Abstract
The interaction of genes and environmental exposures influences the development of asthma and determines asthma severity. This review focuses on recent developments in genetic studies of asthma onset and progression. Genome-wide association studies (GWAS) are currently the most effective approach to study genetics of complex diseases. There have been two large meta-analyses of asthma susceptibility, GABRIEL and EVE, which identified the same four chromosomal regions, many of which had also been identified in previous GWAS: loci in the ORMDL3 region of 17q21, IL1RL/IL18R genes on chromosome 2q, the TSLP gene region on 5q22, and IL33 on chromosome 9p24. These regions were associated with asthma in individuals of different ethnic backgrounds. EVE also identified a novel asthma susceptibility locus, PYHIN1, in individuals of African descent. Genome-wide screens for asthma susceptibility in Asian adults and children both identified genetic variants in the major histocompatiblity complex gene region (HLA region) on chromosome 6p21 as highly associated with asthma risk. This locus was one of the first candidate genes identified for asthma and has been a significant predictor of asthma risk in several GWAS.
There is also a need to understand asthma disease heterogeneity as different phenotypes may reflect several pathogenic pathways. Genes that are associated with phenotypes including lung function, biomarker levels and asthma therapeutic responses provide insight into mechanisms of asthma severity progression. For example, the HHIP gene is a significant predictor of pulmonary function changes in asthma and in the normal population. A joint model of risk variants in lung function genes were highly associated with lower FEV1 and increased asthma severity criteria. In addition, a genome-wide screen to discover pharmacogenetic associations related to response to inhaled glucocorticoids identified two correlated SNPs in the GLCCI1 gene that confer a significant lung function response to this asthma therapy.
Future genetic studies for asthma susceptibility and severity will incorporate exome or whole-genome sequencing to identify common and rare genetic variants. Using these variants identified in comprehensively phenotyped asthmatics will lead to the development of personalized therapy in individuals with asthma.
Keywords: Asthma, genetics, susceptibility, severity, personalized medicine, therapy, lung function
Introduction
Asthma is a heterogenous disease with a complex etiology. The interaction of genes and environmental exposures influences the development of asthma and determines the expression or progression of the disease (Fig. 1) [1]. An overall aim of genetic studies in a complex disease such as asthma is to identify a group of genetic variants that will predict risk for development (susceptibility) or progression (severity) of asthma. Genetic factors related to asthma susceptibility and severity are not limited to a single gene but are due to a number of gene variants that each contribute to the risk architecture.
Fig. 1.
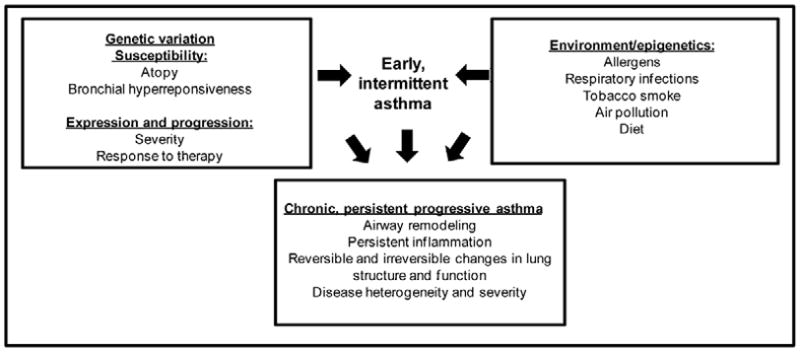
Gene-environment interactions in asthma susceptibility and severity (adapted from Slager et al., 2011).
There has been major progress in determining the genetic factors that are associated with asthma susceptibility, using genome wide association study (GWAS) approaches [2–11]. GWAS genes such as IL13 (interleukin 13), IL33 (interleukin 33), its receptor IL1RL1 (interleukin 1 receptor-like 1 isoform 1) and TSLP (thymic stromal lymphopoietin) have been linked to asthma and several other allergic phenotypes, suggesting dysregulation of shared inflammatory pathways. In addition, the major histocompatiblity complex region, which was one of the first asthma susceptibility loci identified [12], appears to contribute to asthma and allergen sensitization [2]. Regulatory T cell signaling may also play a role in asthma, as the SMAD3 (SMAD family member 3) gene, which encodes a transcriptional modulator related to transforming growth factor β, has also been identified in several genome-wide screens for asthma. A number of these susceptibility genes and their biologic pathways have been replicated in some but often not all asthma populations suggesting that there may be heterogeneity in the genetic risk in populations of different ethnic backgrounds [7, 13]. Results from major GWAS studies of asthma are reviewed in this article and are summarized in Table 1.
Table 1.
Significant asthma susceptibility GWAS variants from meta-analyses or replicated studies.
| Gene(s) | Chromosomal region | Ethnic background(s) | References |
|---|---|---|---|
| ORMDL3/GSDML | 17q21 | All | Moffat 2007, 2010, Torgersen 2011 |
| IL1RL1/IL18R1 | 2q11 | All | Moffatt 2010, Torgerson 2011 |
| TSLP | 5q22 | All | Moffatt 2010, Torgerson 2011, Hirota 2011 |
| IL33 | 9p24 | All | Moffatt 2010, Torgerson 2011 |
| SMAD3 | 15q23 | European | Moffatt 2010 |
| RORA | 15q22 | European | Moffatt 2010 |
| HLA-DQ/DR | 6p21 | All | Moffatt 2010, Li 2010, Hirota 2011, Noguchi 2011 |
| IL13 | 5q31 | European | Moffatt 2010, Li 2010 |
| PYHIN1 | 1q23 | African | Torgerson 2011 |
Many of the large genome-wide screens that have identified genes important to asthma susceptibility were based on a clinical diagnosis of asthma rather than more comprehensive phenotypes that would also evaluate the mechanisms of disease progression and severity. Current studies are now using more extensive characterization to investigate the progression of asthma. Thus, asthma severity may be related to specific subphenotypes, some of which are discussed in this review and summarized in Table 2: 1) genes related to pulmonary function; 2) biomarkers related to asthma progression and risk of exacerbations; 3) pharmacogenetic interactions in which an individual may have reduced responsiveness or be resistant to a specific asthma therapy; or 4) specific gene by environment interactions.
Table 2.
Selected GWAS associations for asthma severity and related traits.
