Background: KAP-1 and HP1-β are involved in DNA repair of heterochromatin via chromatin remodeling.
Results: Chk2-dependent KAP-1 phosphorylation regulates HP1-β mobilization from chromatin following DNA damage.
Conclusion: Regulation of HP1-β mobilization via KAP-1 phosphorylation is required for efficient DNA repair of heterochromatin.
Significance: Understanding the regulation of DNA repair and chromatin remodeling pathways can increase our knowledge of tumorigenesis.
Keywords: Chromatin Remodeling, DNA Damage Response, DNA Repair, Heterochromatin, Signal Transduction
Abstract
The DNA damage response encompasses a complex series of signaling pathways that function to regulate and facilitate the repair of damaged DNA. Recent studies have shown that the repair of transcriptionally inactive chromatin, named heterochromatin, is dependent upon the phosphorylation of the co-repressor, Krüppel-associated box (KRAB) domain-associated protein (KAP-1), by the ataxia telangiectasia-mutated (ATM) kinase. Co-repressors, such as KAP-1, function to regulate the rigid structure of heterochromatin by recruiting histone-modifying enzymes, such HDAC1/2, SETDB1, and nucleosome-remodeling complexes such as CHD3. Here, we have characterized a phosphorylation site in the HP1-binding domain of KAP-1, Ser-473, which is phosphorylated by the cell cycle checkpoint kinase Chk2. Expression of a nonphosphorylatable S473A mutant conferred cellular sensitivity to DNA-damaging agents and led to defective repair of DNA double-strand breaks in heterochromatin. In addition, cells expressing S473A also displayed defective mobilization of the HP1-β chromodomain protein. The DNA repair defect observed in cells expressing S473A was alleviated by depletion of HP1-β, suggesting that phosphorylation of KAP-1 on Ser-473 promotes the mobilization of HP1-β from heterochromatin and subsequent DNA repair. These results suggest a novel mechanism of KAP-1-mediated chromatin restructuring via Chk2-regulated HP1-β exchange from heterochromatin, promoting DNA repair.
Introduction
Chromatin exists in two states, heterochromatin and euchromatin. Euchromatin is less condensed, generally more accessible, and transcriptionally active. In contrast, heterochromatin consists of highly condensed, relatively inaccessible chromatin (e.g. telomeres), which is generally transcriptionally inactive or has the potential to be active but is silenced for a specific purpose (e.g. during development/cellular differentiation or X-chromosome inactivation). The structure of heterochromatin is initiated and maintained by repressors that function to recruit co-repressors such as the KRAB2 domain-associated protein 1 (KAP-1) (1). Co-repressors such as KAP-1 generally act to recruit enzymes that modify histones by removing acetyl groups (some of which promote “open” chromatin and therefore transcription) and by adding methyl groups (some of which promote “closed” chromatin and therefore silence transcription) (2). In addition, specific co-repressors can also regulate nucleosome spacing by the modulation of ATP-dependent nucleosome-remodeling enzymes (such as CHD3 and CHD4) and/or stabilize compacted chromatin by recruiting/loading specific chromodomain-containing proteins (such as HP1), which bind to methylated histones and maintain their closed state (3). These processes, mediated by nuclear co-repressors, form an efficient basis for transcriptional silencing via compaction and stabilization of chromatin to form heterochromatin.
Heterochromatin, aside from disruptions during mitosis and DNA replication, largely exists in a constant, compacted, stable state. One major exception to this is when DNA damage occurs, for example, DNA double-strand breaks induced by ionizing radiation or generated during normal cellular metabolism. Following DNA damage, heterochromatin proximal to the damage site is restructured to allow access to repair machinery and facilitate DNA repair (4, 5). This process involves the DNA damage-induced regulation of two heterochromatin-associated proteins, HP1 and KAP-1 (also known as TIF1b, TRIM28, or KRIP-1) (6, 7). KAP-1 contains two highly conserved domains: an N-terminal RBCC (RING finger, B boxes, coiled-coil), which mediates homo- and heterodimerization (8), and a C-terminal PHD (plant homeodomain) followed by a bromodomain (9). The two C-terminal domains are frequently found in transcriptional cofactors, reflecting the role of KAP-1 as a nuclear co-repressor. In addition to its role in transcription, a role for KAP-1 in the DNA damage response has been revealed. One of the key kinases involved in DNA damage response signaling, ATM, has been shown to phosphorylate KAP-1 on serine 824 following DNA damage, enhancing cell survival and facilitating chromatin relaxation and heterochromatic DNA repair (6, 11).
HP1 is a conserved chromatin-binding protein, composed of an N-terminal chromodomain and a C-terminal chromo shadow domain. The chromodomain of HP1 binds to methylated lysine 9 of histone H3 (H3K9me), making it a major component of heterochromatin required for gene silencing (12). There are three isoforms of HP1: α, β, and γ. HP1-α and -β are mainly considered to be components of constitutive heterochromatin, whereas HP1-γ is found in both euchromatin and heterochromatin (13). KAP-1 interacts with HP1 via its HP1-binding domain, which is required for its gene silencing activity (2).
Several studies have been unable to detect DNA damage-inducible changes in histone modifications that are usually associated with heterochromatin, suggesting that direct epigenetic alteration of heterochromatin does not facilitate DNA repair and that chromatin restructuring following DNA damage occurs via a different mechanism (14, 15). Indeed, a recent study has shown that Ser-824 phosphorylation of KAP-1 disrupts the interaction between CHD3 and KAP-1 and promotes the repair of heterochromatic DNA double-strand breaks (DSBs). This suggests that the restructuring of heterochromatin, through regulation of nucleosome remodeling, can also promote DNA repair (14). In addition, we have previously shown that casein kinase 2 (CK2)-mediated HP1-β phosphorylation following DNA damage is required to mobilize HP1-β within the chromatin, promoting recruitment of DNA repair factors (16, 17). In contrast to this, another group subsequently proposed that over a longer time scale, the three isoforms of HP1 are recruited to sites of DNA damage (18). These apparent discrepancies were reconciled by the proposal of a bimodal model to HP1 kinetics, with HP1 being rapidly mobilized from sites of damaged chromatin followed by slower recruitment (17).
