0 billion ChIP-seq reads in GM12878 and 0.1–1.7 billion ChIP-seq reads in the other nine cell lines that do not have resequencing data available. Together with filtered calls in two additional lymphoblastoid with 1000 Genomes data, we therefore have reliable A-to-G(I) editing candidates in 12 of our 14 cell lines (Supplemental Tables 6–19).
Up to 87% of SNVs that are not SNPs (either in dbSNP or private genomic SNVs) are A-to-G calls; this suggests they are likely to be A-to-I editing candidates. Furthermore, >97% of these candidates are located in introns and 3′ UTRs, which is consistent with what was previously known about RNA editing based on earlier EST surveys and recent reports (Bahn et al. 2012; Peng et al. 2012). Our candidate A-to-G(I) RNA editing sites have a different variant frequency from known SNPs. They tend to cluster predominantly in the 3′ UTR or in introns. In the three cell lines with the least amount of A-to-G(I) editing, there were relatively more C-to-T(U) SNVs, but these were not associated with AU-rich regions, as would have been expected if these are due to APOBEC activity. We also found that individual RNA editing calls are noisy for lowly expressed genes because of depth of coverage requirements for editing calls. If sensitivity to a conservative threshold was the major source of noise from one data set to another, and one replicate to another, then the identity of genes that are edited would be more consistent than the actual edits as long as edits occur in clusters.
Overall, we report 5695 unique candidate A-to-G(I) RNA editing events in 1396 genes, including a subset of 248 genes that were consistently edited across more than five cell types. Ninety-nine percent of the candidate RNA editing calls occurred within known repeats (with 98% of those in Alu elements), and only 24% were annotated in DARNED, which is expected since none of the ENCODE cell lines in this study cover neuronal phenotypes. Comparing our results to previously reported widespread evidence of noncanonical RNA editing besides A-to-G(I) in a different set of HapMap lymphoblastoid cell lines (Li et al. 2011), we found that sequencing and mapping artifacts could account for the vast majority of the unconventional (non-ADAR) variant calls. Moreover, we showed that this is caused, in part, by RNA data generated using older sequencing chemistry.
Genes with at least one edit within all cell types show GO term enrichments in “housekeeping” annotations related to cell division, DNA metabolic process, protein folding, and ribosome, as well as terms relating to viruses and defenses against them. The latter functions are consistent with the view that editing arose first in this context. For example, the protein EIF2AK2 inhibits protein synthesis upon activation by viral RNA and has an average of 13 edits across all cell-types. This meshes well with reports that RNA editing participates in host-viral interactions, such as the editing of the hepatitis C virus (HCV) genome by ADAR (Taylor et al. 2005). Interestingly, some viruses have been able to take advantage of ADAR for their own purposes; endogenous ADAR has been shown to stimulate HIV-1 replication (Doria et al. 2009).
When searching for RNA editing events to create a global map of high-quality candidates, there is a difficult tradeoff between sensitivity (identifying a highly inclusive set of possible edits) and specificity (being more confident that a call is in fact a true RNA edit). We judged it better to have a smaller number of candidate RNA editing events that are highly likely to be true than to have a larger number with an increased percentage of false positives. We undoubtedly lost a substantial number of true, low-level A-to-I RNA editing events in the process, and users of these RNA-Seq data might for some purposes want to build a more inclusive and less certain list. Another caveat that applies to our pipeline is that our method allows for a maximum of three edits per 75 bp within our reads. Thus, if there was a 75-bp window that had significantly more than three edits, our pipeline would only detect the edits on the periphery of this hyper-edited domain. We could allow more mismatches during the alignment step, but then this would exacerbate the problem of incorrectly mapped reads. To date, the RNA-seq field has emphasized aggressive read mapping to maximize sensitivity and new candidate discovery. This inevitably comes at the expense of specificity, and this effect is greater in complex genomes with extensive paralogous gene and repeat families such as those of mammals. Our current view is that there is no single correct threshold for the sensitivity specificity tradeoff: It has to be selected to match the objective of a given study and the future use of each analysis. When an analysis focuses on the small portion of sequence reads that imply differences from the majority that match well to the appropriate genome/transcriptome models, a general trend toward greater conservatism seems justified. We show here that an early sequencing chemistry issue and a problem with accurate read mapping over splice junctions are two specific contributors to false-positive RNA editing candidates. We have greatly reduced these in our pipeline and present a conservative set for the ENCODE cell lines. In doing so, we demonstrated that there is no persuasive evidence in favor of noncanonical editing. A-to-G(I) edits strongly dominated our data except for three cell lines with modest evidence of C-to-T(U) SNVs, and no unknown edit was above the noise level.