| Trait | Gene(s) | References |
|---|---|---|
| Lung function measures (FEV1/FVC or FEV1) | ||
| General population | HHIP, GPR126, ADAM19, AGER/PPT2, FAM13A, PTCH1, PID1, HTR4, GSTCD, TNS1, NOTCH4/AGER/PPT2, THSD4I, NTS12/GSTCD/NPNT, MFAP2, TGFB2, HDAC4, RARB, MECOM, SPATA9, ARMC2, NCR3, ZKSCAN3, CDC123, C10orf11, LRP1, CCDC38, MMP15, CFDP1 and KCNE2 | Wilk 2009, Repapi 2009, Hancock 2009, Artigas 2011 |
| Asthma populations | HHIP, IL6R | Li 2011, Hawkins 2012 |
| Total serum/plasma IgE levels | ||
| General population | STAT6, FCER1A, IL13/RAD50 | Weidinger 2008, Granada 2011 |
| Asthma populations | HLA-DR, STAT6, FCER1A, IL13/RAD50, C11orf30-LRRC32 | Moffatt 2010, Li 2011 |
| Eosinophil levels | ||
| General population | IL1RL1, IKZF2, IL5, SH2B3 | Gudbjartsson 2009 |
| Atopic conditions | TSLP/WDR36 (Eosinophilic esophagitis) WDR36, IL33, MYB (Eosinophil levels and atopic asthma) | Rothberg 2010, Gudbjartsson 2009 |
| Response to asthma therapy | ||
| Inhaled corticosteroids | GLCCI1 | Tantisira 2011 |
Genome wide association studies of asthma susceptibility
Prior to 2007, candidate genes in known asthma and allergy biological pathways were tested for association with asthma susceptibility [7] and family studies and positional cloning techniques identified several putative chromosomal loci related to asthma [6, 14–17]. For example, the T Helper 2 (TH2) immunological pathway is one of the most replicated pathways in human and animal asthma studies [7, 18–22]. However, many of the other positive genetic associations from candidate pathways and family studies have not been consistently replicated, possibly due to relatively small sample sizes, population stratification, inconsistent phenotype definitions, and lack of adequate gene coverage [7]. Following the transition to GWAS, risk variants discovered in these genome-wide screens were more likely to be replicated, at least in populations of similar ethnic background. This review will highlight some of the newer studies and meta-analyses of asthma susceptibility and severity including GWAS in multiple ethnic groups.
GWAS of asthma susceptibility in multiple ethnic groups
Based on a doctor’s diagnosis of asthma, the first asthma susceptibility GWAS was published by Moffatt et al., representing the European GABRIEL consortium in 2007. This report identified genetic markers in the ORMDL3 (ORM1-like 3) on chromosome 17q12–21 as predictors of childhood asthma susceptibility [8]. A large follow-up study in the same European cohort verified this finding, as single nucleotide polymorphisms (SNPs) spanning approximately 380kb in the ORMDL3 genomic region were associated with asthma [3]. Several subsequent GWAS have also confirmed this result, and this is one of the most highly replicated susceptibility loci in genome-wide screens for asthma [9, 11, 23, 24]. However, due to the high degree of linkage disequilibrium (LD) in this region that spans several genes, it is still not clear whether ORMDL3 or a nearby gene is the risk gene with the causative variant(s) [25]. Interestingly, a splice-site mutation which is in strong LD with ORMDL3 but located the adjacent GSDMB (gasdermin-like B) gene was recently identified by the 1000 Genomes Project, indicating this may be a causal risk variant for asthma [26]. This example highlights the importance of new genetic sequencing projects and how these technologies continue to facilitate understanding the genetics of asthma.
While many of the initial asthma GWAS studies were carried out in non-Hispanic white discovery cohorts, there is a need to investigate the genetic pathways that play a role in asthma in individuals of different ethnic backgrounds, especially since minority groups such as African-Americans are more likely to experience higher asthma morbidity and mortality rates than whites [27, 28]. Populations of different ancestry also have different patterns of gene variation and LD that can alter the gene-specific risk variants. Whereas the GABRIEL consortium consisted of European individuals with asthma and unaffected controls, the EVE meta-analysis was established in North America to identify genetic factors that contribute to asthma susceptibility in multiple ethnic groups. Five major susceptibility loci were identified in 5,416 asthma cases and replication was performed in an additional 12,649 individuals of European-American, African-American or African-Caribbean, and Latino ancestry. Four of the chromosomal regions that reached genome-wide significance in EVE had been previously identified in GABRIEL or other studies: loci in the ORMDL3 region of 17q21, the IL1RL/IL18R (interleukin 18 receptor) loci on chromosome 2q, the TSLP gene region on 5q22 and interleukin 33 (IL33) on chromosome 9p24 [11]. Interestingly, these loci are associated with asthma development in all three ethnic groups, while a novel susceptibility locus in individuals of African descent was identified near at the PYHIN1 (pyrin and HIN domain family, member 1) gene on chromosome 1q23 [11]. Prior to the EVE analysis, one of the only asthma GWAS performed in populations of African ancestry was carried out in African-American asthma cases and controls from the United States and African-Caribbean individuals from Barbados. Adjusting for racial admixture, this study identified polymorphisms in ADRA1B (α-1B-adrenergic receptor) on chromosome 5q33, PRNP (prion-related protein) on chromosome 20pter-p12, and DPP10 (dipeptidyl peptidase 10) on chromosome 2q12.3-q14.2 as predictors of asthma [29]. While DPP10 had been identified in earlier positional cloning studies of asthma [14], many of these associations appear to be limited to individuals of African ancestry.
GWAS of asthma in Asian populations
Genome-wide screens for asthma susceptibility in Asian adults and children were published in 2011 and both identified genetic variants in the major histocompatiblity complex gene region (HLA region) on chromosome 6p21 as highly associated with asthma risk. In the Japanese population, 7,171 adult asthma cases and 27,912 unaffected individuals were genotyped in the discovery and replication cohorts, identifying five loci associated with susceptibility to adult asthma, two of which had previously been reported: the HLA region and TSLP/WDR36 (WD repeat domain 36) locus. Three additional genomic regions were also significant at the genome-wide significance level: the USP38 (ubiquitin specific peptidase 38)/GAB1 (GRB2-associated binding protein 1) locus on chromosome 4q31, loci on chromosome 10p14, and a region of chromosome 12q13 [30]. In a separate GWAS of childhood asthma in 938 Japanese pediatric asthma patients and 2,376 controls, SNPs were tested for association with asthma susceptibility and highly significant associations were tested for replication in independent Japanese samples and in Korean samples. This analysis determined that genetic variants in the HLA-DP locus were associated with the risk of pediatric asthma across Asian populations.
Furthermore, a replication study in 710 asthma cases and 656 unaffected controls in the Chinese Han population tested specific variants in the ORMDL3/GSDMB region of chromosome 17q21, providing further evidence that this locus was associated with adult onset asthma risk in multiple ethnic groups [23].
The specific role and functional biology of novel GWAS loci such as ORMDL3 in the pathogenesis of asthma is still unknown. However, there is strong evidence from asthma GWAS of the biologic importance of pathways that communicate epithelial damage to the adaptive immune system, ultimately leading to airway inflammation. Moreover, cytokines derived from epithelial cells such as TSLP and IL33 may promote the TH2 response through activation of receptors such as IL1RL1, on cell types such as mast cells, TH2 cells, and regulatory T cells.
Genetic studies of asthma severity and related phenotypes
Since initial GWAS focused primarily on childhood-onset asthma and asthma susceptibility, it is not well understood whether the same genes that contribute to asthma predisposition also play a role in the progression and severity of disease. In addition, population-based studies often rely on limited phenotypes such as a physician’s diagnosis to accommodate large sample sizes. However, comprehensive studies of asthma severity require more intense, time-consuming, and costly phenotyping which can result in smaller cohort sizes. Asthma severity studies also require different comparison groups than susceptibility studies. Instead of unaffected individuals, subjects with severe asthma should be compared to individuals with mild asthma, which can be an additional complexity for subject recruiting. Analysis of asthma severity can include intermediate phenotypes such as measures of pulmonary function, bronchial hyperresponsiveness (BHR), biomarkers, and response to asthma therapy.