This study demonstrates a novel mechanism that regulates the association of HP1-β with heterochromatin during DNA repair. Using mass spectrometry, we have identified a number of DNA damage-induced phosphorylation sites within KAP-1, including a Chk2 phosphorylation site at serine 473. We show that the ATM-Chk2-dependent phosphorylation of KAP-1 Ser-473 alters the dynamic behavior of HP1-β within heterochromatin and promotes cell survival and repair of heterochromatin-associated DNA damage.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and Cell Lines
Human cell lines were grown in RPMI supplemented with 10% FCS. Mouse embryonic fibroblasts were grown in DMEM supplemented with 10% FCS. The antibodies used were as follows: mouse anti-H2AX Ser-139 (Millipore), mouse anti-cyclin B and rabbit anti-Chk2 (Santa Cruz Biotechnology), mouse anti-β-actin and mouse anti-FLAG M2 antibody (Sigma), rabbit anti-GFP (Molecular Probes), rabbit anti-HP1-β, rabbit anti-trimethyl K9 H3, and mouse anti-KAP-1 antibody (Abcam), mouse anti-ATM Ser-1981 (Cell Signaling), rabbit anti-KAP-1 Ser-824 (Bethyl Laboratories), and rabbit anti-53BP1 (Novus Biologicals). A polyclonal antibody recognizing KAP-1 phosphorylated on Ser-473 was created using a synthetic KAP-1 phospho-peptide (VKRSRpSGEGEV, where pS indicates phospho-serine) conjugated to keyhole limpet hemocyanin (KLH) (Auspep), and inoculations/bleeds were carried out at the Institute of Medical and Veterinary Sciences, Adelaide, Australia. Antibodies were then affinity-purified using nonphosphorylated peptide and phosphorylated peptide columns as described previously (19).
Cloning, Site-directed Mutagenesis, and Expression of KAP-1 Constructs
Full-length KAP-1 was cloned into the HindIII and SalI sites of pFlag-CMV-2 (Fermentas). Mutation of S473A and S473E was carried out by site-directed mutagenesis using a QuikChange Lightning site-directed mutagenesis kit as per the manufacturer's instructions (Stratagene). Expression constructs were transfected into cells using Lipofectamine 2000TM (Invitrogen) as per the manufacturer's instructions, and samples were assayed 24 h after transfection.
siRNA
HP1-β (AGGAAUAUGUGGUGGAAAA) was purchased from GenePharma. Chk2 (GAACCUGAGGAGCCUACCC) and control siRNA were purchased from Invitrogen. siRNAs were transfected into HeLa cells using Lipofectamine 2000TM (Invitrogen) as per the manufacturer's instructions, and samples were analyzed 48 h after transfection.
Mass Spectrometry
KAP-1 phosphorylation sites were identified from immunoprecipitated FLAG-KAP-1 following SDS-PAGE as described previously (20).
Immunoblotting
Immunoblotting was performed as before (21). Briefly, following the indicated treatments, cells were scraped from tissue culture plates and washed once in PBS. Cells were lysed in a buffer containing 20 mm Hepes, pH 8, 150 mm KCl, 5% glycerol, 10 mm MgCl2, 0.5 mm EDTA, 0.02% Nonidet P-40 supplemented with 1 mm NaF, 1 mm Na3VO4, and Complete protease inhibitor mixture (Roche Applied Science). Lysates were sonicated and cleared by centrifugation, and protein concentrations were estimated using the standard Bradford assay (Bradford reagent supplied by Bio-Rad). Typically, 50 μg of protein lysate was separated on a 4–12% SDS-PAGE NOVEX gel (Invitrogen) and immunoblotted with the indicated antibodies.
Immunofluorescence
Cells were seeded onto coverslips the day before siRNA transfection. Following siRNA transfection, cells were allowed to grow for 48 h before treatment or mock treatment with the indicated DNA-damaging agent. After treatment, cells were treated with an extraction buffer (22) for 5 min before fixation in 4% paraformaldehyde. Cells were permeabilized with 0.2% Triton-X for 5 min and blocked in 3% BSA for 30 min. Cells were incubated with the indicated primary antibodies and Alexa Fluor-conjugated secondary antibodies (Molecular Probes) for 1 h each at room temperature. Cells were stained with DAPI before mounting onto slides. γH2AX foci associated within heterochromatin were identified as described initially by Goodarzi et al. (11). Briefly, growth-arrested NIH3T3 cells were transfected with FLAG-KAP-1 (wild type or S473A). After 24 h, cells were treated with 2 Gy of IR and harvested 24 h after IR. Cells were stained with γH2AX (green) and DAPI (blue). High resolution images were captured using a Delta Vision PDV microscope; images were deconvolved using the softWoRx Suite software. Further information on identifying γH2AX foci associated with regions of heterochromatin can be found in supplemental Figs. 3 and 4. For the immunofluorescence shown in supplemental Fig. 5, the following changes were made; cells were not treated with an extraction buffer before fixation and were imaged on a Leica SP5 microscope.
Time-lapse Microscopy, Laser-induced DNA Damage, and Fluorescence Recovery after Photobleaching (FRAP)
Time-lapse microscopy, laser-induced DNA damage, and FRAP were performed as described previously, with some modifications as follows (16). Briefly, A Leica SP5 confocal microscope with a 100× NA 1.4 lens and temperature-controlled incubation unit operating at 37 °C was used, keeping imaging parameters constant between samples. Cells were presensitized with 10 μg/ml Hoechst 33342 by incubation for 5 min at 37 °C. DNA damage was induced in a circle of diameter ∼1 μm using a 405-nm diode laser operating at 100% for 1.2 s. Enhanced green fluorescent protein (EGFP)-HP1-β dynamic behavior was followed using time-lapse imaging using a 488-nm argon laser taking 256 × 256 pixel images at scan speed 700 Hz, producing a picture every ∼0.4 s. Photomultiplier tube detector gain and offset were adjusted so that pixels were not saturated. FRAP photobleaching was performed using 100% of a 488-nm argon laser for 1.2 s in a circular region of size ∼1 μm in diameter. Images were analyzed using Leica LAS AF software to draw regions of interest and export fluorescence intensity data into Microsoft Excel. Corrections for imaging background and bleaching were made in Excel, and graphs were plotted using GraphPad Prism. Curves were fitted in GraphPad Prism using the one-phase association equation
where
 |
to yield t½ and plateau (Ymax) values. FRAP experiments on damaged regions were performed 1 min after damage induction.
Colony-forming Assay
Colony-forming assays were performed as described previously (23).
RESULTS
A Phospho-mapping Screen Identifies Multiple Phosphorylation Sites on KAP-1
KAP-1 function is highly regulated by post-translational modifications including ATM-dependent serine 824 phosphorylation following DNA damage. Therefore, to identify additional DNA damage-induced KAP-1 phosphorylation sites that might regulate KAP-1 function following DNA damage, we used mass spectrometric-based phospho-peptide and phospho-residue mapping (24). A FLAG-KAP-1 fusion protein was transiently expressed in HEK 293T cells that were mock-treated or exposed to 10 Gy of IR. FLAG-KAP-1 was immunoprecipitated using anti-FLAG M2 beads, and immunoprecipitates were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue G250. The FLAG-KAP-1 band was excised from the gel and used for nano-electrospray mass ionization-MS/MS phospho-residue mapping (supplemental Fig. 1, A and B). In total, 14 DNA damage-induced KAP-1 phosphorylation sites were identified (supplemental Fig. 1C). There are 51 reported KAP-1 phospho-sites, 32 of which have been reported more than once (PhosphoSitePlus). However, our subset of 14 detected phospho-sites may be specifically induced by IR.