It is a sobering caution that, even when reads were mapped simultaneously against the genome and known splice junctions, we still had obvious splice-mapping artifacts. We were therefore forced to exclude all calls that mapped within a few bases of splice junctions; this, in turn, dramatically reduced non–A-to-G(I) calls. There remain a few noncanonical calls that pass our criteria, but we expect these to be primarily undetected private SNVs, regions with complicated mapping issues due to paralogous genes and pseudogenes in the genome, or uncharacterized splice junctions. This does not preclude the possibility of some very specific APOBEC-like, noncanonical editing of bases but calls into question their previously reported widespread occurrence (Li et al. 2011), which has been recently called into question (Kleinman and Majewski 2012; Lin et al. 2012; Pickerell et al. 2012).
While GM12878 had been “deeply” resequenced by 2010 standards, we find that filtering with ChIP-seq was actually more effective. While this may seem paradoxical at first, ChIP-seq signal as well as background reads are most likely to come from open chromatin within transcribed genes. Although the 1000 Genomes project has greater coverage throughout the genome, we had in fact greater overall coverage of the candidate RNA SNVs in the pooled ChIP-seq data. While the amount of ChIP-seq data in GM12878 and other ENCODE Tier 1 and Tier 2 cell lines is exceptional, they highlight the issue that really high coverage is necessary to detect a subset of SNVs. The fact that private genomic SNVs need to be accounted for and filtered out for assaying RNA editing suggests that we can use RNA-seq to identify SNVs in expressed genes. In effect, the depth of coverage in RNA-seq over medium to highly expressed genes achieves many of the same benefits of whole-exome sequencing in rare variant discovery. In the future, deconvolving rare genomic variants that are detectable by RNA-seq from true RNA editing events will be done more optimally by simultaneous analysis of the raw reads from RNA-seq and genome-resequencing events at even higher coverage than readily available today.
Methods
Reads for each biological replicate data set were mapped to an expanded genome consisting of the human reference (GRCv37 / UCSC hg19) plus GENCODE v7 splice junctions and added spike sequences using Bowtie, version 0.12.7 (Langmead et al. 2009), with at most three mismatches; reporting in SAM format up to one valid alignment per read; suppressing all alignments for a particular read if more than one reportable alignment exist for it; and using only those alignments that fell into the best stratum. We used Bowtie to map reads instead of TopHat because the SNVs that were called using TopHat did not have a significant difference in SNV distribution when using RNA-seq reads or DNase-seq reads. Read ends were mapped separately and pooled afterward, without taking into account pairing information. The resulting SAM files were then stripped of spliced reads; converted to BAM files using samtools, version 0.1.17; sorted; and indexed; variants were called using the pileup command. We called an SNV when at least three nonidentical reads support a nonreference variant, and the variant is present at a minimum frequency of 10% and is supported by at least one read per strand. We discarded sites with more than one type of SNV call at the same location. In addition to the SNV calls in each full data set, a parallel set of analyses was done with potentially duplicated reads removed using the rmdup option of samtools to create the collapsed set.
The intersection of SNV calls in biological replicates from the full BAM file and the intersection of SNV calls in the collapsed BAM files were intersected to create a list of candidate SNVs. Known SNPs from dbSNP132 that were not annotated as based on cDNA and sites lying outside the “comprehensive set” of GENCODE v7 protein coding gene boundaries were set aside, and the remaining novel SNVs within genic regions were corrected for strand sense. These novel genic candidates were then annotated using ANNOVAR (Wang et al. 2010) with a splicing threshold of 5. Gene Ontology analysis was performed using GREAT, version 1.8.2 (McLean et al. 2010). Sites that were annotated by ANNOVAR as splicing were filtered out. To filter out genomic SNVs, genomic alignments were obtained from the 1000 Genomes project for the CEU GM trio, and ChIP-seq alignments were obtained from ENCODE ChIP-seq data from HudsonAlpha and histone modification data from the Broad Institute. Samtools mpileup was used to look at the nucleotide composition over the SNVs, and sites with any evidence of a genomic SNV were filtered out. Hierarchical clustering for editing frequency of individual sites and number of edits for genes was done using Cluster 3.0 (de Hoon et al. 2004), with the centroid linkage hierarchical clustering option of both sites/genes and cell types. The heatmap was viewed using TreeView-1.1.5 (Page 2002).
Data access
All RNA-seq data are publicly available from the ENCODE repository at the UCSC Genome Browser (http://genome.ucsc.edu/ENCODE/). Details of accession numbers can be found in Supplemental Table 20.