GWAS of severe, persistent asthma
Bleecker and Meyers and colleagues performed a GWAS for asthma susceptibility and severity in a comprehensively phenotyped, longitudinal cohort of 473 non-Hispanic white adult cases in The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens (TENOR) study [31, 32], compared to Illumina general population controls. In this analysis, several variants in the RAD50/IL13 region of chromosome 5q31.1 were identified as predictors of asthma susceptibility. A SNP in intron 2 of RAD50 (adjacent to IL13) was one of the most highly associated with asthma, though strong correlation between variants in this LD block makes it difficult to identify the causal variant [2]. However, this is an important example of a genome-wide study identifying a biologically relevant gene (IL13), which had been observed in several candidate gene studies [7, 33]. IL13 is an important regulator of allergen-induced asthma in mice [34, 35] and also appears to contribute to asthma in this severe allergic cohort. This study also identified variants in the HLA-DR/DQ region on chromosome 6p21.3 associated with asthma, a genomic region which has been replicated in many candidate genes studies [36, 37] and GWAS [3, 30, 38], as discussed above.
Lung function
Extensive phenotypic characterization in the National Heart, Lung, and Blood Institute (NHBLI)-sponsored Severe Asthma Research Program (SARP) cohort indicates that one of the primary determinants of asthma severity is lung function [39, 40]. Therefore identifying the genetic determinants of pulmonary function, a statistically powerful objective measure, represents a relevant approach to define genes that contribute to asthma severity.
There are currently no GWAS powered to test the progression of asthma severity and longitudinal lung function decline. However, two large meta-analyses for normal lung function have been carried out in the European general population. The CHARGE and SpiroMeta consortium both identified the HHIP (hedgehog interacting protein) gene associated with lung function measures forced expiratory volume in 1 second (FEV1) or the FEV1/forced vital capacity (FEV1/FVC) ratio [41, 42], along with eleven other genomic regions for normal pulmonary function (Table 2). To better understand whether these genetic variants also play a role in asthma populations, Li and colleagues tested 14 SNPs in these candidate genes for association with pulmonary function measures in a meta-analysis of three independent asthma cohorts (n=1,441): the NHLBI–sponsored SARP [39, 40] and Collaborative Studies on the Genetics of Asthma (CSGA) studies [43]; and the TENOR study [44]. This analysis identified the HHIP/rs1512288 variant as a significant predictor of FEV1 and FVC in asthma. And in a joint model, an increasing number of risk variants in these lung function genes were highly associated with lower FEV1 and increased asthma severity [45], demonstrating the close relationship between pulmonary function and asthma severity. This additive approach has been used successfully to predict prostate cancer risk [46, 47] and may have important clinical applications to personalized medicine in the future.
In a very large (approximately 90,000 European individuals in the discovery and replication cohorts) follow-up meta-analysis from the CHARGE and SpiroMeta consortia, an additional 16 loci for normal lung function were identified at the genome-wide significance level (Table 2). These genes are related to a number of different physiological pathways including cell growth, signaling and migration. It remains to be determined whether these genes or pathways also contribute to lung function variation in individuals with asthma, however a region of 6p21.33 that contains a non-synonymous SNP in LTA (lymphotoxin a) and a correlated SNP in the promoter region of TNFA (tumor necrosis factor α), were identified in this lung function analysis [48]. These genes have been linked to asthma susceptibility previously [49, 50]. Other plausible asthma-related mechanisms include TGF-β signaling, which can induce mucin expression in bronchial epithelial cells [51].
Interleukin 6 (IL6) is a cytokine that becomes systemically or locally elevated during inflammatory processes [52]. Increased IL6 expression has been observed with lung inflammation and injury [53] and elevated serum IL6 levels have been reported in subjects with asthma [54], chronic obstructive pulmonary disease (COPD) [55–59], and more recently, in a small severe asthma study [60]. For IL6 to have a role in inflammatory lung disease, the IL6 receptor must also be involved, which appears to be the case. In a genome wide association study of 57,800 subjects, the IL6R gene was identified as an asthma susceptibility gene [61], and more recently, the common IL6R coding variation rs2228145 (Asp358Ala) was identified as a potential genetic modifier of lung function in asthma and as a novel genetic marker of asthma [62]. In this last study, subjects of European ancestry with asthma who inherited the minor C allele of rs2228145 (Ala358) had the lowest mean percent predicted FEV1, FEV1/FVC, and the highest mean levels of methacholine responsiveness (PC20). The frequency of the rs2228145 C allele was significantly higher in phenotypic asthma clusters consisting of subjects with more severe asthma [40], and high serum levels of the soluble form of the IL6 receptor (sIL6R) were associated with lower lung function [62]. In cells that do not possess the membrane bound IL6 receptor, the active IL6/sIL6R complex can bind the membrane bound co-receptor glycoprotein 130 (gp130) and initiate IL6 signaling cascade, resulting in phosphorylation and activation of signal transducer and activator of transcription 3 (STAT3) protein, a transcription factor that has been linked to airway inflammation and changes in lung function [63–66]. This extracellular process is termed IL6 transsignaling and has been associated with a range of inflammatory diseases [52, 67], including rheumatoid arthritis [68], Crohn’s disease [69], and inflammatory bowel disease [70]. Long term exposure to IL6 transsignaling may induce morphological changes in smooth muscle structure and may be an important component in long term development of airway remodeling in obstructive lung disease. Anti-IL6R therapy is currently being used to control disease progression in inflammatory diseases such as rheumatoid arthritis [71–75], and thus there is the possibility that anti-IL6R therapy could also be important in the treatment of airway diseases such as asthma.
Biomarkers
Biomarkers such as serum IgE levels and blood or sputum eosinophil levels may be important predictors of asthma severity or risk of exacerbations. Therefore many asthma cohorts as well as large population-based studies have evaluated genetic factors related to these biomarkers (Table 2). The GABRIEL cohort performed a genome-wide study for genetic factors associated with total serum IgE levels in 7087 subjects with asthma and 7667 controls, and identified one novel locus in the class II region of MHC which was significant at the genome-wide level [3]. This study also observed several genetic variants associated with IgE levels in the FCER1A (Fc fragment of IgE, high affinity I), IL13 and STAT6 (signal transducer and activator of transcription) genes. These regions were also reported in previous GWAS for IgE in the general population [76], including a recent analysis by the Framingham cohort [77]. In GABRIEL, genes that were associated with asthma susceptibility generally did not overlap with those associated with IgE. Therefore the authors concluded that loci strongly associated with IgE levels may not play an important role in the development of asthma but may contribute to severity or progression of the disease [3]. In order to identify additional genes associated with IgE levels in non-Hispanic white asthma populations, Li et al. tested SNPs on chromosome 11q13.5 between the C11orf30 (open reading frame 30) and LRRC32 (leucine-rich repeat containing 32) genes, which had previously been identified in genetic analyses of related inflammatory conditions such as atopic dermatitis [78], childhood eczema [79] and Crohn’s disease [80]. Four SNPs in this region were significantly associated with total serum IgE levels in after adjustment for multiple testing, suggesting a common genetic regulation for IgE levels in atopic diseases.
Asthma genes identified through GWAS have also been identified as predictors of blood eosinophil levels, an inflammatory biomarker likely related to asthma pathogenesis and progression. For example, a large population-based GWAS of 9,392 Icelanders identified an IL1RL1 variant which was associated with eosinophil counts and asthma, and SNPs in WDR36, IL33 and MYB that were nominally associated with eosinophil counts and atopic asthma [81]. The 5q22 genomic region spanning TSLP and WDR36 genes, which has been associated with asthma in several reports [3, 11], is also associated with pediatric eosinophilic esophagitism, an allergic disorder characterized by excess eosinophils in the esophagus. Based on expression studies, the authors conclude that TSLP is the most likely causative gene in this region [82].