The surrounding sequence of the KAP-1 phospho-serine 473 (467GVKRSRpSGEGEVSG480) matched that of a Chk1/Chk2 consensus phosphorylation motif, RXXpS (where pS indicates phospho-serine) (25). As Chk1 and Chk2 are both activated following DNA damage (by ATR (ATM and Rad3-related) and ATM, respectively), we postulated that this site might represent a Chk1/Chk2 phosphorylation site within KAP-1. Given the critical role played by many DNA damage-induced phosphorylation-signaling events, a large proportion of these phosphorylation target sequences is evolutionarily conserved. Indeed, the Chk1/Chk2 consensus sequence found within human KAP-1 at Ser-473 is evolutionarily conserved throughout KAP-1 orthologues identified from humans to Drosophila (Fig. 1A). We therefore sought to characterize this phosphorylation site in the context of KAP-1 function following DNA damage.
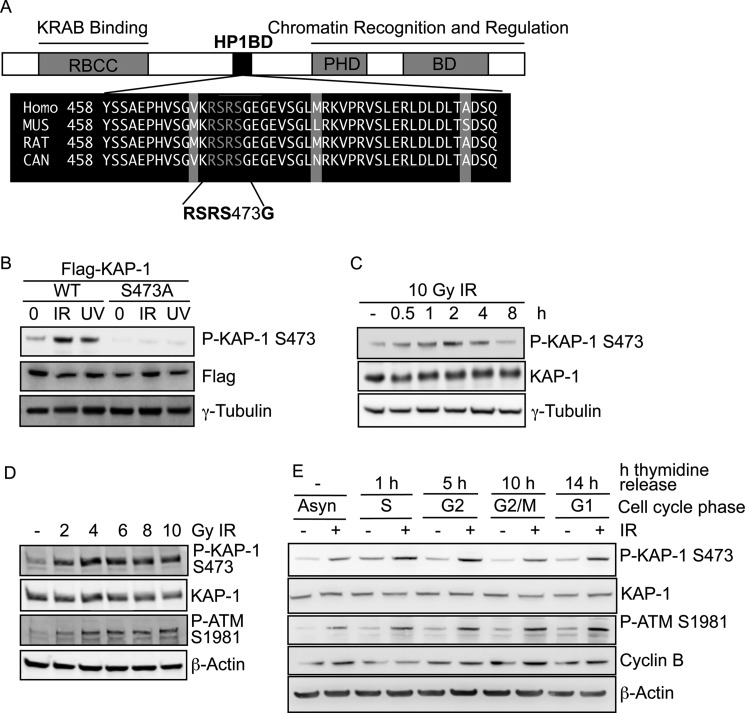
FIGURE 1.
KAP-1 is phosphorylated on Ser-473 following DNA damage. A, a multiple sequence alignment of the KAP-1 HP1-binding domain (HP1BD) in different species. B, 293T cells transfected with wild-type FLAG-KAP-1-WT or FLAG-KAP-1-S473A were mock-irradiated or treated with IR (10 Gy) or UVC (50 J/m2) (as indicated) and harvested 1 h after treatment. Cellular protein extracts were prepared and immunoblotted with the indicated antibodies. P-KAP-1, phospho-KAP-1. C, 293T cells were mock-irradiated or treated with IR (10 Gy) and harvested at the indicated time points. Cellular protein extracts were prepared and immunoblotted with the indicated antibodies. D, HeLa cells were mock-irradiated or treated with the indicated dose of IR and harvested 1 h after irradiation. Cellular protein extracts were prepared and immunoblotted with the indicated antibodies. P-ATM, phospho-ATM. E, HeLa cells were synchronized in S-phase using thymidine. Cells were treated with 6 Gy of IR at the indicated times after thymidine release, and extracts were taken after 1 h. Extracts were immunoblotted with the indicated antibodies.
Phosphorylation of KAP-1 on Ser-473 Is Induced by DNA Damage
To investigate the function of the KAP-1 Ser-473 phosphorylation site, we generated a phospho-specific antibody against this site. The specificity of the pKAP-1-S473 antibody was confirmed using cells transfected with exogenous FLAG fusion proteins of wild-type KAP-1 or a nonphosphorylatable S473A KAP-1 mutant (designated FLAG-KAP-1 WT and FLAG-KAP-1 S473A, respectively) (Fig. 1B). Consistent with other studies (26, 27), we observed that both IR and UVC could stimulate the phosphorylation of KAP-1 on Ser-473 but not the S473A phospho-site mutant (Fig. 1B). Nevertheless a higher proportion of KAP-1 appeared to be phosphorylated on Ser-473 following IR treatment, leading us to focus our studies on KAP-1 phospho-Ser-473 function using this DNA-damaging agent. Furthermore, immunoblotting of lysates from cells harvested at the indicated times following IR revealed that KAP-1 Ser-473 is phosphorylated in a time-dependent manner, with phosphorylation occurring rapidly (<30 min) after IR treatment and appearing maximal at around 2 h and reduced to pre-damage levels by 8 h after IR treatment, mimicking the kinetics of ATM-dependent KAP-1 Ser-824 phosphorylation (6) (Fig. 1C). KAP-1 Ser-473 phosphorylation was also induced in a dose-dependent manner with maximal phosphorylation detected at 4 Gy (Fig. 1D). To examine whether the phosphorylation of KAP-1 Ser-473 was cell cycle-dependent, HeLa cells were synchronized using double thymidine block, and cell extracts were harvested at various time points following mock treatment or treatment with IR. KAP-1 Ser-473 was phosphorylated in all phases of the cell cycle following IR, mirroring the phosphorylation of ATM Ser-1981 (Fig. 1E). We also observed that 1 h after release from thymidine (corresponding to S-phase), KAP-1 Ser-473 was phosphorylated in the absence of IR, as shown previously (28). Like others, we also found that KAP-1 Ser-473 phosphorylation is increased following irradiation, but that its localization appeared diffuse via immunofluorescence (data not shown) (29).
Phosphorylation of KAP-1 on Ser-473 Is ATM- and Chk2-dependent
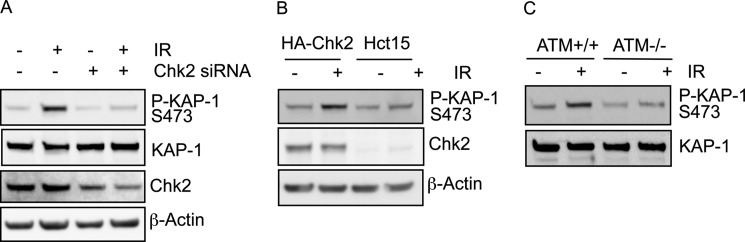
Because the 3 amino acid residues prior to the Ser-473 phosphorylation site match the consensus sequence for a Chk2 phosphorylation site, we next examined the requirement of Chk2 for KAP-1 Ser-473 phosphorylation. HeLa cells were depleted of Chk2 using siRNA and mock-treated or treated with IR. Depletion of Chk2 demonstrated a defect in KAP-1 Ser-473 phosphorylation (Fig. 2A). To further demonstrate the requirement of Chk2 for Ser-473 phosphorylation, we also utilized the Chk2-deficient HCT15 cell line and an isogenic counterpart stably expressing exogenous HA-tagged Chk2. IR-induced KAP-1 Ser-473 phosphorylation was defective in HCT15 cells. However, this defect was rescued in the Chk2-complemented cells, further demonstrating the requirement of Chk2 for KAP-1 Ser-473 phosphorylation following IR (Fig. 2B). Because ATM is required for efficient Chk2 activation and kinase activity following DNA damage (30), we next examined Ser-473 phosphorylation in an ATM-defective cell line. As expected, IR-induced KAP-1 Ser-473 phosphorylation was defective in ATM-null (L3) cells in comparison with ATM wild-type (C3ABR) cells following DNA damage (Fig. 2C). Given the dependence on ATM for KAP-1 Ser-473 phosphorylation, we also assessed whether KAP-1 Ser-473 phosphorylation is dependent on ATM-dependent KAP-1 Ser-824 phosphorylation. This revealed no co-dependence of phosphorylation of KAP-1 on Ser-473 or Ser-824 (supplemental Fig. 2A).
