Background: HIGM-2 syndrome results from mutations spanning AID.
Results: AID mutations are characterized biochemically and analyzed using a surrogate Apo3G structure.
Conclusion: Catalytically active mutants retain salient enzymatic properties of WT AID; catalytically inactive mutants retain salient ssDNA binding properties of WT AID.
Significance: We identify four structural classes of mutants and discuss the catalytic consequences of each mutation.
Keywords: Crystal Structure, DNA Enzymes, Immunology, Protein-DNA Interaction, Protein Structure, APOBEC, Class-switch Recombination, Deoxycytidine Deaminase, HiGM, Somatic Hypermutation
Abstract
Hyper-IgM syndrome type 2 stems from mutations in activation-induced deoxycytidine deaminase (AID) that abolish immunoglobulin class-switch recombination, causing an accumulation of IgM and absence of IgG, IgA, and IgE isotypes. Although hyper-IgM syndrome type 2 is rare, the 23 missense mutations identified in humans span almost the entire gene for AID resulting in a recessive phenotype. Using high resolution x-ray structures for Apo3G-CD2 as a surrogate for AID, we identify three classes of missense mutants as follows: catalysis (class I), substrate interaction (class II), and structural integrity (class III). Each mutant was expressed and purified from insect cells and compared biochemically to wild type (WT) AID. Four point mutants retained catalytic activity at 1/3rd to 1/200th the level of WT AID. These “active” point mutants mimic the behavior of WT AID for motif recognition specificity, deamination spectra, and high deamination processivity. We constructed a series of C-terminal deletion mutants (class IV) that retain catalytic activity and processivity for deletions ≤18 amino acids, with ΔC10 and ΔC15 having 2–3-fold higher specific activities than WT AID. Deleting 19 C-terminal amino acids inactivates AID. WT AID and active and inactive point mutants bind cooperatively to single-stranded DNA (Hill coefficients ∼1.7–3.2) with microscopic dissociation constant values (KA) ranging between 10 and 250 nm. Active C-terminal deletion mutants bind single-stranded DNA noncooperatively with KA values similar to wild type AID. A structural analysis is presented that shows how localized defects in different regions of AID can contribute to loss of catalytic function.
Introduction
Hyper-IgM syndrome type 2 (HIGM-2)2 is a genetic disorder caused by mutations in AID, a B-cell-specific protein (1–3). AID possesses C→U deamination activity preferentially targeting WRC (W = (A/T); R = purine) hot spot motifs on ssDNA and transcribed dsDNA (4–6). In activated B-cells, AID acts at the variable (V) and switch regions of Ig genes initiating two tightly regulated mechanisms of Ab diversification, class switch recombination (CSR), and somatic hypermutation (SHM) (7–9). CSR produces Abs of isotypes IgG, IgA, and IgE by region-specific recombination during which the Ig heavy Cμ exon of IgM is exchanged for a downstream C exon, such as Cγ, Cα, or Cϵ (9). SHM generates point mutations in V regions at an extremely high rate of 10−3 to 10−4 per cell division, resulting in the production of Abs with high affinity for antigen (8).
Clinical studies of families with HIGM-2 syndrome showed that the affected individuals usually have normal or elevated serum levels of IgM but low serum levels of IgG, IgA, and IgE, suggesting a common deficiency in CSR (1–3). To date, at least 39 different naturally occurring AID mutations have been identified in HIGM-2 patients (10–12). Although many mutations are nonsense or deletions, 23 missense HIGM-2 mutations were found at 20 distinct AID loci (Fig. 1). In most cases, HIGM-2 mutations are autosomal recessive, and the patients exhibit deficiency in both CSR and SHM (1–3, 13). A rare autosomal dominant form of HIGM-2 has been described for several patients who appeared to have defective CSR but not SHM (3, 14). Two patients with a dominant form of HIGM-2 had loss of 9 or 17 C-terminal amino acids (14). Clinical features of naturally occurring HIGM AID mutations have been well characterized (1–3, 13). In contrast, except for nonsense and deletion HIGM-2 mutations that lead to the production of truncated proteins, and for AID mutations in the C-deaminase active site, the underlying biochemical causes of HIGM-2 missense mutations have not been studied.
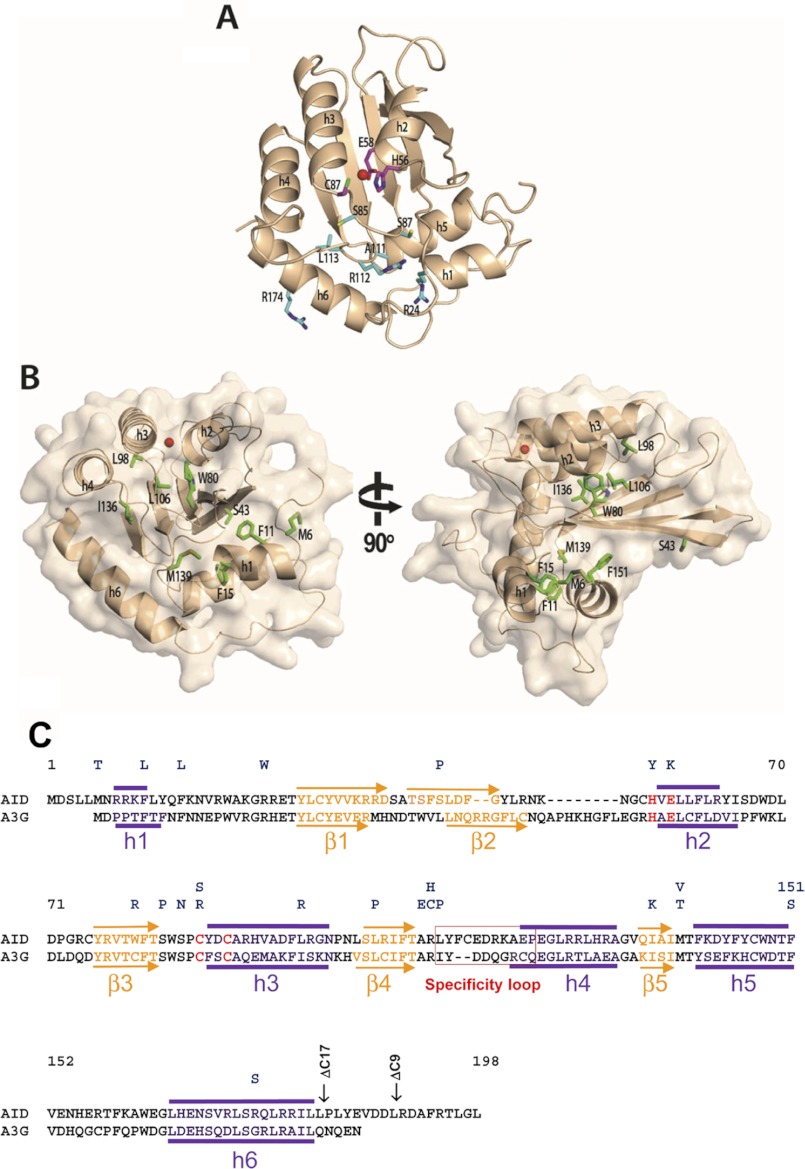
FIGURE 1.