Asthma therapy
Another genetic mechanism that can produce more severe or difficult to manage asthma is response to asthma therapy. This section focuses on new developments in pharmacogenetics research (Table 2).
Corticosteroid (glucocorticoid) pathway
Glucocorticoid (GCs) steroids are currently the most common anti-inflammatory asthma therapy. For the most part, regular use of steroids is effective and reduces mortality due to asthma [83]. However, chronic steroid use can result in side effects which may be alleviated by targeted, inhaled drug delivery to the lung. There is subset of severe asthma patients that requires high-doses of inhaled and oral steroids to control symptoms and asthma exacerbations [39, 40]. However, there is also considerable variability in response to corticosteroids, with a significant number of patients who have no response. Relatively little is known about the pharmacogenetic interactions of this complex pathway [84, 85] though there is likely a genetic component to the variability in corticosteroid responses in childhood and adult asthma [86].
The Hop protein (encoded by the STIP1 gene) is involved in activation of the glucocorticoid receptor and is a novel therapeutic target in the glucocorticoid pathway. In a pharmacogenetic analysis, STIP1 single nucleotide and haplotypic variation was related to changes in FEV1 in response to treatment with inhaled corticosteroids (ICS). There was a heterogeneous response to steroids, as approximately half of STIP1 haplotypes were associated with reduced corticosteroid response and the other half with greater sensitivity to ICS [87].
A recent genome-wide study by Tantisira and colleagues evaluated four asthma populations (n=935) to identify pharmacogenetic associations related to response to inhaled glucocorticoids. A significant, replicated association was found in two correlated SNPs, rs37972 and rs37973, in the GLCCI1 (glucocorticoid-induced transcript 1) gene, which confer a lung function response to inhaled glucocorticoids and are also associated with reduced GLCCI1 expression and luciferase reporter activity. Using data pooled from all treatment trials, the overall mean (±SE) increase in FEV1 for ICS treated subjects homozygous for the rs37973 variant allele was significantly less than for subjects homozygous for the common allele (3.2±1.6% vs. 9.4±1.1% respectively). In this analysis, genotype was estimated to account for 6.6% of inhaled glucocorticoid response variability. This is an example of a replicated pharmacogenetic analysis with functional verification [88]. In addition, Tantisira et al. have also shown that variation in the corticotrophin-releasing hormone receptor 1 gene (CRHR1) is associated with improved lung function response to corticosteroids in three asthma clinical trial populations [86].
Beta2-adrenergic pathway
Beta adrenergic receptor agonists or beta agonists are commonly prescribed medications for relief of bronchoconstriction and long-term symptom treatment in asthma. Short-acting beta agonists (SABA) and long-acting β-agonist (LABA) target beta2-adrenergic receptors on the surface of airway smooth muscle cells and other lung or inflammatory cells [83]. Many pharmacogenetic studies have explored whether ADRB2 (beta-2 adrenergic receptor) gene variation could explain differences in bronchodilator response among asthma patients or identify a subgroup of patients with reduced response [83, 89]. Many of these studies have focused on the common functional nonsynonymous Gly16Arg and Gln27Glu mutations [90, 91]. For example, reduced pharmacologic responses to use of SABA but not to LABA in individuals homozygous for the ADBR2 Arg16 variant have been reported [92–96]. However, there are additional rare variants in ADRB2 that may also play a role in response to therapy. Extensive resequencing of the ADRB2 locus in African-Americans and non-Hispanic whites with asthma revealed four rare ADRβ2 polymorphisms which were analyzed for association with asthma severity outcomes: 25bp insertion-deletion at nucleotide -376, Asn15Ser (rs33973603), Thr164Ile (rs1800888, also identified in Liggett et al. [97]), and Ser220Cys (rs3729943) [98]. African-American cases with rare ADRB2 alleles treated with LABA demonstrated greater percentage of sputum eosinophils when compared to African-American cases with common alleles (28% versus 7%; P=0.02) or those with rare alleles not treated with LABA (28% versus 0.4%; P=0.01) [99]. Additionally, non-Hispanic white asthma cases with rare alleles treated with LABA demonstrated a significantly increased number of urgent care and emergency department visits for asthma in the past year compared to asthma patients with ADRB2 common alleles [99]. These analyses suggest that rare and common gene variation may modulate response to asthma therapy.
T Helper 2 pathway
There are several novel therapies currently under development targeting molecules in the T Helper 2 (TH2) pathway. Two recent clinical trials of these biologics identified specific population subgroups with improved therapeutic responses based on biomarker levels or specific genotypes. Corren et al. hypothesized that anti–IL13 therapy would benefit asthma patients with a baseline profile consistent with IL13 activity [100]. In a randomized, double-blind, placebo-controlled study of lebrikizumab, a monoclonal antibody to IL13, patient subgroups were prespecified according to TH2 status (based on total IgE level and blood eosinophil counts) and serum periostin level. After 12 weeks, the mean increase in FEV1 was 5.5 percentage points higher in the anti-IL13 group compared to the placebo group in the overall trial (P=0.02). However, among patients in the high-periostin or high fraction of exhaled nitric oxide (FeNO) subgroup, the increase from baseline FEV1 was 8.2 percentage points higher in the anti-IL13 group than placebo [100].
Additionally, Wenzel and colleagues reported results from a clinical trial of the anti-interleukin 4 receptor antagonist pitrakinra. In an ICS-withdrawal, double-blind, randomized, placebo-controlled, multicenter trial in 534 patients with uncontrolled, moderate to severe asthma, participants were randomized to 1 mg (n=132), 3 mg (n=137), 10 mg (n=128) pitrakinra or placebo (n=137) twice daily for a 12-week treatment period. In the overall study population, efficacy was not demonstrated, however statistically significant efficacy was observed in pre-specified subpopulations including: 1) subjects with increased blood eosinophil levels (peripheral blood count >350 cells/mm3, n=125), 2) upper tertile fraction of exhaled nitric oxide (range 20–122 ppb, n=64), and 3) individuals with GG genotype in IL-4 receptor (IL4R) variant rs8832 (n=134 non-Hispanic whites). Of these three subgroups, the greatest relative reduction in incidence of asthma exacerbation at the highest anti-interleukin 4 receptor dose compared to placebo (88%) was observed for IL4R/rs8832 GG subjects. Participants with the IL4R/rs8832GG genotype randomized to active anti-interleukin 4 receptor therapy had decreased asthma exacerbations, decreased nocturnal awakenings and activities limited by asthma. There was also a dose-dependent reduction in exacerbations in these subjects as the frequency of exacerbations in the placebo, 1 mg, 3 mg, and 10 mg treatment groups was 25%, 16%, 12%, and 3% respectively [101–103]. This analysis represents one of the largest pharmacogenetic investigations of the TH2 pathway in patients with uncontrolled, moderate to severe asthma, identifying an asthma subgroup with improved response to this therapy.
Issues such as statistical power, generalizability and functional characterization that are critical for all genetic studies are especially important in pharmacogenetic studies. Identifying appropriate replication populations also remains a challenge, due to the highly specific treatment regimens specified in clinical trials. However these studies may represent an important step in personalized medicine, as clinical decisions regarding appropriate therapy for asthma patients may rely on these types of studies in the future.3
Gene/environment interactions and asthma
Despite the importance of environmental factors to the development of asthma (Fig. 1), very few gene/environment interactions have been consistently identified and replicated for asthma severity. Challenges in genetic association studies such as adequate sample size, population stratification, gene coverage, and adjustment for false positives can be compounded in gene/environment interaction studies and accurately measuring environmental factors represents additional obstacles.
