Failure to inactivate APC/CCdhA at the G1–S boundary of the cell cycle as a result of a γ-tubulin mutation that disrupts the APC/CCdhA localization prevents cell cycle progression.
Abstract
A γ-tubulin mutation in Aspergillus nidulans, mipA-D159, causes failure of inactivation of the anaphase-promoting complex/cyclosome (APC/C) in interphase, resulting in failure of cyclin B (CB) accumulation and removal of nuclei from the cell cycle. We have investigated the role of CdhA, the A. nidulans homologue of the APC/C activator protein Cdh1, in γ-tubulin–dependent inactivation of the APC/C. CdhA was not essential, but it targeted CB for destruction in G1, and APC/CCdhA had to be inactivated for the G1–S transition. mipA-D159 altered the localization pattern of CdhA, and deletion of the gene encoding CdhA allowed CB to accumulate in all nuclei in strains carrying mipA-D159. These data indicate that mipA-D159 causes a failure of inactivation of APC/CCdhA at G1–S, perhaps by altering its localization to the spindle pole body, and, thus, that γ-tubulin plays an important role in inactivating APC/CCdhA at this point in the cell cycle.
Introduction
γ-Tubulin localizes to polar microtubule–organizing centers (PMTOCs) such as centrosomes and spindle pole bodies (SPBs), and γ-tubulin complexes play a critical role in nucleating microtubule assembly (Wiese and Zheng, 1999; Oakley, 2000; Job et al., 2003). Analyses of γ-tubulin mutations have revealed that γ-tubulin has additional essential functions that, as yet, have been incompletely determined (Lange, 2002; Cuschieri et al., 2007).
A great deal of data indicates that PMTOCs and PMTOC proteins, including γ-tubulin complex proteins, play a key role in regulating the cell cycle and, in particular, the G1–S transition (Maniotis and Schliwa, 1991; Vardy and Toda, 2000; Hinchcliffe et al., 2001; Rieder et al., 2001; Balczon et al., 2002; Gromley et al., 2003; Matsumoto and Maller, 2004; Doxsey et al., 2005; Sluder, 2005; Srsen et al., 2006; Cuschieri et al., 2007; Mikule et al., 2007; Cheng et al., 2008; Ferguson and Maller, 2010; Nayak et al., 2010). The mechanisms by which the PMTOCs regulate the G1–S transition are incompletely understood, but it has been hypothesized that PMTOCs act as signaling centers that “bind cell cycle regulatory molecules in a way that activates their function or raises their local concentration to the point that essential reactions occur in a timely fashion” (Hinchcliffe et al., 2001).
In Aspergillus nidulans, only the cells at the tips of growing hyphae go through the cell cycle. These cells are multinucleate, and the nuclei within a tip cell go through the cell cycle in synchrony. Cyclin B (CB) and cyclin-dependent kinase 1 (Cdk1) accumulate in nuclei in the S and G2 phases of the cell cycle and disappear from nuclei in mitosis (Nayak et al., 2010). CB is required for both S phase and mitosis in A. nidulans (O’Connell et al., 1992; Osmani et al., 1994; Nayak et al., 2010). Nayak et al. (2010) found that at restrictive temperatures, the cold-sensitive γ-tubulin allele mipA-D159 caused individual nuclei within a tip cell to fail to accumulate CB, Cdk1, and the phosphatase Ancdc14 in interphase. As a result, these nuclei were permanently taken out of the cell cycle, whereas other nuclei in the same cell continued to cycle. After each mitosis, more nuclei became noncycling. Extensive experimentation revealed that in the noncycling nuclei, a key cell cycle regulatory complex, the anaphase-promoting complex/cyclosome (APC/C), was constitutively active, continuously targeting CB for destruction and preventing entry into S phase. These data revealed that γ-tubulin plays an unexpected but important role in inactivating the APC/C at some point between late mitosis and S phase.
The APC/C is activated by binding to two proteins, Cdc20 and Cdh1 (Acquaviva and Pines, 2006; Thornton and Toczyski, 2006; van Leuken et al., 2008). Cdh1 activates the APC/C in G1 preventing accumulation of cyclins required for S phase, and APC/CCdh1 must be inactivated for the cell cycle to proceed beyond G1 (Li and Zhang, 2009). Based on the data of Nayak et al. (2010), one model for the continuous activation of the APC/C is that mipA-D159 causes a failure of inactivation of APC/CCdh1 at the G1–S boundary. If true, this would indicate that γ-tubulin plays an important role in inactivation of APC/CCdh1, and it might provide a link between G1–S regulation and PMTOCs. Here, we report our tests of this model.