A structural model of AID. A, AID model (wheat) was generated using Apo3G-CD2 as a template. Class I residues H56Y, E58K, and C87R/C87S (magenta sticks) are important for catalysis. His-56 and Cys-87 are involved in coordinating the zinc ion (red sphere), and Glu-58 is involved in proton shuffling. Residues in class II (R24W, S83P, S85N, A111E, R112C/R112H, L113P, and R174S, cyan sticks) are likely involved in interaction with the ssDNA substrate. B, class III residues (Met-6, Phe-11, Phe-15, Trp-80, Leu-98, Leu-106, Ile-136, Met-139, and Phe-151, green sticks) are important for maintaining the structural integrity of AID. C, alignment of AID and Apo3G-CD2. HIGM-2 base substitution mutations and C-terminal deletion mutations are indicated on top. Secondary structure predictions based on the AID model are indicated above the AID sequence. α-Helices and β-strands depicted below the A3G-CD2 sequence are from its x-ray crystal structure (17). Active site residues are highlighted in red.
AID belongs to the APOBEC family of cytidine and deoxycytidine deaminases with diverse cellular functions, including the regulation of adaptive and innate immunity and the modulation of protein expression (15). High resolution structures of the deaminase domain of APOBEC enzymes have been determined for Apo2 (16) and for the C-terminal catalytic domain (CD2) of Apo3G (17–20). Overall, these structures showed a highly conserved deaminase domain with a five-stranded β-sheet core flanked by six α-helices (Fig. 1). Alignment of AID to the Apo2 and the CD2 domain of Apo3G showed that missense HIGM-2 mutations are distributed throughout AID without concentrating in any specific region of the protein (Fig. 1). We have identified three classes of missense mutants, “catalysis” (class I), “substrate interaction” (class II), and “structural integrity” (class III). C-terminal deletions (class IV) are grouped separately.
In this study, we provide a biochemical characterization of HIGM-2 missense and C-terminal deletion AID mutants. Most HIGM-2 missense mutations abolish AID activity, but four retain partial C deamination activity. The partially active mutants are compared with WT AID in terms of deamination motif specificity, mutation spectra, ssDNA binding, and processivity (5, 21). The C-terminal region of AID, which is needed to bind cofactors essential for class switching (22), has not been studied systematically to determine its contribution to functionality. We have measured AID deamination activity for a series of C-terminal deletion mutants to determine the minimal number of amino acids required for AID to remain active. We investigate each of the HIGM-2 point mutants and C-terminal deletions using the Apo3G-CD2 structure (17) to infer how localized structural defects in AID can cause a loss of AID function.
EXPERIMENTAL PROCEDURES
Enzymes and Substrates
Point mutation and C-terminal deletion mutant AID were constructed by site-directed mutagenesis (QuikChange site-directed mutagenesis kit, Stratagene) using the pAcG2T-AID vector (4) as the template. Recombinant baculoviruses encoding WT and mutant AID were generated according to the recommended protocol (BD Biosciences). Wild type and mutant GST-AID proteins were expressed and purified as described previously (5, 23). AID proteins were dialyzed in a buffer containing 20 mm Tris-HCl (pH 7.5), 250 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, and 10% glycerol and stored at −80 °C. Unmodified and fluorescein-dT oligonucleotide substrates were synthesized and purified by denaturing PAGE. M13mp2 gapped DNA substrate was prepared as described (5, 23).
Deamination Assay to Measure AID-specific Activity on ssDNA
Activities of WT and mutants AID proteins were measured using 32P-labeled 36-nt ssDNA 5′-AGAAAAGGGGAAAGCAAAGAGGAAAGGTGAGGAGGT-3′. Deamination reactions (30-μl volume) were carried out in a buffer containing 20 mm Tris (pH 8.0), 1 mm dithiothreitol, 1 mm EDTA, 25 nm of the substrate DNA, 200 ng of RNase A. Following incubation at 37 °C, the reactions were quenched by a double extraction with phenol/chloroform/isoamyl alcohol (25:24:1), and the deamination product was analyzed as described previously (4). WT AID (10 nm) was used to measure C deamination activity. AID mutants were screened for C deamination activity using protein concentrations in a 10–90 nm range using a 60-min reaction time. Specific activities, calculated as picomoles of deaminated ssDNA substrate/min/μg of enzyme, were determined in the linear range of protein concentration and incubation times. The following protein concentrations were used in calculating AID specific activities: WT AID, S43P, and C-terminal deletion mutants (10 nm); L98R (75 nm); and R174S (79 nm). The linear time range for WT, S43P, and C-terminal deletions was ≤5 min, L98R ≤10 min, and R174S ≤20 min.
Analysis of AID-targeted C Deamination on lacZa-gapped Substrate
Deaminations of WT and mutant AID on the lacZa target were measured using the following reaction conditions: 30-μl volume, 20 mm Tris (pH 8.0), dithiothreitol (1 mm), EDTA (1 mm), gapped DNA (500 ng), RNase A (0.2 μg), and appropriate amount of WT or mutant AID. Following incubations for 5 min at 37 °C, the reactions were quenched by a double extraction with phenol/chloroform/isoamyl alcohol (25:24:1). Conversions of C→U on the DNA substrate were detected as white or light blue plaques indicating C→T mutations in a lacZα target gene after transfection into uracil glycosylase-deficient (ung−) Escherichia coli (5, 21). Mutation spectra and mutability indices were calculated as described previously (5, 21, 23).
Steady-state Rotational Anisotropy Binding Assay
WT and mutant AID binding to ssDNA was monitored by changes in steady-state fluorescence depolarization (rotational anisotropy). A fluorescein-labeled (FdT) ssDNA substrate containing a single C was used to measure AID binding: 5′-AGAAAAGGGGAAAGCAAAGAGGAAAGG(FdT)GAGGAGGT-3′. Reaction mixture (200 μl), containing fluorescein-labeled DNA (10 nm) in buffer (20 mm Tris-HCl (pH 8.0), 1 mm DTT, 1 mm EDTA, 4 μg of RNase A), was incubated with increasing concentrations (0–600 nm) of AID at room temperature. Rotational anisotropy was measured using a QuantaMaster QM-1 fluorometer (Photon Technology International) with a single emission channel. Samples were excited with vertically polarized light at 494 nm, and both vertical and horizontal emission were monitored at 520 nm (7 nm band pass). The microscopic dissociation constant (KA) is defined as the concentration of AID at which half of the total ssDNA is bound.
The KA and Hill coefficient parameters were calculated by fitting the data to either a rectangular hyperbola for noncooperative binding or to a sigmoidal curve for cooperative binding using Sigma Plot 10.0 software. The data were calculated from three independent experiments.
Structural Modeling
The 3D-JIGSAW comparative modeling program (24) was used to generate the AID model using the crystal structure of Apo3G-CD2 (Protein Data Bank code 3IQS) as a template. The root mean square deviation value was determined by superposition of the AID model with the Apo3G-CD2 structure in PyMOL (25). Structure figures were created using PyMOL.