There have been several gene/environment studies to evaluate the interaction between genes and smoke exposure, including a genetic linkage analysis in 200 Dutch families. Linkage signals for asthma and BHR were observed on chromosomes 3p and 5q, though passive smoke exposure accounted for BHR linkage to 5q [15]. Similar results were observed in US populations [104]. Additional studies in candidate genes such as ADAM33 (A disintegrin and metalloprotease 33) have also demonstrated the role of smoke exposure on increased risk of asthma. The ADAM33 gene was identified through positional cloning [6] and replicated as an asthma susceptibility gene in several candidate gene studies. ADAM33 variants have also been tested in 200 Dutch asthma cases with more than 25 years of longitudinal data and the S_2 polymorphism was a significant predictor of increased decline in FEV1 [105]. This gene has also been associated with risk of COPD in a Dutch cohort [106] and in a cross-sectional population of 880 long-term tobacco smokers [107], suggesting that this gene may be related to disease progression in asthma and other respiratory diseases.
Investigators in the GABRIELA Study Group conducted a genome-wide interaction analysis for asthma and atopy testing 500,000 SNPs and rural farm-related exposures in 1708 European children. Overall, no significant interactions were identified and the authors conclude that common genetic polymorphisms were unlikely to moderate the influence of the farming environment on childhood asthma, though rare variants may play a role [108]. This analysis emphasizes that new statistical and measurement tools are needed to address these issues genome-wide [109, 110].
There have been other studies in families to investigate gene/environment interaction on a genome-wide basis or in candidate gene analysis. The Childhood Asthma Management Program conducted a genome-wide study of genes and modulation of vitamin D levels related to asthma exacerbations using population-based and family-based approaches in 403 individuals and trios. Three common variants in the CRTAM gene (class I MHC–restricted T cell–associated molecule) were associated with an increased rate of asthma exacerbations based on low vitamin D levels; these results were replicated in 584 children from a Costa Rican cohort. Functional studies then explored the interaction of vitamin D and the nonsynonymous coding polymorphism rs2272094 on CRTAM expression. The results suggest that maintaining adequate vitamin D levels may be especially important in subsets of asthma patients based on genotype [111]. In another candidate gene analysis, variants in the P2Y12 (purinergic receptor P2Y) gene, which is necessary for leukotriene E4-induced inflammation, were tested for interaction with house dust mite exposure and association with lung function. Five P2RY12 SNPs were predictors of multiple lung function measures (P=0.006–0.025) in 422 children with asthma and their parents. Individuals homozygous for minor alleles in P2RY12 exposed to house dust mite had lower lung function than those unexposed (P interaction=0.0028–0.040) [112].
Despite the obvious challenges involved in gene/environment interaction studies, these types of analyses may account for some of the “missing heritability” that GWAS variants do not explain in common diseases such as asthma [113]. But perhaps even more challenging than systematic and comprehensive evaluation of gene/environment interactions is defining how environmental exposures lead to disease. It is likely that epigenetic mechanisms are an important component for understanding the development of asthma. Several studies are now focusing on understanding how epigenetic modifications such as methylation regulate gene expression and may ultimately affect disease susceptibility and severity [114]. Interaction studies can also provide insight into which environmental exposures that contribute to asthma severity and risk of exacerbations and may guide interventions in the future [110, 115].
Summary and future studies
More than 10 years have passed since the first human genome was sequenced in 2001 [116, 117] and GWAS is currently the most effective approach to study genetics of human diseases. Several genes/regions have been consistently associated with asthma susceptibility through this method: ORMDL3/GSDMB, IL33, IL1RL1, RAD50/IL13, HLA-DR/DQ, TSLP, and SMAD3 (Table 1) [3, 11]. In many cases, the effect of each individual genetic variant is relatively small [113], suggesting that the additive effect of multiple risk variants should be taken into account. An important goal of the genetic approach in complex disease is to identify a group of variants that will reliably predict risk of the development (susceptibility) or progression (severity) of disease. Improved phenotyping approaches will also improve our ability to link genotype and phenotype. There is a need to understand asthma disease heterogeneity as different phenotypes may reflect several pathogenic pathways that have different underlying genetic architecture [40].
Genes that contribute to phenotypes such as lung function and biomarker levels provide insight into mechanisms of asthma progression (Table 2). However, additional studies evaluating genes and environmental factors leading to disease severity are needed. This will ultimately lead to greater understanding of the biologic pathways that contribute to disease progression. Future genetic studies in asthma severity will also incorporate whole-genome or exome-specific sequencing to identify more common and rare genetic variants. Exome and complete genome sequencing will provide better genomic coverage than existing genotyping platforms and will discover biologically relevant causal variants. For example, resequencing the IL4 gene in African-American identified an excess of private noncoding SNPs in asthma cases compared with unaffected individuals (P=0.031) [118]. New genotyping technologies and comprehensive phenotyping should elucidate the genetics of asthma severity in the future. Understanding the functional biology of these novel variants discovered through genome-wide sequencing will also become increasing important. Using these variants identified in comprehensively phenotyped studies, we may more effectively develop personalized therapy for all individuals with asthma.
Synopsis.
This article summarizes major findings in genome-wide studies of asthma susceptibility and severity. Two large meta-analyses identified four chromosomal regions which were consistently associated with development of asthma. Genes that are associated with asthma subphenotypes such as lung function, biomarker levels and asthma therapeutic responses can provide insight into mechanisms of asthma severity and disease progression. Future genetic studies will incorporate sequencing in comprehensively phenotyped asthmatics to lead to the development of personalized therapy.
Key points.
Genes and environmental exposures interact to influence risk of asthma susceptibility and severity
Two large meta-analyses of asthma susceptibility, GABRIEL and EVE, identified four chromosomal regions which were associated with asthma in individuals of different ethnic backgrounds: loci in the ORMDL3 region of 17q21, IL1RL/IL18R on chromosome 2q, TSLP on 5q22, and IL33 on chromosome 9p24
Genome-wide screens for asthma susceptibility in Asian populations identified genetic variants in the major histocompatiblity complex gene region (HLA region) on chromosome 6p21 associated with asthma risk; this locus has been a significant predictor of asthma susceptibility in several genetic studies
Genes that are associated with asthma subphenotypes such as lung function, biomarkers levels and asthma therapeutic responses can provide insight into mechanisms of asthma severity progression
A joint model of risk variants in lung function genes identified in the general population were highly associated with lower lung function and increased severity in asthma populations
A pharmacogenetic genome-wide screen identified two correlated genetic variants in the GLCCI1 gene related to response to inhaled glucocorticoids
Future genetic studies for asthma susceptibility and severity will incorporate exome or whole-genome sequencing in comprehensively phenotyped asthmatics which will contribute to personalized asthma therapy
Acknowledgments
This work was funded by NHLBI grants HL69167and HL101487. RES was supported by HL089992 and HL101487.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Rebecca E. Slager, Email: rslager@wakehealth.edu.
Gregory A. Hawkins, Email: ghawkins@wakehealth.edu.
Xingnan Li, Email: xinli@wakehealth.edu.
Dirkje S. Postma, Email: d.s.postma@umcg.nl.
Deborah A. Meyers, Email: dmeyers@wakehealth.edu.
Eugene R. Bleecker, Email: ebleeck@wakehealth.edu.