Acknowledgments
We thank Wendy Lee and Alicia Rogers for assistance. The work of A.M. and E.P. on this manuscript was supported by the UC Irvine Center for Complex Biological Systems and U.S. National Institutes of Health (NIH) P50 GM076516, and the work of B.W. and B.J.W. was supported by the Beckman Foundation, the Donald Bren Endowment, and NIH grant U54 HG004576.
Footnotes
[Supplemental material is available for this article.]
Article and supplemental material are at http://www.genome.org/cgi/doi/10.1101/gr.134957.111.
Freely available online through the Genome Research Open Access option.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranat L, Raitskin O, Sperling J, Sperling R 2008. The editing enzyme ADAR and the mRNA surveillance protein hUpf1 interact in the cell nucleus. Proc Natl Acad Sci 105: 5028–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391 doi: 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn JH, Lee JH, Li G, Greer C, Peng G, Xiao X 2012. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res 22: 142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308 [DOI] [PubMed] [Google Scholar]
- Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O'Connell MA, Gallo A 2008. Down-regulation of RNA editing in pediatric astrocytomas: ADARB1 editing activity inhibits cell migration and proliferation. J Biol Chem 283: 7251–7260 [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S 2004. Open Source Clustering Software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Doria M, Neri F, Gallo A, Farace MG, Michienzi A 2009. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR stimulates viral infection. Nucleic Acids Res 37: 5848–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Keller W 2001. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci 26: 376–384 [DOI] [PubMed] [Google Scholar]
- Iizasa H, Nishikura K 2009. A new function for the RNA-editing enzyme ADAR. Nat Immunol 10: 16–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci 91: 11457–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A 2004. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res 14: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran A, Baranov PV 2010. DARNED: A DAtabase of RNa EDiting in humans. Bioinformatics 26: 772–776 [DOI] [PubMed] [Google Scholar]
- Kleinman CL, Majewski J 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome.” Science 335: 1302 doi: 10.1126/science.1209658 [DOI] [PubMed] [Google Scholar]
- Kumar M, Carmichael GC 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc Natl Acad Sci 94: 3542–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG 2011. Widespread RNA and DNA sequence differences in the human transcriptome. Science 333: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Piskol R, Tan MH, Li JB 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome.” Science 335: 1302 doi: 10.1126/science.1210624 [DOI] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S 2004. RNA editing of a miRNA precursor. RNA 10: 1174–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Kawahara Y, Tamburro KM, Nishikura K 2006. A-to-I RNA editing and human disease. RNA Biol 3: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Larch RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nishikura K 2006. Editor meets silencer: Crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol 7: 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 79: 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K, Yoo C, Kim U, Murray JM, Estes PA, Cash FE, Liebhaber SA 1991. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J 10: 3523–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD 2002. Visualizing phylogenetic trees using TreeView. Curr Protoc Bioinformatics 6.2.1–6.2.15. [DOI] [PubMed] [Google Scholar]
- Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. 2007. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 17: 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng PL, Zhong X, Tu W, Soundarapandian MM, Molner P, Zhu D, Lau L, Liu S, Liu F, Lu Y 2006. ADARB1-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron 49: 719–733 [DOI] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. 2012. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. doi: 10.1038/nbt.2122 [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Gilad Y, Pritchard JK 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome.” Science 335: 1302 doi: 10.1126/science.1210484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson AG, Bass BL 1994. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J 13: 5701–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL 2005. Regulating gene expression through RNA nuclear retention. Cell 123: 249–263 [DOI] [PubMed] [Google Scholar]
- Schrider DR, Gout JF, Hahn MW 2011. Very few RNA and DNA sequence differences in the human transcriptome. PLoS ONE 6: e25842 doi: 10.1371/journal.pone.0025842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg PH, Higuchi M, Sprengel R 1998. RNA editing of brain glutamate receptor channels: Mechanism and physiology. Brain Res Brain Res Rev 26: 217–229 [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K 2001. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM 2005. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR. J Virol 79: 6291–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RW, Smith JE, Cooperman BS, Nishikura K 1989. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci 86: 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura D, Weiss M, Nishikura K 2004. Stress-induced apoptosis associated with null mutation of ADAR RNA editing deaminase gene. J Biol Chem 279: 4952–4961 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Z, Blackwell K, Carmichael GG 2005. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol 15: 384–391 [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H 2010. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164 doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Fu TB, Lazinski D, Taylor J 1992. Editing on the genomic RNA of human hepatitis delta virus. J Virol 66: 4693–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]