RESULTS
There are 23 base substitution mutations occurring at 20 distinct sites in AID that have been identified in patients suffering from HIGM-2 syndrome (10–12). Three of the mutated sites contain two different amino acids. These missense mutations are distributed over the entire gene for AID (Fig. 1). Two C-terminal deletion mutants of AID, involving the loss of either 9 or 17 amino acids, have also been found in individuals with HIGM-2 syndrome (Fig. 1) (14). AID exhibits strong homology with the catalytically active CD2 domain of Apo3G. We have used the x-ray crystal structure of Apo3G-CD2 (17) as a surrogate to identify the corresponding locations of each mutation in AID (Fig. 1) and to explore how catalysis and ssDNA binding might be influenced. The structural analysis allows us to group the HIGM-2 point and C-terminal deletion mutants into four classes that reflect the contributions of each of the mutants to catalysis (class I), AID-DNA substrate interactions (class II), structural integrity (class III), and C-terminal deletions (class IV).
A Biochemical Analysis of HIGM-2 AID Amino Acid Substitution Mutants
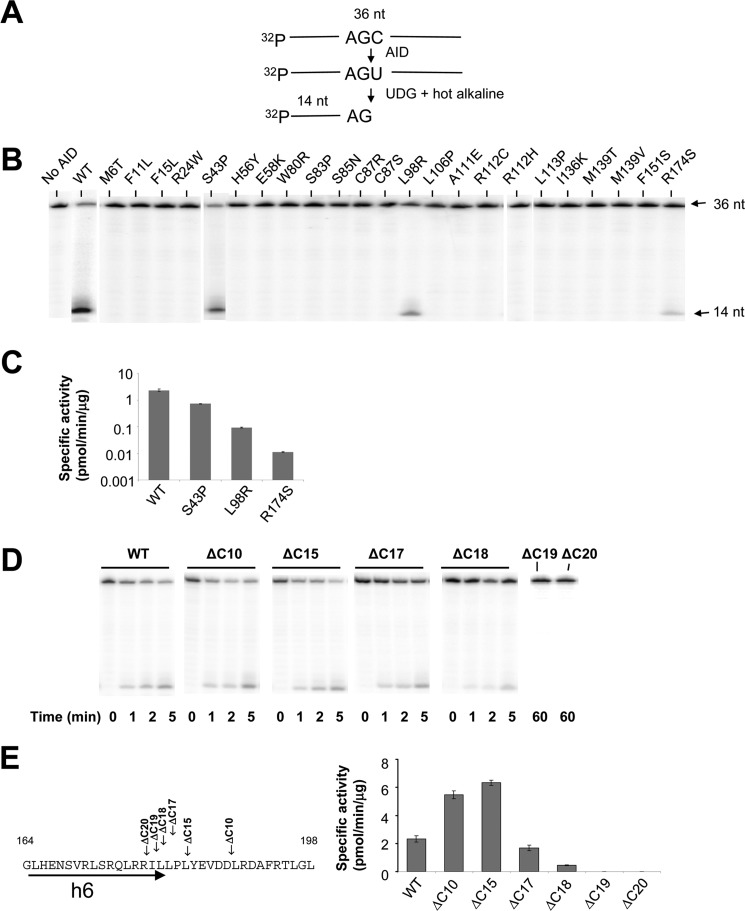
Using a 32P-based polyacrylamide gel assay to measure AID-catalyzed deamination of C→U (see under “Experimental Procedures” and Fig. 2A), we found that of the 23 HIGM-2 point mutants, 18 appear to be catalytically inactive, having no detectable C-deamination activity in an AGC hot motif (Fig. 2B). The five catalytically active mutants are S43P, L98R, R174S, F15L, and R112C. Of these, S43P, L98R, and R174S show activity in a 32P gel-based assay (Fig. 2B). S43P (13) has about ⅓ the specific activity of WT AID (Fig. 2C), in agreement with previous data (21). L98R and R174S have not been characterized previously. L98R is 25-fold less active than WT AID, and R174S is 200-fold less active than WT AID (Fig. 2C). F15L and R112C, which appear to be inactive in the 32P-based gel assay (Fig. 2B), have detectable deamination activity in a high sensitivity M13 lacZ mutation reporter assay (“Experimental Procedures”) (Table 1) (5, 21). C→T mutations initiated by AID-catalyzed C→U deaminations in lacZ were measured in all 16 triplet hot (WRC), cold (SYC), and neutral motifs (Table 2). The concentrations of AID and ssDNA and incubation times were chosen so that ratio of mutant (white)/WT(blue) plaques was <5% (5, 21). These conditions were used to ensure that each substrate is acted on by no more than a single AID molecule, as prescribed by Poisson statistics (26). DNA clones isolated from individual mutant plaques were sequenced to determine the number of deaminated motifs per clone and the frequency of deaminations occurring in hot, cold, and neutral motifs in lacZ.
FIGURE 2.
Analysis of deamination activity of WT and mutant AID on an ssDNA oligonucleotide substrate. A, schematic representation of the C-deamination assay. A 32P-labeled 36-nt substrate was incubated with AID followed by UDG and hot alkali treatment to generate a 14-nt fragment product. B, screening for deamination activity for AID HIGM-2 missense mutants. WT AID (10 nm) or mutant AID (10 to 100 nm) was incubated with ssDNA substrate for 60 min at 37 °C. The appearance of a 14-nt fragment product demonstrates that three AID missense mutants (S43P, L98R, and R174S) retain deamination activity with specific activities shown in C. D and E, analysis of ssDNA deamination (D) and specific activity (E) of C-terminal deletion AID mutants. AID protein concentrations and incubation times used to calculate specific activities are described under “Experimental Procedures.” Error bars represent ±1 S.E. derived from ≥3 independent measurements.
TABLE 1.
AID-catalyzed deaminations in a lacZa reporter gene on an M13-gapped DNA
WT and mutant AID activity on lacZa gap assay were monitored as an increase in frequency of mutated (white or light blue) plaques over the background mutant frequency without AID (5).
| AID mutant | No. of mutant plaques | Total plaques | Mutant frequency |
|---|---|---|---|
| WT | 126 | 6104 | 206 × 10−4 |
| Class I (catalysis) | |||
| H56Y | 0 | 7546 | <1.3 × 10−4 |
| E58K | 0 | 9947 | <1.0 × 10−4 |
| C87R | 3 | 7177 | 4.2 × 10−4 |
| C87S | 0 | 5011 | <2.0 × 10−4 |
| Class II (substrate interaction) | |||
| R24W | 0 | 12,634 | <0.8 × 10−4 |
| S83P | 0 | 13,143 | <0.8 × 10−4 |
| S85N | 0 | 14,383 | <0.7 × 10−4 |
| A111E | 6 | 29,437 | 2.0 × 10−4 |
| R112C | 13 | 6989 | 18.6 × 10−4 |
| R112H | 1 | 9488 | 1.1 × 10−4 |
| L113P | 0 | 9290 | <1.1 × 10−4 |
| R174S | 46 | 13,203 | 34.8 × 10−4 |
| Class III (structural integrity) | |||
| M6T | 0 | 5636 | <1.8 × 10−4 |
| F11L | 1 | 8415 | 1.2 × 10−4 |
| F15L | 56 | 13,741 | 40.8 × 10−4 |
| W80R | 2 | 16,623 | 1.2 × 10−4 |
| L98R | 368 | 20,851 | 176.5 × 10−4 |
| L106P | 0 | 4768 | <2.1 × 10−4 |
| I136K | 0 | 17,526 | <0.6 × 10−4 |
| M139T | 0 | 11,400 | <0.9 × 10−4 |
| M139V | 0 | 7935 | <1.3 × 10−4 |
| F151S | 0 | 10,511 | <1.0 × 10−4 |
| S43P | 73 | 8332 | 87.6 × 10−4 |
| Class IV (C-terminal deletion) | |||
| ΔC10 | 101 | 11,840 | 85.3 × 10−4 |
| ΔC15 | 226 | 23,232 | 97.3 × 10−4 |
| ΔC19 | 3 | 11,300 | 2.7 × 10−4 |
| ΔC20 | 0 | 15,304 | <0.7 × 10−4 |
| No AID | 4 | 8271 | 4.8 × 10−4 |
TABLE 2.