Results and discussion
Identification and localization of an A. nidulans cdh1 orthologue
BLAST searches revealed that the predicted product of the A. nidulans gene AN2965 (using the Aspergillus Genome Database gene numbering system) has strong homology to Cdh1 (3e-157 vs. the human homologue). Our localizations and functional analyses (see following paragraphs) indicate strongly, moreover, that AN2965 is the A. nidulans cdh1 orthologue. We designate AN2965 cdhA and its product CdhA.
We fused the GFP coding sequence in frame with the 3′ end of the cdhA gene. The fusion gene was the only copy of the cdhA gene in the genome, and it was fully functional, supporting normal growth at temperatures from 20–42°C (Fig. S1). To correlate CdhA-GFP localization with cell cycle and mitotic stages, we created strains expressing both CdhA-GFP and histone H1–monomeric RFP (mRFP; strains listed in Table S1). In mitosis, CdhA-GFP began to be visible at the SPB in anaphase (Fig. 1), and the signal increased during telophase. Long-term time-lapse datasets captured at 25°C revealed that it remained visible at the SPB for 39 ± 12 min (mean ± SD) after mitosis (Fig. 2, A–C). It then disappeared from the SPB but remained faintly visible in the nucleoplasm (Fig. 2, G–I). The same datasets revealed that the cell cycle was 199 ± 49 min at this temperature, so after mitosis, CdhA is visible at the SPB for ∼20% of the cell cycle. At 32°C, G1 takes ∼15% of the cell cycle (Bergen and Morris, 1983), so the disappearance of CdhA from the SPB corresponds reasonably well to the expected time of the G1–S transition. CdhA reappeared at the SPB 48 ± 16 min before mitosis (n = 36; Fig. 2, J–L). This corresponds to 74.2% of the cell cycle if one takes mitotic exit as one’s starting point. At 32°C, G1 and S have been calculated to occupy ∼55% of the cell cycle (Bergen and Morris, 1983), so CdhA reappears at the SPB in G2. It disappeared from the SPB 4 ± 2 min before mitotic chromosomal condensation was detectable (n = 26). We were able to verify the SPB localization of CdhA because it colocalized with Cdk1-mCherry fusion (Fig. S1), and the SPB location of Cdk1-mCherry has previously been established (Nayak et al., 2010). Interestingly, the localization pattern of CdhA is different from that of the APC/C. The APC/C is concentrated in the nucleus throughout the cell cycle in A. nidulans and does not relocate to and from the SPB (Nayak et al., 2010). This indicates that CdhA has its own localization determinants, and its localization is not dependent on the APC/C. These data also indicate that the association of CdhA with the APC/C is transient because, if it were stable, the APC/C and CdhA would show similar localization patterns, at least from late mitosis to the end of G1, when CdhA activates the APC/C.
Figure 1.
Localization of CdhA-GFP in mitosis. (A–I) Images are projections of z stacks collected at 1-min intervals at 25°C. At 0 min (metaphase), CdhA-GFP is barely detectable (A–C). At anaphase (D–F), it is visible at three out of four SPBs (arrows) and between separating chromatin masses (arrowheads in D and F). This spindlelike localization was seen in less than a quarter of the nuclei observed, perhaps because the localization is very brief. At the 2-min time point (telophase; G–I), CdhA-GFP is present at the SPBs of each daughter nucleus.
Figure 2.
Localization of CdhA-GFP throughout the cell cycle. (A–L) Images are z-stack projections from a time-lapse dataset collected at 25°C. At the M–G1 transition (0 min, bottom two nuclei; A–C), it is present on SPBs but not evident in the nucleoplasm. In early G1 (D–F), it is at the SPB and faintly visible in the nucleoplasm. The SPB signal is gone in S phase (G–I). CdhA-GFP is only faintly visible in the nucleoplasm. In G2, CdhA-GFP reappears at SPBs (J–L).
cdhA is not essential
We deleted cdhA by replacing it with the Aspergillus fumigatus pyrG gene (AfpyrG; confirmed by diagnostic PCR and Southern hybridizations; Fig. S2). Thus, the cdhA gene is not essential. cdhA deletion (cdhAΔ) strains grew well at temperatures from 20 to 42°C but showed a slight reduction in growth at 42°C (Fig. S2).