FIGURE 2.
KAP-1 Ser-473 phosphorylation is dependent upon Chk2 and ATM. A, 293T cells transfected with control or Chk2 siRNA were mock-irradiated or treated with IR (10 Gy) and harvested 1 h after treatment. Cellular protein extracts were prepared and immunoblotted with the indicated antibodies. P-KAP-1, phospho-KAP-1. B, HCT15 cells stably expressing HA-Chk2 or HA alone were mock-irradiated or treated with IR (10 Gy) and harvested 1 h after treatment. Cellular protein extracts were prepared and immunoblotted with the indicated antibodies. C, C3ABR (ATM+/+) or L3 (ATM−/−) cells were mock-irradiated or treated with IR (10 Gy) and harvested 1 h after treatment. Cellular protein extracts were prepared and immunoblotted with the indicated antibodies.
Cells Expressing KAP-1 S473A Are Sensitive to DNA Damage
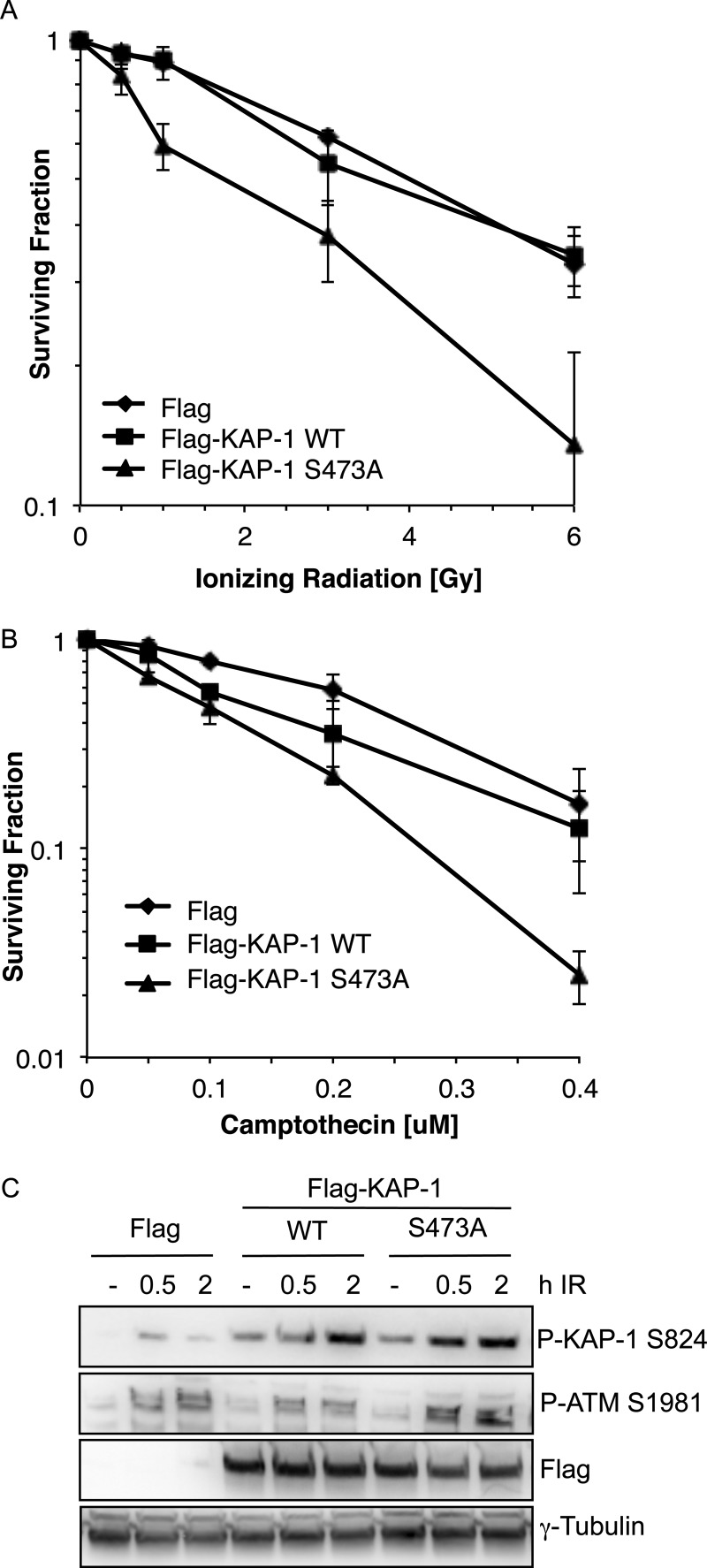
To further characterize the role of Chk2-dependent KAP-1 Ser-473 phosphorylation, we examined the sensitivity of cells expressing either wild-type KAP-1 (FLAG-KAP-1 WT) or S473A KAP-1 (FLAG-KAP-1 S473A) to IR-induced or topoisomerase II inhibitor camptothecin-induced DNA damage (Fig. 3, A and B, and supplemental Fig. 2B). This revealed that cells expressing FLAG-KAP-1 S473A were 2.6-fold more sensitive to 6 Gy of IR and 5-fold more sensitive to 0.4 μm camptothecin when compared with cells expressing FLAG-KAP-1 WT, suggesting a role for KAP-1 Ser-473 phosphorylation in DNA damage repair or signaling. An early event in the cellular response to DNA damage is the activation of ATM via autophosphorylation on serine 1981 and the subsequent phosphorylation of downstream targets such as KAP-1 Ser-824 (6, 30). We found that phosphorylation of KAP-1 on Ser-473 was not required for ATM-mediated signaling events, as measured by ATM autophosphorylation and ATM-dependent KAP-1 phosphorylation on Ser-824 following IR treatment, suggesting that expression of FLAG-KAP-1 S473A does not impair ATM activation or its downstream signaling function following DNA damage (Fig. 3C). Consistent with this, this phosphorylation event was also dispensable for ATM -and Chk2-dependent maintenance of the G2/M checkpoint after IR (data not shown).
FIGURE 3.