Mutability index for WT and AID mutants
The mutability index is defined as the number of times a given trinucleotide motif within a segment of DNA contains a mutation, divided by the number of times the oligonucleotide would be expected to be mutated for a mechanism with no sequence bias (5, 40).
| Motif | Mutability index |
||||||
|---|---|---|---|---|---|---|---|
| WT | F15L | S43P | L98R | R174S | ΔC15 | ΔC10 | |
| Hot spots | |||||||
| AAC | 1.75 | 1.65 | 1.87 | 1.72 | 1.84 | 1.65 | 2.21 |
| AGC | 1.61 | 1.55 | 1.97 | 1.48 | 1.74 | 1.59 | 2.15 |
| TAC | 2.05 | 2.10 | 1.84 | 2.38 | 2.26 | 2.20 | 2.13 |
| TGC | 1.97 | 1.93 | 2.01 | 2.16 | 1.90 | 2.06 | 2.96 |
| MI average (WRC) | 1.85 | 1.81 | 1.92 | 1.94 | 1.94 | 1.88 | 2.36 |
| Cold spots | |||||||
| CCC | 0.50 | 0.59 | 0.40 | 0.59 | 0.56 | 0.42 | 0.32 |
| CTC | 0.68 | 0.53 | 0.46 | 0.54 | 0.48 | 0.53 | 0.33 |
| GCC | 0.32 | 0.15 | 0.13 | 0.17 | 0.27 | 0.13 | 0.02 |
| GTC | 0.53 | 0.31 | 0.46 | 0.29 | 0.45 | 0.30 | 0.46 |
| MI average (SYC) | 0.51 | 0.40 | 0.36 | 0.40 | 0.44 | 0.35 | 0.28 |
| Intermediates | |||||||
| ACC | 0.88 | 0.94 | 0.62 | 0.82 | 0.94 | 0.91 | 0.56 |
| ATC | 1.10 | 1.44 | 0.86 | 1.61 | 2.11 | 0.98 | 0.59 |
| CAC | 0.99 | 1.08 | 0.91 | 0.92 | 1.01 | 1.09 | 0.81 |
| CGC | 1.09 | 1.00 | 1.52 | 1.01 | 1.03 | 1.25 | 1.47 |
| GAC | 0.43 | 0.44 | 0.58 | 0.44 | 0.19 | 0.65 | 0.59 |
| GGC | 0.72 | 0.76 | 0.77 | 0.77 | 0.56 | 0.81 | 0.42 |
| TCC | 1.03 | 1.02 | 0.89 | 0.99 | 0.63 | 0.84 | 0.64 |
| TTC | 0.76 | 0.98 | 0.71 | 0.93 | 0.82 | 0.83 | 0.62 |
| MI average | 0.88 | 0.96 | 0.86 | 0.94 | 0.91 | 0.92 | 0.71 |
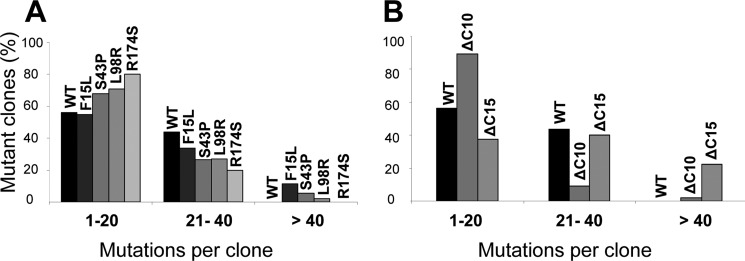
We measured the clonal distribution of C→T lacZ mutations for WT AID and four AID mutants S43P, L98R, R174S, and F15L (Fig. 3A). The average number of mutations/clone is about the same for WT AID and for each AID mutant, WT AID (15 mutations), L98R (15 mutations), R174S (12 mutations), and F15L (21 mutations). WT AID generates 1–20 mutations in 55% of the clones and 21–40 mutations in 45% of the clones. A similar broad mutational distribution also occurs for each AID mutant (Fig. 3A). For L98R, 70% of the clones have 1–20 mutations and 30% have 21–50 mutations; for R174 S, 80% of the clones have 1–20 mutations and 20% have 21–40 mutations; for F15L, 55% of the clones have 1–20 mutations and 45% have 21–60 mutations. Thus, once bound to an ssDNA substrate, each AID mutant catalyzes processive multiple deaminations for the entire length of lacZ and has a spatial distribution statistically similar to WT AID (supplemental Fig. S1). Representative clones for each mutant class contain combinations of singletons and clusters of mutations suggesting repetitive sliding and hopping/jumping movement on ssDNA (26) (supplemental Fig. S2).
FIGURE 3.
Distribution of WT and mutant AID-catalyzed deaminated M13 phage clones in a lacZa target gene. AID deaminations in lacZa are detected as C→T mutations in white or light blue M13 plaques. The distribution of clones with C→T mutations is tabulated as a percentage of clones containing 1–20, 21–40, or >40 mutations. A, mutant clones (%) for WT AID and F15L, S43P, L98R, and R174S HIGM-2 AID missense mutants, and B, mutant clones (%) for WT AID and ΔC10 and ΔC15 C-terminal deletion mutants.
A calculation of the mutability index (27), which is defined as the observed frequency that each trinucleotide motif (XYC) is deaminated compared with the expected frequency with no sequence bias, provides quantitative comparison of mutant and WT AID deamination specificities (5, 21). We have determined the MI values in the 16 trinucleotide motifs for WT AID and four of the point mutants, L98R, R174S, F15L, and S43P (Table 2). WT AID and three AID mutants have about the same average MI for WRC hot (∼1.9), intermediate (∼0.9), and SYC (∼0.4 to 0.5) cold motifs (Table 2). The results for S43P agree with previous data (21). Because there are few mutants in each motif, differences in WT and mutant AID MI values for individual motifs, typically less than 2-fold (Table 2), most likely result from statistical fluctuations. Although L98R, R174S, and F15L show large reductions in activity compared with WT AID, they nevertheless catalyze processive deaminations with mutation spectra and motif specificities similar to WT AID (Table 2; supplemental Fig. S1).