References
- 1.Slager RE, Li X, Meyers DA, et al. Recent developments in the genetics of asthma susceptibility and severity. In: Chung KF, Bel EH, Wenzel SE, editors. Difficult-to-Treat Severe Asthma. Sheffield: European Respiratory Society Journals; 2011. pp. 82–96. [Google Scholar]
- 2.Li X, Howard TD, Zheng SL, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125:328–335. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes and immunity. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 5.Ober C, Tan Z, Sun Y, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–91. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Eerdewegh P, Little RD, Dupuis J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 7.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–82. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 8.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 9.Himes BE, Hunninghake GM, Baurley JW, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Gen. 2009;84:581–93. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reijmerink NE, Postma DS, Bruinenberg M, et al. Association of IL1RL1, IL18R1, and IL18RAP gene cluster polymorphisms with asthma and atopy. J Allergy Clin Immunol. 2008;122:651–4. e8. doi: 10.1016/j.jaci.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 11.Torgerson DG, Ampleford EJ, Chiu GY, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffatt MF, Schou C, Faux JA, et al. Association between quantitative traits underlying asthma and the HLA-DRB1 locus in a family-based population sample. Eur J Hum Gen. 2001;9:341–6. doi: 10.1038/sj.ejhg.5200636. [DOI] [PubMed] [Google Scholar]
- 13.Meyers DA. Genetics of asthma and allergy: what have we learned? J Allergy Clin Immunol. 2010;126:439–46. doi: 10.1016/j.jaci.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen M, Heinzmann A, Noguchi E, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Gen. 2003;35:258–63. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- 15.Meyers DA, Postma DS, Stine OC, et al. Genome screen for asthma and bronchial hyperresponsiveness: interactions with passive smoke exposure. J Allergy Clin Immunol. 2005;115:1169–75. doi: 10.1016/j.jaci.2005.01.070. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Meyers DA, Ober C, et al. Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three U.S. populations: Collaborative Study on the Genetics of Asthma. Am J Hum Genet. 2001;68:1437–46. doi: 10.1086/320589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koppelman GH, Meyers DA, Howard TD, et al. Identification of PCDH1 as a novel susceptibility gene for bronchial hyperresponsiveness. Am J Resp Crit Care Med. 2009;180:929–35. doi: 10.1164/rccm.200810-1621OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershey GK, Friedrich MF, Esswein LA, et al. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med. 1997;337:1720–5. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- 19.Ober C, Leavitt SA, Tsalenko A, et al. Variation in the interleukin 4-receptor alpha gene confers susceptibility to asthma and atopy in ethnically diverse populations. Am J Hum Genet. 2000;66:517–26. doi: 10.1086/302781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard TD, Koppelman GH, Xu J, et al. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet. 2002;70:230–6. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabesch M, Schedel M, Carr D, et al. IL-4/IL13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006;117:269–74. doi: 10.1016/j.jaci.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Ericksen MB, Levin LS, et al. Functional effect of the R110Q IL13 genetic variant alone and in combination with IL4RA genetic variants. J Allergy Clin Immunol. 2004;114:553–60. doi: 10.1016/j.jaci.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 23.Fang Q, Zhao H, Wang A, et al. Association of genetic variants in chromosome 17q21 and adult-onset asthma in a Chinese Han population. BMC Med Gen. 2011;12:133. doi: 10.1186/1471-2350-12-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock DB, Romieu I, Shi M, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genetics. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wjst M. ORMDL3--guilt by association? Clin Exp Allergy. 2008;38:1579–81. doi: 10.1111/j.1365-2222.2008.03086.x. [DOI] [PubMed] [Google Scholar]
- 26.Durbin RM, Abecasis GR, Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenzel SE, Busse WW. Severe asthma: lessons from the Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:14–21. doi: 10.1016/j.jaci.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 28.Gupta RS, Carrion-Carire V, Weiss KB. The widening black/white gap in asthma hospitalizations and mortality. J Allergy Clin Immunol. 2006;117:351–8. doi: 10.1016/j.jaci.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 29.Mathias RA, Grant AV, Rafaels N, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–346. e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirota T, Takahashi A, Kubo M, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Gen. 2011;43:893–6. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolan CM, Fraher KE, Bleecker ER, et al. Design and baseline characteristics of the epidemiology and natural history of asthma: Outcomes and Treatment Regimens (TENOR) study: a large cohort of patients with severe or difficult-to-treat asthma. Ann Allergy Asthma Immunol. 2004;92:32–9. doi: 10.1016/S1081-1206(10)61707-3. [DOI] [PubMed] [Google Scholar]
- 32.Haselkorn T, Fish JE, Zeiger RS, et al. Consistently very poorly controlled asthma, as defined by the impairment domain of the Expert Panel Report 3 guidelines, increases risk for future severe asthma exacerbations in The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens (TENOR) study. J Allergy Clin Immunol. 2009;124:895–902. e1–4. doi: 10.1016/j.jaci.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 33.Howard TD, Whittaker PA, Zaiman AL, et al. Identification and association of polymorphisms in the interleukin-13 gene with asthma and atopy in a Dutch population. Am J Respir Cell Mol Biol. 2001;25:377–84. doi: 10.1165/ajrcmb.25.3.4483. [DOI] [PubMed] [Google Scholar]
- 34.Grunig G, Warnock M, Wakil AE, et al. Requirement for IL13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–3. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wills-Karp M, Luyimbazi J, Xu X, et al. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–61. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 36.Booth M, Shaw MA, Carpenter D, et al. Carriage of DRB1*13 is associated with increased posttreatment IgE levels against Schistosoma mansoni antigens and lower long-term reinfection levels. J Immunol. 2006;176:7112–8. doi: 10.4049/jimmunol.176.11.7112. [DOI] [PubMed] [Google Scholar]
- 37.Munthe-Kaas MC, Carlsen KL, Carlsen KH, et al. HLA Dr-Dq haplotypes and the TNFA-308 polymorphism: associations with asthma and allergy. Allergy. 2007;62:991–8. doi: 10.1111/j.1398-9995.2007.01377.x. [DOI] [PubMed] [Google Scholar]