Cells expressing FLAG-KAP-1 S473A are sensitive to DNA damage. A and B, colony survival in HeLa cells. Cells were transfected with FLAG-CMV, FLAG-KAP-1 WT, or FLAG-KAP-1 S473A and treated with the indicated dose of IR (A) or camptothecin (B). Cells were left to form colonies for 10 days before staining with methylene blue. The data shown represent the average and S.E. of three independent experiments. C, HeLa cells were transfected with FLAG-CMV, FLAG-KAP-1 WT, or FLAG-KAP-1 S473A and treated with IR (6 Gy). Cell extracts were taken at 0.5 and 2 h after IR and immunoblotted with the indicated antibodies. P-KAP-1, phospho-KAP-1; P-ATM, phospho-ATM.
Cells Expressing S473A Display Defective Foci Resolution Following DNA Damage
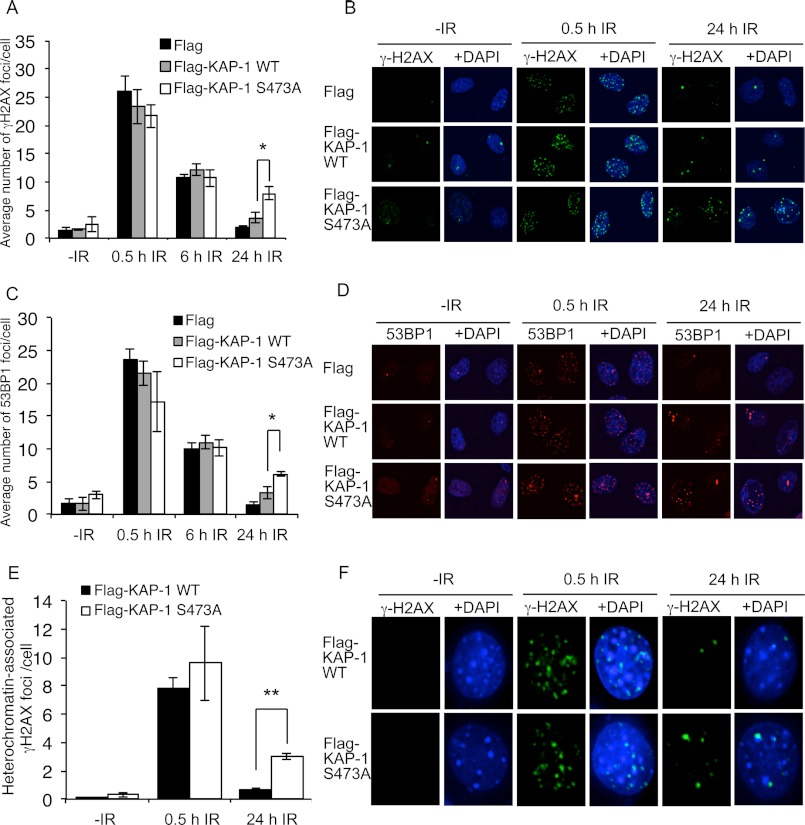
γH2AX is a major marker of DNA damage and can be used to measure the kinetics of DNA repair (31). To investigate the function of KAP-1 Ser-473 phosphorylation in DNA repair, we next examined the resolution of γH2AX foci in cells expressing wild-type and S473A KAP-1. We found that cells expressing FLAG-CMV, FLAG-KAP-1 WT, and FLAG-KAP-1 S473A all displayed a similar number of γH2AX foci at 0.5 and 6 h after IR treatment (Fig. 4, A and B). However, 24 h after IR, significantly more γH2AX foci were present in cells expressing FLAG-KAP-1 S473A when compared with the cells transfected with FLAG-CMV or FLAG-KAP-1 WT. To confirm that this was a repair defect and not a defect in γH2AX dephosphorylation, we next performed staining with 53BP1, another protein that is recruited to DNA repair foci following DNA damage. Similar to γH2AX, persistence of 53BP1 foci occurred in cells expressing FLAG-KAP-1 S473A, suggesting that the remaining foci were the result of a DNA repair defect (Fig. 4, C and D). The differences in the number of γH2AX foci in FLAG-KAP-1 WT- and FLAG-KAP-1 S473A-expressing cells at the 24-h time point were not due to differences in cell cycle distribution (data not shown).
FIGURE 4.
Cells expressing FLAG-KAP-1 S473A display defective foci resolution in heterochromatin following DNA damage. A–D, HeLa cells were transfected with FLAG-CMV, FLAG-KAP-1 WT, or FLAG-KAP-1 S473A and treated with 2 Gy of IR before fixation and staining with γH2AX (A and B) or 53BP1 (C and D) antibodies at the indicated times. Representative images are shown in B and D, and graphical representation of data is shown in A and C. The results shown represent the average and S.E. of three independent experiments. The foci in 50–100 cells were counted in each independent experimental condition. * indicates a two-tailed p value of < 0.01. E and F, γH2AX foci that persist in S473A-expressing cells are associated with heterochromatin. Growth-arrested NIH3T3 cells were transfected with FLAG-KAP-1 WT or S473A. After 24 h, cells were mock-treated or treated with 2 Gy of IR and harvested at the indicated times after IR. Cells were stained with γH2AX (green) and DAPI (blue). Foci were scored as heterochromatin-associated positive or negative. The results shown represent the average and S.E. of three independent experiments. The association between γH2AX foci and heterochromatin in 40–50 cells was quantified in each independent experimental condition. ** indicates a two-tailed p value of < 0.0001.
γH2AX Foci That Persist in S473A-expressing Cells Are Associated with Heterochromatin
We have shown that Chk2-mediated KAP-1 Ser-473 phosphorylation also requires ATM. Because ATM has previously been shown to be required to repair DNA double-strand breaks in regions of heterochromatin, we hypothesized that the γH2AX and 53BP1 foci remaining in cells expressing S473A may be unrepaired DSBs within heterochromatin (11). To examine this, we utilized growth-arrested NIH3T3 murine cells that contain dense DAPI staining in “chromocenter” regions, representing pericentric and centromeric heterochromatin (32). To ensure that these dense DAPI-stained areas in NIH3T3 cells contained heterochromatin, under our experimental conditions, we carried out immunofluorescence studies using antibodies directed against K9 trimethylated histone H3, a marker of heterochromatin. Aligning with previous investigations, the dense DAPI-stained regions co-stained with K9 trimethylated histone H3, whereas euchromatin was relatively unstained (supplemental Fig. 3) (11, 32). Similarly to the γH2AX foci that persisted in cells treated with an ATM inhibitor (11), the γH2AX foci that persisted in cells expressing FLAG-KAP-1 S473A were also found to be associated with heterochromatin (Fig. 4, E and F). In cells expressing FLAG-KAP-1 WT, an average of 0.69 γH2AX foci/per cell (40% of foci) remaining at 24 h after IR was found to be associated with regions of heterochromatin. In contrast, in cells expressing FLAG-KAP-1 S473A, an average of three γH2AX foci/cell (68% of foci) remaining at 24 h after IR treatment was associated with heterochromatin, suggesting that KAP-1 Ser-473 phosphorylation is required for efficient repair of heterochromatin-associated DNA damage (Fig. 4, E and 4F, and supplemental Figs. 3 and 4).