Biochemical Properties of AID C-terminal Deletion Mutants
The C-terminal region binds proteins targeting AID to Ig switch region DNA (28, 29). Two naturally occurring mutations causing a loss of 9 and 17 C-terminal amino acids of AID (ΔC9 and ΔC17) were found in HIGM-2 patients (14); the patient with ΔC17 also has an insertion in the C-terminal region (14). A ΔC10 mouse also has an IgM phenotype (30). In these examples, SHM is almost normal (14, 30), suggesting that AID retains significant C deamination activity when 17 C-terminal amino acids are deleted. To determine the effect of deleting C-terminal amino acids on AID activity, we have constructed C-terminal deletion mutants (ΔC10, ΔC15, ΔC17, ΔC18, ΔC19, and ΔC20). The ΔC10 and ΔC15 mutants are ∼2.5-fold more active than WT AID, and ΔC17 is slightly less active (Fig. 2, D and E). AID is inactivated by removal of 19 C-terminal amino acids (Fig. 2, D and E). The C-terminal deletion mutants retain high processivity. For example, ΔC10 generates 1–20 mutations in 40% of the clones and 21–80 mutations in 60% of the clones (Fig. 3B). The average MI for the C-terminal deletion mutants is similar to WT AID for the hot and neutral motifs, but fewer deaminations are found in GCC causing an overall lower average MI for cold motifs (Table 2).
Binding WT and Mutant AID to ssDNA
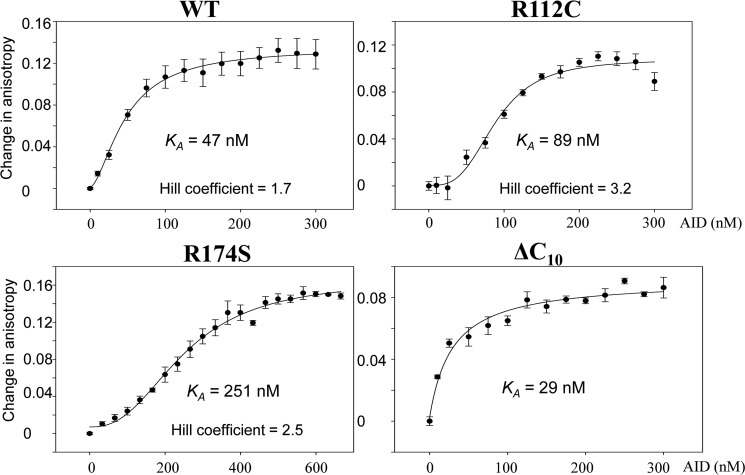
Binding of AID to fluorescein-labeled DNA was measured by an increase in steady-state rotational anisotropy (Fig. 4 and supplemental Fig. S3). WT AID binds to ssDNA with an apparent KA ∼47 nm (Fig. 4). A least squares fit to a sigmoidal curve (Hill coefficient ∼1.7) is much better than to rectangular hyperbola, suggesting the possibility that WT AID might bind cooperatively as a dimer.
FIGURE 4.
WT and mutant AID binding to ssDNA. AID-ssDNA binding was measured by rotational anisotropy using a fluorescein-labeled 36-nt oligonucleotide substrate. The changes in rotational anisotropy with increasing AID concentrations were fit to either a sigmoidal or hyperbolic binding curve. Values for each data point represent the mean ± S.E. determined from three independent measurements.
The active site mutants H56Y and E58K bind ssDNA (supplemental Fig. S3A). Thus, the absence of the detectable C deamination activity in both assays (Fig. 1B and Table 1) is unlikely to result from an inability to bind ssDNA. We note, however, that a complete saturation binding curve was not obtainable for the class I mutants (supplemental Fig. S3) because of limitations in obtaining sufficiently high concentrations of purified proteins (<10 μg/ml).
There are eight HIGM-2 class II (substrate interaction) point mutants. Only one, R174S, has measurable deamination activity in the 32P gel-based assay (1/200th compared with WT AID, see Fig. 2, B and C) and is active in the M13 plaque assay (Table 1). R174S binds to ssDNA cooperatively but with about 5.3-fold lower apparent affinity compared with WT AID (Fig. 4). R112C has a low level of activity above background in the plaque assay but is inactive in the 32P biochemical assay (Fig. 2B). R112C also binds ssDNA cooperatively having a 2-fold reduced affinity compared with WT AID (Fig. 4). R112H and L113P are inactive yet bind ssDNA cooperatively with similar affinities as WT AID (supplemental Fig. S3B).
The class III structural integrity mutant S43P is the most active of the HIGM-2 point mutants, ⅓ as active as WT AID in the 32P assay (Fig. 2, B and C). S43P binds ssDNA with a 4-fold higher apparent affinity (KA ∼12 nm, see supplemental Fig. S4) compared with WT AID (KA ∼47 nm, see Fig. 4). Binding is cooperative with a Hill coefficient ∼1.5, similar to WT AID (Hill coefficient ∼1.7). L98R is 25-fold less active than WT AID in the 32P assay (Fig. 2, B and C). The low activity L98R mutant does not bind DNA cooperatively (supplemental Fig. S4), whereas F15L, which is weakly active in the lacZ plaque assay, binds DNA cooperatively as does inactive W80R (supplemental Fig. S4), having apparent affinities similar to WT AID. Catalytically inactive L106P binds DNA with relatively high affinity and cooperativity (KA ∼6.9 nm and Hill coefficient ∼2.9, see supplemental Fig. S4).
The active C-terminal deletion mutants (ΔC10, ΔC15, and ΔC18) bind ssDNA about the same as WT AID (KA ∼29–42 nm, see Fig. 4 and supplemental Fig. S5). However, unlike WT AID, binding is not cooperative. Catalytically inactive ΔC20 binds ssDNA with a much lower apparent affinity (KA ∼137 nm), yet exhibits clear sigmoidal binding, with a Hill coefficient ∼2 (supplemental Fig. S5).
The data suggest that the absence of activity for the various classes of point mutants does not reflect a failure to bind ssDNA nor does cooperative binding appear to play a major role. There is also no correlation of activity with binding affinity or cooperativity for the deletion mutants. Instead, insights into the biochemical properties of the missense and deletion mutants might reflect structural perturbations in AID that can be revealed through comparative homology modeling with Apo3G-CD2.
Structural Evaluation of HIGM-2 Point Mutants and C-terminal Deletion CSR Mutants Using Comparative Homology Modeling
We previously solved the crystal structures of truncated Apo2 and the C-terminal domain of Apo3G (Apo3G-CD2) (16, 17). Four other structures (three NMR and one crystal) of Apo3G-CD2 have also been solved (17–20). Apo2 and Apo3G-CD2 show substantial overlap of an overall core structure containing a five β-strand sheet surrounded by six α-helices. All APOBEC family proteins share considerable sequence homology between the family members. AID shares 44% homology (31.1% identity and 44% similarity) with Apo2 and 54% homology (39.4% identity and 54.3% similarity) with Apo3G-CD2.
Because of the high sequence homology between AID and Apo-3G-CD2, the x-ray crystal structure of Apo3G-CD2 was used to generate a comparative homology model of AID (Fig. 1). Mapping the HIGM-2 and C-terminal mutations onto the AID model can provide insight into the structural consequences of AID mutations on biochemical function. Superposition of the “homology AID model” onto the A3G-CD2 structure gives a low global root mean square deviation of 0.173 Å, providing validation that the modeling is likely to provide an accurate picture of salient biochemical properties of AID, subject to the caveat that at present there is no APOBEC protein-nucleic acid co-crystal structure. Each AID mutant was examined in terms of how a substituted amino acid (HIGM-2 point mutant) or loss of amino acids (C-terminal deletions) might influence catalytic and ssDNA interaction properties. The AID mutants lie in four distinct classes as follows: catalysis (class I), ssDNA substrate interactions (class II), structural integrity (class III), and C-terminal deletions (class IV).