- 38.Noguchi E, Sakamoto H, Hirota T, et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genetics. 2011;7:e1002170. doi: 10.1371/journal.pgen.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore WC, Bleecker ER, Curran-Everett D, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore WC, Meyers DA, Wenzel SE, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–23. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Gen. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Repapi E, Sayers I, Wain LV, et al. Genome-wide association study identifies five loci associated with lung function. Nat Gen. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyers DA, Wjst M, Ober C. Description of three data sets: Collaborative Study on the Genetics of Asthma (CSGA), the German Affected-Sib-Pair Study, and the Hutterites of South Dakota. Genet Epidemiol. 2001;21 (Suppl 1):S4–8. doi: 10.1002/gepi.2001.21.s1.s4. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Ampleford EJ, Howard TD, et al. The C11orf30-LRRC32 region is associated with total serum IgE levels in asthmatic patients. J Allergy Clin Immunol. 2012;129:575–8. doi: 10.1016/j.jaci.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Howard TD, Moore WC, et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127:1457–65. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Zheng SL, Isaacs SD, et al. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. PNAS. 2010;107:2136–40. doi: 10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 48.Soler Artigas M, Loth DW, Wain LV, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Gen. 2011;43:1082–90. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruse CE, Hill MC, Tobin M, et al. Tumour necrosis factor gene complex polymorphisms in chronic obstructive pulmonary disease. Resp Med. 2007;101:340–4. doi: 10.1016/j.rmed.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 50.Wu H, Romieu I, Sienra-Monge JJ, et al. Parental smoking modifies the relation between genetic variation in tumor necrosis factor-alpha (TNF) and childhood asthma. Env Health Perspectives. 2007;115:616–22. doi: 10.1289/ehp.9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu HW, Balzar S, Seedorf GJ, et al. Transforming growth factor-beta2 induces bronchial epithelial mucin expression in asthma. Am J Path. 2004;165:1097–106. doi: 10.1016/s0002-9440(10)63371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Febbraio MA, Rose-John S, Pedersen BK. Is interleukin-6 receptor blockade the Holy Grail for inflammatory diseases? Clin Pharm Ther. 2010;87:396–8. doi: 10.1038/clpt.2010.1. [DOI] [PubMed] [Google Scholar]
- 53.Ammit AJ, Moir LM, Oliver BG, et al. Effect of IL-6 trans-signaling on the pro-remodeling phenotype of airway smooth muscle. Am J Phys Lung Cell Mol Phys. 2007;292:L199–206. doi: 10.1152/ajplung.00230.2006. [DOI] [PubMed] [Google Scholar]
- 54.Yokoyama A, Kohno N, Fujino S, et al. Circulating interleukin-6 levels in patients with bronchial asthma. Am J Resp Crit Care Med. 1995;151:1354–8. doi: 10.1164/ajrccm.151.5.7735584. [DOI] [PubMed] [Google Scholar]
- 55.Garrod R, Marshall J, Barley E, et al. The relationship between inflammatory markers and disability in chronic obstructive pulmonary disease (COPD) Primary Care Resp J. 2007;16:236–40. doi: 10.3132/pcrj.2007.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schols AM, Buurman WA, Staal van den Brekel AJ, et al. Evidence for a relation between metabolic derangements and increased levels of inflammatory mediators in a subgroup of patients with chronic obstructive pulmonary disease. Thorax. 1996;51:819–24. doi: 10.1136/thx.51.8.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karadag F, Karul AB, Cildag O, et al. Biomarkers of systemic inflammation in stable and exacerbation phases of COPD. Lung. 2008;186:403–9. doi: 10.1007/s00408-008-9106-6. [DOI] [PubMed] [Google Scholar]
- 58.Broekhuizen R, Wouters EF, Creutzberg EC, et al. Raised CRP levels mark metabolic and functional impairment in advanced COPD. Thorax. 2006;61:17–22. doi: 10.1136/thx.2005.041996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee TM, Lin MS, Chang NC. Usefulness of C-reactive protein and interleukin-6 as predictors of outcomes in patients with chronic obstructive pulmonary disease receiving pravastatin. Am J Cardio. 2008;101:530–5. doi: 10.1016/j.amjcard.2007.09.102. [DOI] [PubMed] [Google Scholar]
- 60.Morjaria JB, Babu KS, Vijayanand P, et al. Sputum IL-6 concentrations in severe asthma and its relationship with FEV1. Thorax. 2011;66:537. doi: 10.1136/thx.2010.136523. [DOI] [PubMed] [Google Scholar]
- 61.Ferreira MA, Matheson MC, Duffy DL, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hawkins GA, Robinson MB, Hastie AT, et al. IL6R variation Asp358Ala is a potential modifier of lung function in asthma. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.03.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simeone-Penney MC, Severgnini M, Tu P, et al. Airway epithelial STAT3 is required for allergic inflammation in a murine model of asthma. J Immunol. 2007;178:6191–9. doi: 10.4049/jimmunol.178.10.6191. [DOI] [PubMed] [Google Scholar]
- 64.Litonjua AA, Tantisira KG, Lake S, et al. Polymorphisms in signal transducer and activator of transcription 3 and lung function in asthma. Resp Res. 2005;6:52. doi: 10.1186/1465-9921-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao H, Guo RF, Speyer CL, et al. Stat3 activation in acute lung injury. J Immunol. 2004;172:7703–12. doi: 10.4049/jimmunol.172.12.7703. [DOI] [PubMed] [Google Scholar]
- 66.Gao H, Ward PA. STAT3 and suppressor of cytokine signaling 3: potential targets in lung inflammatory responses. Expert Op Ther Targets. 2007;11:869–80. doi: 10.1517/14728222.11.7.869. [DOI] [PubMed] [Google Scholar]
- 67.Rose-John S, Waetzig GH, Scheller J, et al. The IL-6/sIL-6R complex as a novel target for therapeutic approaches. Expert Op Ther Targets. 2007;11:613–24. doi: 10.1517/14728222.11.5.613. [DOI] [PubMed] [Google Scholar]
- 68.Plushner SL. Tocilizumab: an interleukin-6 receptor inhibitor for the treatment of rheumatoid arthritis. Ann Pharmacother. 2008;42:1660–8. doi: 10.1345/aph.1L268. [DOI] [PubMed] [Google Scholar]
- 69.Brulhart L, Nissen MJ, Chevallier P, et al. Tocilizumab in a patient with ankylosing spondylitis and Crohn’s disease refractory to TNF antagonists. Joint Bone Spine. 2010;77:625–6. doi: 10.1016/j.jbspin.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 70.Rose-John S, Mitsuyama K, Matsumoto S, et al. Interleukin-6 trans-signaling and colonic cancer associated with inflammatory bowel disease. Curr Pharm Des. 2009;15:2095–103. doi: 10.2174/138161209788489140. [DOI] [PubMed] [Google Scholar]
- 71.De Bandt M, Saint-Marcoux B. Tocilizumab for multirefractory adult-onset Still’s disease. Ann Rheum Dis. 2009;68:153–4. doi: 10.1136/ard.2008.088179. [DOI] [PubMed] [Google Scholar]
- 72.Hagihara K, Kawase I, Tanaka T, et al. Tocilizumab ameliorates clinical symptoms in polymyalgia rheumatica. J Rheumatol. 2010;37:1075–6. doi: 10.3899/jrheum.091185. [DOI] [PubMed] [Google Scholar]
- 73.Kawabata H, Tomosugi N, Kanda J, et al. Anti-interleukin 6 receptor antibody tocilizumab reduces the level of serum hepcidin in patients with multicentric Castleman’s disease. Haematologica. 2007;92:857–8. doi: 10.3324/haematol.10794. [DOI] [PubMed] [Google Scholar]
- 74.Kluger N, Bessis D, Guillot B. Tocilizumab as a potential treatment in Schnitzler syndrome. Med Hypotheses. 2009;72:479–80. doi: 10.1016/j.mehy.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka T, Ogata A, Narazaki M. Tocilizumab for the treatment of rheumatoid arthritis. Expert Rev Clin Immunol. 2010;6:843–54. doi: 10.1586/eci.10.70. [DOI] [PubMed] [Google Scholar]