APC/CCdhA targets CB for destruction in G1
APC/CCdh1 destroys S-phase cyclins in G1 in many organisms, preventing the premature initiation of S (Li and Zhang, 2009). The timing of CdhA localization to the SPB in G1 suggested it might have the same function in A. nidulans. In A. nidulans, CB is required for S phase as well as mitosis (Osmani et al., 1994), and it is destroyed during mitosis (De Souza et al., 2009; Nayak et al., 2010). It remains undetectable in G1 nuclei but becomes visible in nuclei during S and remains visible in nuclei in G2 (Nayak et al., 2010). As mentioned, only tip cell nuclei go through the cell cycle in A. nidulans, and they go through it in synchrony. The fraction of tip cells in which CB is absent from nuclei is, thus, proportional to the length of G1 (Nayak et al., 2010). If APC/CCdhA targets CB for destruction in G1, deletion of cdhA should allow CB to accumulate earlier in the cell cycle, and the percentage of tip cells with nuclei lacking CB should be reduced. To determine whether this was the case, we grew cdhA+ and cdhAΔ strains expressing histone H1–mRFP and CB-GFP for 21 h at 25°C. We then captured z-series stacks of random fields over a 1-h period and scored tip cells for the presence of CB-GFP in nuclei. In agreement with previous data (Nayak et al., 2010), the mean percentage of tip cells with nuclei with visible CB was 60.0 ± 5.4% (six experiments, two strains). In a cdhAΔ strain, 87.2 ± 3.5% of hyphal tips had nuclei with visible CB-GFP (three experiments). The difference was highly significant (P = 0.00008, unpaired t test). Thus, deletion of cdhA shortens the period in which CB is absent from nuclei. Given that mitosis occupies ∼5% of the cell cycle, in cdhAΔ strains, CB is undetectable in nuclei in only 7–8% of interphase tip cells. It may be accumulating in these nuclei but at undetectable levels. These data indicate that CdhA plays a key role in preventing the accumulation of CB in G1 in A. nidulans. We can reasonably conclude, based on these data and abundant data on the function of Cdh1 in other organisms, that APC/CCdhA targets CB for destruction in G1, preventing the onset of S.
After mitosis, CdhA is located at the SPB for ∼20% of the cell cycle, but CB does not become detectable until several minutes later, ∼40% of the way through the cell cycle. This apparent timing discrepancy is undoubtedly a result, in part, of the fact that after APC/CCdhA is inactivated, it takes a few minutes for CB to accumulate to levels that we can detect microscopically. There may also be some residual APC/CCdhA activity for a few minutes after CdhA-GFP is no longer detectable at the SPB.
Time-lapse imaging revealed that the cell cycle time in a cdhAΔ strain (LO2019) was 178 ± 39 min versus 199 ± 49 min in a cdhA+ control strain. In spite of overlapping SDs, the difference in cell cycle length (21 min) is statistically significant (P = 0.005, unpaired t test), and this reveals that deletion of cdhA shortens the cell cycle, consistent with the observed reduction in the length of G1. The large SDs reflect cell-to-cell variation in cell cycle length rather than experimental imprecision.
To determine whether mipA-D159 altered the timing of CB accumulation in cdhA+ and cdhAΔ strains, we grew strains at 25°C (a restrictive temperature for mipA-D159) for 21 h and captured z-series stacks of random fields over a 1-h period. In a cdhA+ mipA-D159 strain, 60.7 ± 11.4% (three experiments) of hyphal tips contained nuclei with CB. This was essentially identical to the mipA+ strain. In these multinucleate hyphal tip cells, some nuclei do not accumulate CB because of the nuclear autonomous, constitutive activation of the APC/C caused by mipA-D159. We scored tip cells as positive if any nuclei contained CB. In a cdhAΔ mipA-D159 strain, 91 ± 3.6% (three experiments) of hyphal tip cells was CB positive (CB+), similar to mipA+ cdhAΔ strains. Thus, mipA-D159 does not appear to affect the timing of the accumulation of CB. Rather, it simply prevents some nuclei from accumulating it at all.