Phosphorylation of KAP-1 Ser-473 Is Required for Mobility of HP1-β Following DNA Damage
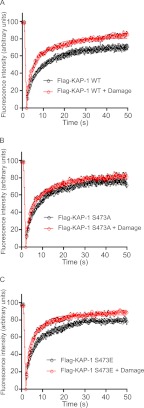
Because the KAP-1 Ser-473 phosphorylation site is located in the region of KAP-1 required for interaction with the heterochromatin protein HP1-β, we next considered whether phosphorylation of this site might be required to regulate HP1-β. Previous studies have shown that phosphorylation of KAP-1 on Ser-473 leads to dissociation of KAP-1 from HP1-β (27, 28); however, other studies were unable to detect any differences in binding of KAP-1 Ser-473 phospho-mimic to HP1-β (26, 29). We have also been unable to detect any binding differences between KAP-1 S473A and S473E mutants to HP1-β (data not shown). This prompted us to hypothesize that KAP-1 Ser-473 phosphorylation may regulate another aspect of HP1-β function, such as its localization. In unperturbed cells, HP1-β is localized to the heterochromatin by binding to histone H3 methylated on lysine 9 (H3K9me) (aside from in cells in S-phase and mitosis), where it promotes the condensed heterochromatic structure. As described previously, we used an EGFP-HP1-β fusion protein to monitor changes in the dynamic behavior of HP1-β at DNA break sites (16). Stable cell lines were generated that express EGFP-HP1-β and wild-type or S473A FLAG-KAP-1 (supplemental Fig. 5). Consistent with our previous work in cells expressing wild-type KAP-1, dynamic changes in HP1-β mobility (assessed by FRAP) occurred rapidly after laser-induced DNA damage (Fig. 5A and supplemental Fig. 6). In contrast, cells expressing nonphosphorylatable FLAG-KAP-1 S473A showed a less marked alteration in HP1-β dynamics after DNA damage, suggesting that phosphorylation of KAP-1 on Ser-473 was required for changes in HP1-β mobility on chromatin after DNA damage (Fig. 5B). HP1-β mobilization kinetics were also examined in cells expressing the phospho-mimic mutant KAP-1-S473E (supplemental Fig. 5) and were found to be similar to cells expressing wild-type KAP-1, suggesting that KAP-1 phosphorylation alone may not be sufficient to mobilize HP1-β from chromatin (Fig. 5C).
FIGURE 5.
Phosphorylation of KAP-1 Ser-473 is required for HP1-β mobility following DNA damage. A–C, FRAP analysis of EGFP-HP1-β in heterochromatin, with and without laser-induced DNA damage, in cells stably expressing FLAG-KAP-1 WT (A), FLAG-KAP-1 S473A (B), or FLAG-KAP-1 S473E (C). Data show mean recovery curves (symbols) ± S.E. (n = 12), fitted with a one-phase association equation (solid lines; see “Experimental Procedures” for Equations 1 and 2).
We have previously shown that DNA damage-induced phosphorylation of HP1-β, by CK2 (Thr-51), results in altered HP1-β dynamic behavior around chromatin. This altered dynamic behavior of CK2-phosphorylated HP1-β is the result of disruption of hydrogen bonds that fold its chromodomain around H3K9me (16). The phosphorylation and subsequent dynamic changes of HP1-β on chromatin promote the early phosphorylation of γH2AX and therefore the DNA damage response. Given the above, we next examined whether phosphorylation of KAP-1 was required for phosphorylation of HP1-β on Thr-51. Using immunofluorescence, we were unable to detect a significant defect in the number of phospho-HP1-β Thr-51 foci in IR-treated cells expressing nonphosphorylatable KAP-1 S473A when compared with cells expressing FLAG-CMV alone (data not shown). This suggests that phosphorylation of KAP-1 on Ser-473 is not required for phosphorylation of HP1-β, but that both KAP-1 Ser-473 phosphorylation and HP1-β Thr-51 phosphorylation are required for the mobilization of HP1-β following DNA damage.
HP1-β Knockdown Alleviates the Requirement for KAP-1 Ser-473 Phosphorylation
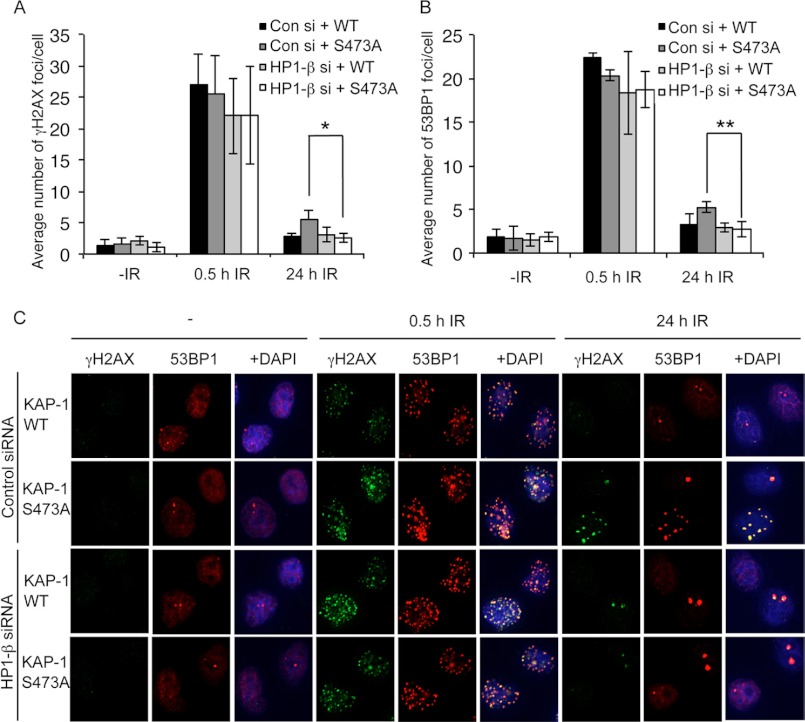
Because we have shown that KAP-1 phosphorylation is required for HP1-β mobilization and subsequent DNA repair in regions of heterochromatin, we next wanted to assess whether removal of HP1-β from cells could bypass the requirement for KAP-1 Ser-473 phosphorylation in DNA repair. Endogenous HP1-β was depleted from cells ectopically expressing FLAG-CMV, FLAG-KAP-1 WT, or FLAG-KAP-1 S473A using siRNA, and the kinetics of γH2AX and 53BP1 foci resolution following IR treatment were assessed to measure DNA repair (Fig. 6 and supplemental Fig. 7). Notably, although the effects were modest, HP1 depletion reproducibly and significantly rescued the DNA repair defect observed in cells expressing KAP-1 S473A, supporting our evidence that phosphorylation of KAP-1 on Ser-473 is required for removal of HP1-β from heterochromatin facilitating efficient DNA repair (Fig. 6).
FIGURE 6.