- 76.Weidinger S, Gieger C, Rodriguez E, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genetics. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Granada M, Wilk JB, Tuzova M, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol. 2012;129:840–45. doi: 10.1016/j.jaci.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Esparza-Gordillo J, Weidinger S, Folster-Holst R, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Gen. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 79.O’Regan GM, Campbell LE, Cordell HJ, et al. Chromosome 11q13.5 variant associated with childhood eczema: an effect supplementary to filaggrin mutations. J Allergy Cin Immunol. 2010;125:170–4. e1–2. doi: 10.1016/j.jaci.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Gen. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gudbjartsson DF, Bjornsdottir US, Halapi E, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Gen. 2009;41:342–7. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 82.Rothenberg ME, Spergel JM, Sherrill JD, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Gen. 2010;42:289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pascual RM, Bleecker ER. Pharmacogenetics of asthma. Curr Opin Pharmacol. 2010;10:226–35. doi: 10.1016/j.coph.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 84.Adcock IM, Barnes PJ. Molecular mechanisms of corticosteroid resistance. Chest. 2008;134:394–401. doi: 10.1378/chest.08-0440. [DOI] [PubMed] [Google Scholar]
- 85.Barnes PJ, Adcock IM. Glucocorticoid resistance in inflammatory diseases. Lancet. 2009;373:1905–17. doi: 10.1016/S0140-6736(09)60326-3. [DOI] [PubMed] [Google Scholar]
- 86.Tantisira KG, Lake S, Silverman ES, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13:1353–9. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 87.Hawkins GA, Lazarus R, Smith RS, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123:1376–83. e7. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365:1173–83. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lima JJ, Blake KV, Tantisira KG, et al. Pharmacogenetics of asthma. Curr Opin Pulm Med. 2009;15:57–62. doi: 10.1097/MCP.0b013e32831da8be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Green SA, Turki J, Innis M, et al. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33:9414–9. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 91.Green SA, Turki J, Bejarano P, et al. Influence of beta 2-adrenergic receptor genotypes on signal transduction in human airway smooth muscle cells. Am J Respir Crit Care Med. 1995;13:25–33. doi: 10.1165/ajrcmb.13.1.7598936. [DOI] [PubMed] [Google Scholar]
- 92.Bleecker ER, Nelson HS, Kraft M, et al. Beta2-receptor polymorphisms in patients receiving salmeterol with or without fluticasone propionate. Am J Respir Crit Care Med. 2010;181:676–87. doi: 10.1164/200809-1511OC. [DOI] [PubMed] [Google Scholar]
- 93.Bleecker ER, Postma DS, Lawrance RM, et al. Effect of ADRB2 polymorphisms on response to longacting beta2-agonist therapy: a pharmacogenetic analysis of two randomised studies. Lancet. 2007;370:2118–25. doi: 10.1016/S0140-6736(07)61906-0. [DOI] [PubMed] [Google Scholar]
- 94.Israel E, Chinchilli VM, Ford JG, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364:1505–12. doi: 10.1016/S0140-6736(04)17273-5. [DOI] [PubMed] [Google Scholar]
- 95.Israel E, Drazen JM, Liggett SB, et al. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000;162:75–80. doi: 10.1164/ajrccm.162.1.9907092. [DOI] [PubMed] [Google Scholar]
- 96.Wechsler ME, Kunselman SJ, Chinchilli VM, et al. Effect of beta2-adrenergic receptor polymorphism on response to longacting beta2 agonist in asthma (LARGE trial): a genotype-stratified, randomised, placebo-controlled, crossover trial. Lancet. 2009;374:1754–64. doi: 10.1016/S0140-6736(09)61492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liggett SB, Wagoner LE, Craft LL, et al. The Ile164 beta2-adrenergic receptor polymorphism adversely affects the outcome of congestive heart failure. J Clin Inv. 1998;102:1534–9. doi: 10.1172/JCI4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hawkins GA, Tantisira K, Meyers DA, et al. Sequence, haplotype, and association analysis of ADRbeta2 in a multiethnic asthma case-control study. Am J Respir Crit Care Med. 2006;174:1101–9. doi: 10.1164/rccm.200509-1405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ortega VE, Hastie A, Sadeghnejad A, et al. Rare beta2-adrenergic receptor gene polymorphisms In asthma cases and controls from the Severe Asthma Research Program. Am J Respir Crit Care Med. 2011;183:A1357. [Google Scholar]
- 100.Corren J, Lemanske RF, Hanania NA, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–98. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 101.Slager RE, Hawkins GA, Otulana BA, et al. Interleukin 4 receptor polymorphisms predict therapeutic pitrakinra treatment response in moderate to severe asthma. Am J Respir Crit Care Med. 2011;183:A6178. [Google Scholar]
- 102.Wenzel SE, Ind PW, Otulana BA, et al. A phase 2b study of inhaled pitrakinra, an IL-4/IL13 antagonist, successfully identified responder subpopulations of patients with uncontrolled asthma. Am J Respir Crit Care Med. 2011;183:A6179. [Google Scholar]
- 103.Slager RE, Otulana BA, Hawkins GA, et al. Interleukin 4 receptor polymorphisms predict reduction in asthma exacerbations during response to an anti-interleukin 4 alpha receptor antagonist. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.03.030. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Colilla S, Nicolae D, Pluzhnikov A, et al. Evidence for gene-environment interactions in a linkage study of asthma and smoking exposure. J Allergy Clin Immunol. 2003;111:840–6. doi: 10.1067/mai.2003.170. [DOI] [PubMed] [Google Scholar]
- 105.Jongepier H, Boezen HM, Dijkstra A, et al. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy. 2004;34:757–60. doi: 10.1111/j.1365-2222.2004.1938.x. [DOI] [PubMed] [Google Scholar]
- 106.van Diemen CC, Postma DS, Vonk JM, et al. A disintegrin and metalloprotease 33 polymorphisms and lung function decline in the general population. Am J Respir Crit Care Med. 2005;172:329–33. doi: 10.1164/rccm.200411-1486OC. [DOI] [PubMed] [Google Scholar]
- 107.Sadeghnejad A, Ohar JA, Zheng SL, et al. Adam33 polymorphisms are associated with COPD and lung function in long-term tobacco smokers. Resp Res. 2009;10:21, e1–9. doi: 10.1186/1465-9921-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ege MJ, Strachan DP, Cookson WO, et al. Gene-environment interaction for childhood asthma and exposure to farming in Central Europe. J Allergy Clin Immunol. 2011;127:138–44. 144, e1–4. doi: 10.1016/j.jaci.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 109.Khoury MJ, Wacholder S. Invited commentary: from genome-wide association studies to gene-environment-wide interaction studies--challenges and opportunities. Am J Epi. 2009;169:227–30. doi: 10.1093/aje/kwn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends in Gen. 2011;27:107–15. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Du R, Litonjua AA, Tantisira KG, et al. Genome-wide association study reveals class I MHC-restricted T cell-associated molecule gene (CRTAM) variants interact with vitamin D levels to affect asthma exacerbations. J Allergy Clin Immunol. 2012;129:368–73. doi: 10.1016/j.jaci.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bunyavanich S, Boyce JA, Raby BA, et al. Gene-by-environment effect of house dust mite on purinergic receptor P2Y12 (P2RY12) and lung function in children with asthma. Clin Exp Allergy. 2012;42:229–37. doi: 10.1111/j.1365-2222.2011.03874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Gen. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ho SM. Environmental epigenetics of asthma: an update. J Allergy Clin Immunol. 2010;126:453–65. doi: 10.1016/j.jaci.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vercelli D. Gene-environment interactions: the road less traveled by in asthma genetics. J Allergy Clin Immunol. 2009;123:26–7. doi: 10.1016/j.jaci.2008.11.031. [DOI] [PubMed] [Google Scholar]
- 116.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 117.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 118.Haller G, Torgerson DG, Ober C, et al. Sequencing the IL4 locus in African Americans implicates rare noncoding variants in asthma susceptibility. J Allergy Clin Immunol. 2009;124:1204–9. e9. doi: 10.1016/j.jaci.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]