Class I active center residues are responsible for catalysis (Fig. 1A, magenta sticks). The point mutants (H56Y, E58K, and C87R/C87S) abolish C deaminase activity (Fig. 2A). The H56Y and C87R/C87S mutants presumably are no longer able to coordinate the catalytic Zn2+ ion in the enzyme active center, whereas E58K may lose the ability to mediate proton shuffling needed for C deamination (16–20).
Class II mutants (R24W, S83P, S85N, A111E, R112C/R112H, L113P, and R174S, Fig. 1A, cyan sticks) contain amino acid residues located on or near the protein surface; these mutants are predicted to alter the AID interaction with the DNA substrate. Of the seven predicted substrate interaction mutants, six (R24W, S83P, S85N, A111E, R112C, and L113P) showed no detectable activity in the 32P C deamination assay (Fig. 2B), nor did they give rise to C→T mutations in the highly sensitive lacZa mutational assay (Table 1). Arg-24, conserved for the two available APOBEC structures, Apo2 (16) and Apo3G-CD2 catalytic domain (17), is located on the putative flexible active center loop 1 (AC-loop 1), and has been shown to be essential for DNA binding (31). R24W shows no measurable C deamination activity (Fig. 2B). S83P and S85N are located next to the conserved Trp-84 that may be involved in positioning the target cytidine into the active center; therefore, mutations in this surrounding area likely inhibit deamination activity. The “specificity loop” (loop 7) has previously been shown to contain residues that determine an APOBEC-specific hot spot deamination motif within the ssDNA target substrate (32–34). Mutations in this loop (A111E, R112C, and L113P) may alter the stability of the specificity loop, leading to the inability of AID to properly position the substrate in the active center.
We were able to purify and concentrate a sufficient amount of protein to measure ssDNA binding for two class II mutants, R174S and R112C (Fig. 4). R174S shows strongly reduced but detectable deamination activity in the 32P assay (Fig. 2, B and C). R174S binds to ssDNA weakly, but cooperatively, having a 5-fold higher apparent KA (250 nm) compared with WT AID (47 nm) (Fig. 4). In contrast, R112C, which has no measurable deamination activity in either assay (Fig. 2B, Table 1), nevertheless binds cooperatively to ssDNA (Hill coefficient = 3.2, KA = 89 nm; Fig. 4). Although substrate binding is reduced for both mutants, the substantial loss in activity may result from improper motif alignment in the active cleft. In the case of the conserved residue Arg-112, its role may be to maintain the structural integrity of the motif specificity loop (loop-7), where Arg-112 is located just below the active center at the N terminus of loop-7 (Fig. 1) (33). Arg-174 is located on α-helix 6 (Fig. 1) (33), which has also been shown to be important in ssDNA binding and deamination (17). We refer here to an “equivalency” of AID residue Arg-174 with Apo3G Arg-374 and Arg-376 (Fig. 1) (33), where we have shown that the Apo3G-CD2 R374D/R376D mutant shows a reduced binding and catalytic activity (17), similar to that shown by AID R174S.
Class III structural integrity mutants are composed of nine conserved hydrophobic amino acid residues (Met-6, Phe-11, Phe-15, Trp-80, Leu-98, Leu-106, Ile-136, Met-139, and Phe-151) located in the interior of AID (Fig. 1B, green sticks) (33) and one amino acid (S43P) that likely disrupts the stability of the β2-strand. The introduction of charged residues in the hydrophobic core of an enzyme typically can result in a major disruption of protein folding and an attendant loss of catalytic activity. Based on the structures for Apo2 (16) and Apo3G-CD2 (17), W80R, I136K, and L106P are predicted to destabilize α-helices 2 and 3 (Fig. 1). None of these mutants show detectable activity in either assay (Fig. 2B and Table 1). M139V and F151S are predicted to destabilize α-helix 5, which packs on one side of the five-stranded β-sheet core structure, by disrupting hydrophobic packing of α-helix 5 (Fig. 1). These two mutants are also catalytically dead in both assays (Fig. 2B and Table 1). A complete loss of deamination function was also observed for Apo3G when mutations were introduced at residues corresponding to Met-139 and Phe-151 (M338A and F350A) (35).
α-Helix 1 is likely to be destabilized by mutants F11L and F15L, present in the helix, and by M6T, which is located nearby on the N-terminal loop of helix 1 (33). α-Helix 1 is the only helix that varies in position between the Apo2 and A3G-CD2 structures, probably because these structures are N-terminal deletion constructs. Nevertheless, in Apo2 and all but one of the A3G-CD2 structures (NMR Protein Data Bank code 2KBO is different), α-helix 1 is anchored to the structure through hydrophobic bond interactions with α-helix 5 and the C-terminal region of the β1-strand. The stabilization of α-helix 1 also helps stabilize the open conformation of AC-loop 1, which has previously been predicted to be part of the APOBEC substrate groove (17). Mutations on AC-loop 1 of AID and Apo3G inhibit deaminase activity, most likely by disrupting the AC-loop 1 conformation and substrate-binding groove (17, 31). HIGM-2 mutants F11L, F15L, and M6T may disrupt α-helix 1 hydrophobic packing to the structure resulting in destabilization of the AC-loop 1 and disruption of the AID substrate groove shown by a complete loss of deamination activity for F11L and M6T in both assays (Fig. 2B and Table 1). However, F15L, although it has no detectable catalytic activity in the 32P substrate cleavage assay (Fig. 2B), is active in the more sensitive M13-lacZ assay (Table 1). Remarkably, DNA sequenced from the mutant plaques shows that F15L catalyzes highly processive C deaminations similar to WT AID (Fig. 3) suggesting that perhaps about 2% of the F15L mutants may retain sufficient structural integrity of the AC-loop 1 to permit an essentially normal deamination pattern. However, the absence of a detectable 32P cleavage band (Fig. 2B) implies that the specific activity of F15L is far less than 1/50th of WT AID. The detection limit for the assay is ∼1/200th of WT AID (see, e.g. R174S, Fig. 2B).
Class IV mutants consist of a series C-terminal deletions (ΔC10, ΔC15, ΔC17, ΔC18, ΔC19, and ΔC20). It has been suggested that the C terminus is essential for binding proteins such as UNG, MSH2, 14-3-3, and DNA-PKcs required to target AID to switch-region (28, 29, 36). For the HIGM-2 C-terminal mutants, although CSR is absent, SHM is almost normal (14, 30), suggesting that AID retains a considerable level of C deamination activity when as many as 17 C-terminal amino acids have been removed. We find that ΔC10 and ΔC15, are ∼2.5-fold more active than WT AID (Fig. 2E), accompanied by ∼2-fold tighter binding and a loss of binding cooperativity (Fig. 4). A reduction in activity compared with WT AID begins to be apparent for ΔC17, is much more pronounced for ΔC18, and activity is abolished for ΔC19 and ΔC20 (Fig. 2, D and E). An alignment with APOBEC single domains indicates that AID has an extended C-terminal tail (Fig. 1C) that differs from all other APOBEC proteins and therefore is not visualized in any of the previously solved APOBEC structures. A secondary structure prediction (PredictProtein) (37) of this region suggests that residues within this 13-amino acid tail likely form a helix (“putative” α-helix 7). The C terminus of α-helix 6 is near the predicted substrate groove. The loss of activity for ΔC19 may result from a partial truncation of the C-terminal portion of the structured α-helix 6, which could destabilize the helix. Helix 6 contains residues important for AID (Arg-174) and Apo3G (Arg-374/ Arg-376) DNA binding (see DNA binding data, Fig. 4) (17).