HP1-β knockdown alleviates the requirement for KAP-1 Ser-473 phosphorylation. A–C, HeLa cells were transfected with HP1-β siRNA for 24 h and then transfected with FLAG-CMV, FLAG-KAP-1 WT, or FLAG-KAP-1 S473A. Transfected cells were treated with 2 Gy of IR 48 h after siRNA transfection before fixation and staining with γH2AX (A and C) or 53BP1 (B and C) antibodies at the indicated times. The data shown represent the average and S.E. of three independent experiments. * indicates a two-tailed p value of < 0.04. ** indicates a two-tailed p value of < 0.02. Con si, control siRNA.
DISCUSSION
Here, we present evidence that Chk2-mediated phosphorylation of the transcriptional co-repressor KAP-1 is required for the efficient repair of DNA within heterochromatin. We initially identified the KAP-1 Ser-473 phosphorylation site following a phospho-mapping screen. Consistent with the work of other groups, we confirmed that KAP-1 Ser-473 was an ATM- and DNA damage-dependent Chk2 phosphorylation site following ionizing radiation (26, 27). Cells expressing a nonphosphorylatable S473A mutant of KAP-1 were found to be more sensitive to DNA-damaging agents, suggesting that phosphorylation of KAP-1 on Ser-473 may be required for DNA repair. Subsequent examination of repair foci kinetics in S473A-expressing cells confirmed a repair defect associated with heterochromatic regions. Further study of the KAP-1 Ser-473 phosphorylation site revealed that it was required for the rapid change in the dynamic behavior of HP1-β around chromatin following DNA damage. This event was confirmed to be required for DNA damage repair as HP1-β knockdown alleviated the requirement for KAP-1 phosphorylation in DSB repair.
These data support emerging evidence for different mechanisms of DNA repair in euchromatin and heterochromatin. Due to the relaxed or open structure of euchromatin, DNA repair factors can act to repair DNA lesions rapidly and efficiently. However, in heterochromatin, where chromatin is more tightly condensed, remodeling is required to allow activity of repair factors to facilitate efficient repair. The efficient DNA repair of heterochromatin is governed by multiple, redundant mechanisms, with each independent mechanism contributing toward DNA repair. The Chk2-mediated phosphorylation of KAP-1 Ser-473 is one of many such mechanisms that together contribute to efficient and accurate heterochromatic DNA repair (33, 34).
A question that remains to be answered is what is the precise mechanism by which KAP-1 mediated mobilization of HP1-β within chromatin promotes DNA repair in heterochromatin. It is tempting to speculate that the rigid heterochromatic structure conferred by binding of HP1-β is not compatible with a specific event during DNA repair (for example, resection, strand invasion, or ligation, depending upon the type of DSB repair). However, further investigation is required to fully understand the requirement of HP1-β and its dynamic interaction with chromatin for DNA repair.
We have previously reported that HP1-β phosphorylation on Thr-51 was required to disrupt its affinity for H3K9me, leading to changes in its dynamic behavior around chromatin (16). Here, we further show that phosphorylation of KAP-1 on Ser-473 is also required for the mobilization of HP1-β. Because expression of a nonphosphorylatable KAP-1 S473A mutant does not cause any detectable changes in HP1-β Thr-51 phosphorylation following DNA damage, we can only speculate as to the function of Ser-473 phosphorylation. It is possible that Ser-473 phosphorylation leads to a conformational change in KAP-1, which can, in combination with HP1-β Thr-51 phosphorylation, further disrupt the interaction of HP1-β with H3K9me residues or partner proteins, leading to its mobilization. In addition, we also cannot exclude the possibility that defects in HP1-β Thr-51 phosphorylation do occur at heterochromatin-specific DNA damage sites in cells expressing KAP-1 S473A, but are below detectable levels in our current experimental conditions.
It has recently been suggested that another HP1 isoform, HP1-α, is required for homologous recombination-mediated DNA repair (35). However, our data showing that knockdown of HP1-β corrects the heterochromatic DNA repair defect observed in cells expressing S473A suggests that the presence HP1-β may not be required for DNA repair (and could in fact be inhibitory to DNA repair) under our experimental conditions. Another study also found that HP1-α/β/γ were not required for repair of DNA lesions induced by IR, which supports our data presented here and from our previous studies that HP1-β needs to be mobilized around heterochromatin for efficient DNA repair to take place (11, 16).
Considering the data presented here and the data of others (6, 11, 16, 17, 26–28), we propose the following model for KAP-1- and HP1-β-mediated heterochromatin remodeling and DNA repair. Following DSB induction, ATM is activated and phosphorylates downstream proteins such as Chk2 (30). Chk2 then phosphorylates KAP-1 on Ser-473, promoting dynamic changes in HP1-β mobilization within heterochromatin. This leads to alterations in heterochromatin that promote recruitment of factors to facilitate DNA repair.
Several groups have attempted to examine chromatin remodeling at sites of DSBs, and data suggest that modifications of histones surrounding DNA DSBs in heterochromatin either are below currently detectable levels or do not occur. The evidence we have presented here along with our previous work on HP1-β mobility support the notion that chromatin organization can occur via targeting of histone-binding proteins rather than alteration of the histone code itself (16, 36). The role of ATM-dependent phosphorylation of KAP-1 on Ser-824 in the repair of heterochromatin-associated DSBs has recently been extensively characterized (6, 10, 11, 14). Induction of Ser-824-phosphorylated KAP-1 following DSBs leads to chromatin relaxation via removal of the nucleosome remodeler CHD3 from chromatin (14). We suggest that Chk2-dependent phosphorylation of KAP-1 on Ser-473 offers an additional mechanism for chromatin relaxation via mobilization of HP1-β from chromatin, promoting DNA repair protein activity and repair of the DNA lesion.
Supplementary Material
Acknowledgments
We thank members of the Khanna laboratory for comments and support during this study.
This work was supported by a discovery grant from the Australian Research Council and by a grant from the United Kingdom Medical Research Council (to A. R. V.).

This article contains supplemental Figs. 1–7.
- KRAB
- Krüppel-associated box
- KAP-1
- KRAB domain-associated protein 1
- HP1-β
- heterochromatin protein 1β
- DSB
- double-strand break
- ATM
- ataxia telangiectasia-mutated
- CK2
- casein kinase 2
- FRAP
- fluorescence recovery after photobleaching
- EGFP
- enhanced green fluorescent protein
- Gy
- gray
- IR
- ionizing radiation.