Cdh1 is required to prevent CB accumulation in subapical nuclei
In A. nidulans, subapical cells are blocked in G1 with no CB accumulating in nuclei (Nayak et al., 2010). They resume mitotic activity when side branches emerge from them. Little is known about the mechanism of G1 blockage in subapical nuclei. To determine whether CdhA is required to prevent the accumulation of CB in subapical cells, we examined CB-GFP accumulation in subapical cells in a cdhAΔ strain (Fig. 3). CB-GFP was detectable in nuclei of 98.6 ± 2.4% (three experiments, n = 80 cells) of subapical cells in randomly chosen fields, whereas in a cdhA+ strain, CB-GFP was present in nuclei of only 2.9 ± 2.6% of subapical cells (n = 90, three experiments). This implies that APC/CCdhA is constitutively active in nuclei in subapical cells and helps to keep them in G1. Interestingly, even though nuclei in subapical cdhAΔ cells do accumulate CB-GFP, time-lapse imaging revealed that they do not undergo mitosis until a new side branch forms. There must be an additional, currently unknown cell cycle blocking mechanism in these cells. CB accumulated at SPBs in these nuclei (Fig. 3 A) and in tip cell nuclei in cdhAΔ strains, so CdhA is not required for localization of CB to the SPB.
Figure 3.
Deletion of cdhA results in the accumulation of CB in subapical cells at 25°C. (A) The arrows in the brightfield images indicate septa, showing the boundaries of each cell and revealing that each cell is subapical (i.e., has no hyphal apex). In the cdhA+ strain, CB-GFP did not accumulate in this cell or other subapical cells, but in the cdhAΔ strain, it did accumulate. CB accumulated at SPBs in the cdhAΔ strain (arrowheads; brightfield images are single focal plane, and others are projections). (B) Quantitation of the accumulation of CB-GFP in subapical cells of cdhA+ and cdhAΔ strains (mean of three experiments for each strain; error bars are SDs; n = 90 for cdhA+; n = 80 for cdhAΔ).
γ-Tubulin plays a key role in inactivating APC/CCdhA
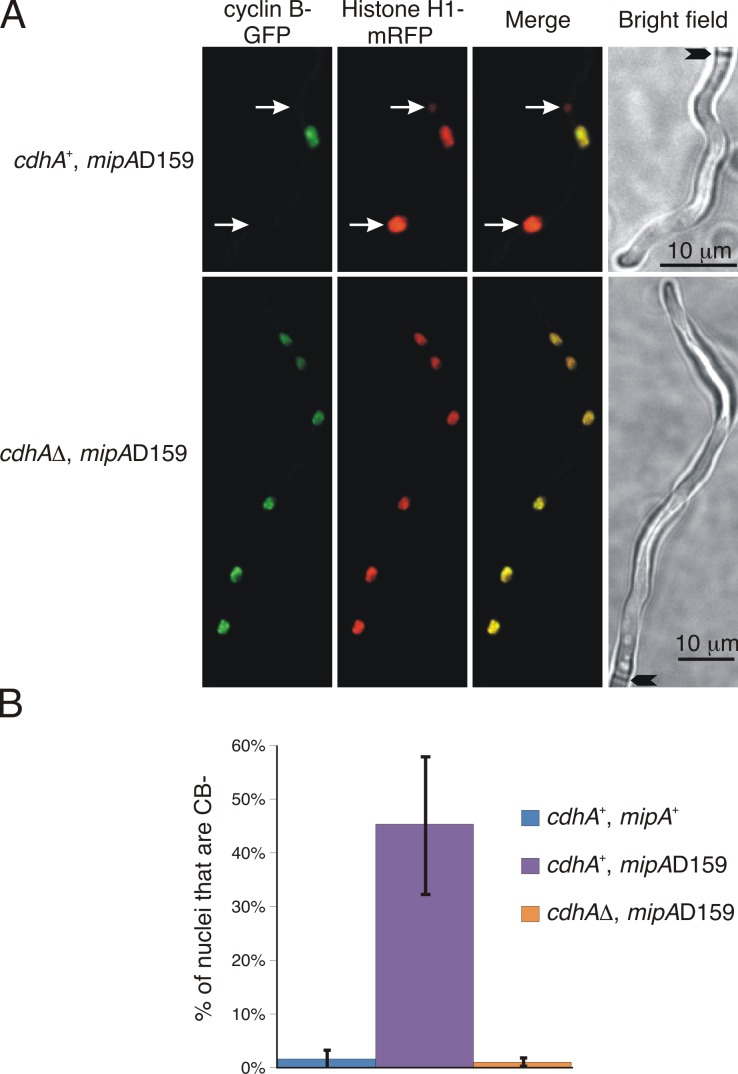
If mipA-D159 causes constitutive APC/C activity by causing a failure of inactivation of APC/CCdhA, deletion of cdhA would reverse the constitutive activation. This should reduce or eliminate nuclei that fail to accumulate CB (CB− nuclei) at restrictive temperatures. To test this possibility, we created strains carrying mipA-D159 or mipA+ and cdhAΔ or cdhA+ and expressing CB-GFP and histone H1–mRFP. As mentioned, nuclei go through the cell cycle in synchrony. In mipA+ cells, if CB is present in one nucleus of a tip cell, it is present in all nuclei in the same cell; but, in mipA-D159 tip cells at restrictive temperatures, some nuclei are CB− because of nuclear autonomous APC/C activity, whereas other nuclei in the same cell are CB+ (Nayak et al., 2010). We grew the strains at 25°C (a restrictive temperature for mipA-D159) for 21 h and collected z-series stacks of random fields over a 1-h period. We determined the percentage of CB− nuclei in tip cells in which at least one nucleus contained visible CB. With two cdhA+ mipA+ strains, only 1.5 ± 1.6% of nuclei was CB− (Fig. 4). In a cdhA+ mipA-D159 strain, 45.2 ± 13% of nuclei was CB− (three experiments). The CB− nuclei are those with constitutive APC/C activity resulting in the destruction of CB. In three cdhAΔ mipA-D159 strains, only 0.8 ± 0.7% of nuclei was CB− (nine experiments). Thus, deletion of cdhA reduces the numbers of CB− nuclei to wild-type (WT) levels at restrictive temperatures in strains carrying mipA-D159.