DISCUSSION
Following low affinity Ab formation by V(D)J recombination (38), AID initiates Ab diversity by deaminating C→U, during transcription of Ig variable and switch regions DNA (8, 9). The absence of a repertoire of high affinity Abs resulting from a loss of either SHM or CSR, although not fatal, nonetheless results in a severe loss of immune response (39). HIGM-2 syndrome, characterized by an inability to carry out CSR needed to convert IgM to IgG, IgA, or IgE, is caused predominantly by missense mutations in AID that occur over almost the entire gene. HIGM-2 missense mutations are deficient in SHM and CSR. In contrast, the C-terminal deletions, ΔC9 and ΔC17, cause a loss of CSR, but SHM is virtually normal (14). The widespread distribution of HIGM-2 mutations encompassing the entire AID gene (Fig. 1) provided the impetus to explore the functional biochemical properties for these mutants in relation to the predicted structural properties of AID.
The “holy grail,” a high resolution structure for AID either alone or bound to ssDNA, is not available. However, there are two high resolution apo-APOBEC protein structures, one for Apo2 (16) and another for Apo3G-CD2 (17), which offer a next-best strategy to model AID. Apo2 and Apo3G-CD2 show strong homologies with AID (33), and both structures have served successfully as surrogates to predict how mutations in conserved corresponding amino acid residues in AID abolish catalysis directly or by inhibiting binding to DNA substrates or via improper positioning of the deamination motif relative to the catalytic zinc atom (17, 33).
We have placed the 23 missense mutations at 20 sites in AID into three classes based on the structural analysis, and four of these retain partial catalytic activity. None of the four class I active center mutants have detectable C-deaminase activity in either the 32P-cleavage assay (Fig. 2) or lacZ reporter assay (Table 1). One of eight class II DNA substrate interaction mutants, R174S, is active at a level of 1/200th compared with WT AID (Fig. 2, B and C). Two of 11 class III structural integrity mutants are active in the 32P assay, S43P having ⅓ and L98R with 1/20th the activity of WT AID, respectively (Fig. 2, B, and C). Class III F15L, inactive in the 32P assay, is active in the lacZ M13 reporter assay (Table 1).
An inability to bind ssDNA would account a priori for an absence of activity. Yet many of the catalytically inactive mutants bind ssDNA with apparent affinities comparable with WT AID. For example, two of the dead class II substrate interaction mutants, R112H (KA ∼40 nm) and L113P (KA ∼35 nm), bind ssDNA with apparent affinities indistinguishable from WT AID (KA ∼47 nm), each showing cooperative binding with a Hill coefficient of about 2 (Fig. 4 and supplemental Fig. S3B). Inactive class III structural integrity mutants also mimic WT AID-ssDNA binding, F15L (KA ∼31 nm, Hill coefficient ∼2.2) and W80R (KA ∼35 nm, Hill coefficient ∼2.9) (supplemental Fig. S4). The inactive L106P class III mutant binds cooperatively to ssDNA with about 7-fold higher affinity than WT AID (KA ∼6.9 nm, Hill coefficient ∼3) (supplemental Fig. S4). The catalytically active S43P, which has ⅓ the specific activity of WT AID, binds ssDNA with about a 4-fold higher affinity (supplemental Fig. S4).
WT AID-catalyzed deaminations are targeted to trinucleotide motifs, with WRC hot motifs favored over SYC cold motifs (5, 26). The HIGM-2 point mutants and the C-terminal deletion mutants all have motif specificities similar to WT AID, with relatively small differences for individual motif MIs for the various mutants (Table 2). A distinctive property of WT AID is its ability to catalyze multiple deaminations processively, with a single molecule sliding and hopping/jumping along a single ssDNA molecule, prior to switching to a different DNA substrate (5, 21, 23, 26). S43P and L98R, which have ⅓ and 1/20th the specific activity of WT AID, respectively, have about the same distributions of mutations/clone as WT AID, with about 2–5% exceeding 40 mutations (Fig. 3). The least active of the HIGM-2 AID missense mutants, R174S (1/200th compared with WT AID) (Fig. 2, B and C) and F15L, with no detectable activity in the 32P gel-based assay (Fig. 2B), are processive; remarkably, about 12% of F15L clones have more than 40 mutations (Fig. 3). The combined data show that, except for having reduced deamination activities, the “active” missense mutants exhibit properties similar to WT AID with respect to motif specificity, substrate binding, processivity, mutation spectra (supplemental Fig. S1), and spatial deamination patterns (supplemental Fig. S2).
It is well established that the C-terminal region is needed for binding proteins that target AID to Ig switch regions DNA (28, 29, 36). Deletions in this region cause a loss of CSR, but not SHM (14, 30). Our biochemical data show that significant deamination activity is retained for deletions of as many as 18 C-terminal amino acids, whereas removal of 19 amino acid residues abolished activity. Because deletions of 19 or more amino acids lead to a partial truncation of the structured α-helix 6, our data suggested that whole α-helix 6 is required for the structural integrity of the core deaminase domain of AID, as well as of APOBEC proteins. With the exception of a replacement of amino acids located directly in the catalytic active site (H56Y, E58K, and C87R/C87S), it was not clear why the remaining 19 HIGM-2 missense mutants were unable to support either CSR or SHM. Our surrogate structural model for AID suggests how a wide variety of single amino acid substitutions over almost the entire gene are able to strongly suppress, and in most instances, completely abolish AID catalytic activity. These seemingly more subtle effects involve perturbations to the specificity loop, hydrophobic core, and secondary structures.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM21422 and ES013192 (to M. F. G.) and GM087986 (to X. S. C.).

This article contains supplemental Figs. S1–S5.
- HIGM-2
- hyper-IgM syndrome type 2
- AID
- activation-induced deoxycytidine deaminase
- Apo2
- Apobec2
- Apo3G-CD2
- C-terminal catalytic domain of Apobec3G
- V
- variable region
- CSR
- class switch recombination
- SHM
- somatic hypermutation
- ssDNA
- single-stranded DNA
- nt
- nucleotide
- MI
- mutability index.