REFERENCES
- 1. Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., 3rd (1996) KAP-1, a novel co-repressor for the highly conserved KRAB repression domain. Genes Dev. 10, 2067–2078 [DOI] [PubMed] [Google Scholar]
- 2. Ryan R. F., Schultz D. C., Ayyanathan K., Singh P. B., Friedman J. R., Fredericks W. J., Rauscher F. J., 3rd (1999) KAP-1 co-repressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell Biol. 19, 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sripathy S. P., Stevens J., Schultz D. C. (2006) The KAP-1 co-repressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell Biol. 26, 8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goodarzi A. A., Jeggo P., Lobrich M. (2010) The influence of heterochromatin on DNA double-strand break repair: getting the strong, silent type to relax. DNA Repair 9, 1273–1282 [DOI] [PubMed] [Google Scholar]
- 5. Zeng W., Ball A. R., Jr., Yokomori K. (2010) HP1: heterochromatin-binding proteins working the genome. Epigenetics 5, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D. C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1-dependent pathway. Nat. Cell Biol. 8, 870–876 [DOI] [PubMed] [Google Scholar]
- 7. Cann K. L., Dellaire G. (2011) Heterochromatin and the DNA damage response: the need to relax. Biochem. Cell Biol. 89, 45–60 [DOI] [PubMed] [Google Scholar]
- 8. Peng H., Feldman I., Rauscher F. J., 3rd (2002) Hetero-oligomerization among the TIF family of RBCC/TRIM domain-containing nuclear cofactors: a potential mechanism for regulating the switch between coactivation and co-repression. J. Mol. Biol. 320, 629–644 [DOI] [PubMed] [Google Scholar]
- 9. Zeng L., Zhou M. M. (2002) Bromodomain: an acetyl-lysine-binding domain. FEBS Lett. 513, 124–128 [DOI] [PubMed] [Google Scholar]
- 10. Noon A. T., Shibata A., Rief N., Löbrich M., Stewart G. S., Jeggo P. A., Goodarzi A. A. (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 12, 177–184 [DOI] [PubMed] [Google Scholar]
- 11. Goodarzi A. A., Noon A. T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P. A. (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 [DOI] [PubMed] [Google Scholar]
- 12. Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., Shabanowitz J., Hunt D. F., Funabiki H., Allis C. D. (2005) Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 13. Dinant C., Luijsterburg M. S. (2009) The emerging role of HP1 in the DNA damage response. Mol. Cell Biol. 29, 6335–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodarzi A. A., Kurka T., Jeggo P. A. (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18, 831–839 [DOI] [PubMed] [Google Scholar]
- 15. Tjeertes J. V., Miller K. M., Jackson S. P. (2009) Screen for DNA damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ayoub N., Jeyasekharan A. D., Bernal J. A., Venkitaraman A. R. (2008) HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453, 682–686 [DOI] [PubMed] [Google Scholar]
- 17. Ayoub N., Jeyasekharan A. D., Venkitaraman A. R. (2009) Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle 8, 2945–2950 [PubMed] [Google Scholar]
- 18. Luijsterburg M. S., Dinant C., Lans H., Stap J., Wiernasz E., Lagerwerf S., Warmerdam D. O., Lindh M., Brink M. C., Dobrucki J. W., Aten J. A., Fousteri M. I., Jansen G., Dantuma N. P., Vermeulen W., Mullenders L. H., Houtsmuller A. B., Verschure P. J., van Driel R. (2009) Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 185, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khanna K. K., Gatei M., Tribbick G. (2006) Phospho-specific antibodies as a tool to study in vivo regulation of BRCA1 after DNA damage. Methods Mol. Med. 120, 441–452 [DOI] [PubMed] [Google Scholar]
- 20. Graham M. E., Anggono V., Bache N., Larsen M. R., Craft G. E., Robinson P. J. (2007) The in vivo phosphorylation sites of rat brain dynamin I. J. Biol. Chem. 282, 14695–14707 [DOI] [PubMed] [Google Scholar]
- 21. Bolderson E., Tomimatsu N., Richard D. J., Boucher D., Kumar R., Pandita T. K., Burma S., Khanna K. K. (2010) Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 38, 1821–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Young D. B., Jonnalagadda J., Gatei M., Jans D. A., Meyn S., Khanna K. K. (2005) Identification of domains of ataxia telangiectasia-mutated required for nuclear localization and chromatin association. J. Biol. Chem. 280, 27587–27594 [DOI] [PubMed] [Google Scholar]
- 23. Richard D. J., Bolderson E., Cubeddu L., Wadsworth R. I., Savage K., Sharma G. G., Nicolette M. L., Tsvetanov S., McIlwraith M. J., Pandita R. K., Takeda S., Hay R. T., Gautier J., West S. C., Paull T. T., Pandita T. K., White M. F., Khanna K. K. (2008) Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 453, 677–681 [DOI] [PubMed] [Google Scholar]
- 24. Larsen M. R., Graham M. E., Robinson P. J., Roepstorff P. (2004) Improved detection of hydrophilic phospho-peptides using graphite powder microcolumns and mass spectrometry: evidence for in vivo doubly phosphorylated dynamin I and dynamin III. Mol. Cell. Proteomics 3, 456–465 [DOI] [PubMed] [Google Scholar]
- 25. O'Neill T., Giarratani L., Chen P., Iyer L., Lee C. H., Bobiak M., Kanai F., Zhou B. B., Chung J. H., Rathbun G. A. (2002) Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J. Biol. Chem. 277, 16102–16115 [DOI] [PubMed] [Google Scholar]
- 26. Blasius M., Forment J. V., Thakkar N., Wagner S. A., Choudhary C., Jackson S. P. (2011) A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 12, R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu C., Zhang S., Gao X., Gao X., Xu X., Lv Y., Zhang Y., Zhu Z., Zhang C., Li Q., Wong J., Cui Y., Zhang W., Ma L., Wang C. (2012) Roles of Krüppel-associated box (KRAB)-associated co-repressor KAP-1 Ser-473 phosphorylation in DNA damage response. J. Biol. Chem. 287, 18937–18952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang C. W., Chou H. Y., Lin Y. S., Huang K. H., Chang C. J., Hsu T. C., Lee S. C. (2008) Phosphorylation at Ser-473 regulates heterochromatin protein 1 binding and co-repressor function of TIF1β/KAP-1. BMC Mol. Biol. 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. White D., Rafalska-Metcalf I. U., Ivanov A. V., Corsinotti A., Peng H., Lee S. C., Trono D., Janicki S. M., Rauscher F. J., 3rd (2012) The ATM substrate KAP-1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Mol. Cancer Res. 10, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S. J. (2000) Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 97, 10389–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Löbrich M., Shibata A., Beucher A., Fisher A., Ensminger M., Goodarzi A. A., Barton O., Jeggo P. A. (2010) γH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations, and optimization. Cell Cycle 9, 662–669 [DOI] [PubMed] [Google Scholar]
- 32. Guenatri M., Bailly D., Maison C., Almouzni G. (2004) Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166, 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murray J. M., Stiff T., Jeggo P. A. (2012) DNA double-strand break repair within heterochromatic regions. Biochem. Soc. Trans. 40, 173–178 [DOI] [PubMed] [Google Scholar]
- 34. Xu Y., Price B. D. (2011) Chromatin dynamics and the repair of DNA double-strand breaks. Cell Cycle 10, 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baldeyron C., Soria G., Roche D., Cook A. J., Almouzni G. (2011) HP1-α recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol. 193, 81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lomberk G., Wallrath L., Urrutia R. (2006) The heterochromatin protein 1 family. Genome Biol. 7, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.