REFERENCES
- 1. Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., Tezcan I., Ersoy F., Kayserili H., Ugazio A. G., Brousse N., Muramatsu M., Notarangelo L. D., Kinoshita K., Honjo T., Fischer A., Durandy A. (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102, 565–575 [DOI] [PubMed] [Google Scholar]
- 2. Minegishi Y., Lavoie A., Cunningham-Rundles C., Bédard P. M., Hébert J., Côté L., Dan K., Sedlak D., Buckley R. H., Fischer A., Durandy A., Conley M. E. (2000) Mutations in activation-induced cytidine deaminase in patients with hyper-IgM syndrome. Clin. Immunol. 97, 203–210 [DOI] [PubMed] [Google Scholar]
- 3. Imai K., Zhu Y., Revy P., Morio T., Mizutani S., Fischer A., Nonoyama S., Durandy A. (2005) Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2. Clin. Immunol. 115, 277–285 [DOI] [PubMed] [Google Scholar]
- 4. Bransteitter R., Pham P., Scharff M. D., Goodman M. F. (2003) Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 100, 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pham P., Bransteitter R., Petruska J., Goodman M. F. (2003) Processive AID-catalyzed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424, 103–107 [DOI] [PubMed] [Google Scholar]
- 6. Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F. W. (2003) Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730 [DOI] [PubMed] [Google Scholar]
- 7. Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA-editing enzyme. Cell 102, 553–563 [DOI] [PubMed] [Google Scholar]
- 8. Peled J. U., Kuang F. L., Iglesias-Ussel M. D., Roa S., Kalis S. L., Goodman M. F., Scharff M. D. (2008) The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 26, 481–511 [DOI] [PubMed] [Google Scholar]
- 9. Stavnezer J., Guikema J. E., Schrader C. E. (2008) Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26, 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Durandy A. (2009) Immunoglobulin class switch recombination. Study through human natural mutants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Durandy A., Peron S., Taubenheim N., Fischer A. (2006) Activation-induced cytidine deaminase. Structure-function relationship as based on the study of mutants. Hum. Mutat. 27, 1185–1191 [DOI] [PubMed] [Google Scholar]
- 12. Kracker S., Durandy A. (2011) Insights into the B cell-specific process of immunoglobulin class switch recombination. Immunol. Lett. 138, 97–103 [DOI] [PubMed] [Google Scholar]
- 13. Zhu Y., Nonoyama S., Morio T., Muramatsu M., Honjo T., Mizutani S. (2003) Type two hyper-IgM syndrome caused by mutation in activation-induced cytidine deaminase. J. Med. Dent. Sci. 50, 41–46 [PubMed] [Google Scholar]
- 14. Ta V. T., Nagaoka H., Catalan N., Durandy A., Fischer A., Imai K., Nonoyama S., Tashiro J., Ikegawa M., Ito S., Kinoshita K., Muramatsu M., Honjo T. (2003) AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 4, 843–848 [DOI] [PubMed] [Google Scholar]
- 15. Chelico L., Pham P., Petruska J., Goodman M. F. (2009) Biochemical basis of immunological and retroviral responses to DNA-targeted cytosine deamination by activation-induced cytidine deaminase and APOBEC3G. J. Biol. Chem. 284, 27761–27765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prochnow C., Bransteitter R., Klein M. G., Goodman M. F., Chen X. S. (2007) The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature 445, 447–451 [DOI] [PubMed] [Google Scholar]
- 17. Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008) Crystal structure of the antiviral APOBEC3G catalytic domain and functional implications. Nature 456, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Furukawa A., Nagata T., Matsugami A., Habu Y., Sugiyama R., Hayashi F., Kobayashi N., Yokoyama S., Takaku H., Katahira M. (2009) Structure, interaction, and real time monitoring of the enzymatic reaction of wild type APOBEC3G. EMBO J. 28, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen K. M., Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., Matsuo H. (2008) Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452, 116–119 [DOI] [PubMed] [Google Scholar]
- 20. Harjes E., Gross P. J., Chen K. M., Lu Y., Shindo K., Nowarski R., Gross J. D., Kotler M., Harris R. S., Matsuo H. (2009) An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J. Mol. Biol. 389, 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pham P., Smolka M. B., Calabrese P., Landolph A., Zhang K., Zhou H., Goodman M. F. (2008) Impact of phosphorylation and phosphorylation-null mutants on the activity and deamination specificity of activation-induced cytidine deaminase. J. Biol. Chem. 283, 17428–17439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shinkura R., Ito S., Begum N. A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. (2004) Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 7, 707–712 [DOI] [PubMed] [Google Scholar]
- 23. Bransteitter R., Pham P., Calabrese P., Goodman M. F. (2004) Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 279, 51612–51621 [DOI] [PubMed] [Google Scholar]
- 24. Bates P. A., Kelley L. A., MacCallum R. M., Sternberg M. J. (2001) Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins Suppl. 5, 39–46 [DOI] [PubMed] [Google Scholar]
- 25. Schrödinger, LLC (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 26. Pham P., Calabrese P., Park S. J., Goodman M. F. (2011) Analysis of a single-stranded DNA-scanning process in which activation-induced deoxycytidine deaminase (AID) deaminates C to U haphazardly and inefficiently to ensure mutational diversity. J. Biol. Chem. 286, 24931–24942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shapiro G. S., Aviszus K., Murphy J., Wysocki L. J. (2002) Evolution of Ig DNA sequence to target specific base positions within codons for somatic hypermutation. J. Immunol. 168, 2302–2306 [DOI] [PubMed] [Google Scholar]
- 28. Ranjit S., Khair L., Linehan E. K., Ucher A. J., Chakrabarti M., Schrader C. E., Stavnezer J. (2011) AID binds cooperatively with UNG and Msh2-Msh6 to Ig switch regions dependent upon the AID C terminus. J. Immunol. 187, 2464–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu Z., Fulop Z., Wu G., Pone E. J., Zhang J., Mai T., Thomas L. M., Al-Qahtani A., White C. A., Park S. R., Steinacker P., Li Z., Yates J., 3rd, Herron B., Otto M., Zan H., Fu H., Casali P. (2010) 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 17, 1124–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barreto V., Reina-San-Martin B., Ramiro A. R., McBride K. M., Nussenzweig M. C. (2003) C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell 12, 501–508 [DOI] [PubMed] [Google Scholar]
- 31. Demorest Z. L., Li M., Harris R. S. (2011) Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. J. Biol. Chem. 286, 26568–26575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kohli R. M., Abrams S. R., Gajula K. S., Maul R. W., Gearhart P. J., Stivers J. T. (2009) A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 284, 22898–22904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prochnow C., Bransteitter R., Chen X. S. (2009) APOBEC deaminases-mutases with defensive roles for immunity. Sci. China C Life Sci. 52, 893–902 [DOI] [PubMed] [Google Scholar]
- 34. Carpenter M. A., Rajagurubandara E., Wijesinghe P., Bhagwat A. S. (2010) Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair 9, 579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen K. M., Martemyanova N., Lu Y., Shindo K., Matsuo H., Harris R. S. (2007) Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 581, 4761–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu X., Geraldes P., Platt J. L., Cascalho M. (2005) The double-edged sword of activation-induced cytidine deaminase. J. Immunol. 174, 934–941 [DOI] [PubMed] [Google Scholar]
- 37. Rost B., Yachdav G., Liu J. (2004) The PredictProtein server. Nucleic Acids Res. 32, W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schatz D. G., Swanson P. C. (2011) V(D)J recombination. Mechanisms of initiation. Annu. Rev. Genet. 45, 167–202 [DOI] [PubMed] [Google Scholar]
- 39. Rajewsky K., Förster I., Cumano A. (1987) Evolutionary and somatic selection of the antibody repertoire in the mouse. Science 238, 1088–1094 [DOI] [PubMed] [Google Scholar]
- 40. Wysocki L. J., Gefter M. L. (1989) Gene conversion and the generation of antibody diversity. Annu. Rev. Biochem. 58, 509–531 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.