Figure 4.
Deletion of cdhA drastically reduces CB− nuclei. (A) Projections of z-series stacks of tip cells in strains carrying mipA-D159 grown at a restrictive temperature of 25°C (brightfield images are single focal plane). The top row (cdhA+) shows a tip cell with three nuclei, although they are different in size as a result of unequal segregation of chromosomes. Two of the nuclei lack CB-GFP (arrows). In the bottom row (cdhAΔ strain), CB-GFP accumulates in all nuclei. Arrowheads in the brightfield images indicate septa, revealing that each cell is a tip cell. (B) Quantification of CB− nuclei. Values are means, and error bars are SD. cdhA+ mipA+ data are from six experiments with two different strains. cdhA+ mipA-D159 data are from three experiments. cdhAΔ mipA-D159 data are from nine experiments with three different strains.
From these data, we can conclude that CdhA is required for the continuous nuclear-autonomous destruction of CB that is caused by mipA-D159 at restrictive temperatures. Based on these data, our previous demonstration that continuous destruction of CB is a result of constitutive activation of the APC/C (Nayak et al., 2010), the fact that Cdh1 activates the APC/C, and our previous data on the timing of the failure of APC/C inactivation (Nayak et al., 2010), we can deduce that in strains carrying a WT cdhA gene, mipA-D159 causes a nuclear-autonomous failure of inactivation of APC/CCdhA at G1–S, resulting in failure of accumulation of CB and other APC/CCdhA substrates necessary for cell cycle progression. It follows that γ-tubulin must have an important role in inactivating APC/CCdhA and, thus, in regulating the G1–S transition.
mipA-D159 alters the localization pattern of CdhA
It has been suggested that Cdh1 is activated at centrosomes (Raff et al., 2002). Our data are consistent with the hypothesis that localization of CdhA at the SPB is necessary, but not sufficient, for APC/CCdhA activity. From late mitosis until the G1–S boundary, when APC/CCdhA is strongly predicted to be active, CdhA is located at the SPB. At the G1–S boundary, when APC/CCdhA becomes inactive, CdhA leaves the SPB but is still present in the nucleoplasm. CdhA comes back to the SPB in G2 when it is not known to be active, but by then, CB and Cdk1 have accumulated, and they inactivate Cdh1 by phosphorylating it (Zachariae et al., 1998).
If the SPB localization of CdhA is important for its activity, mipA-D159 might alter CdhA function by altering its localization. To test this, we needed to be able to image CdhA, nuclei, and CB, or a surrogate, in the same cell. Our best option, based on the intensity and stability of the fluorescent proteins at our disposal, was to use CdhA-GFP, nup49-mCherry, which localizes to the nuclear envelope and allows nuclei to be distinguished (De Souza et al., 2009; Nayak et al., 2010), and Cdk1-mCherry. Cdk1-mCherry is brighter and more stable than CB-mCherry and more amenable to long-term imaging. CB and Cdk1 form a complex, and nuclei that fail to accumulate CB also fail to accumulate Cdk1 (Nayak et al., 2010). CB− nuclei are easily distinguished because nup49-mCherry labels the nuclear periphery, but Cdk1-mCherry does not accumulate in the nucleoplasm, although it accumulates in other nuclei in the same tip cell. We created a strain carrying mipA-D159 and expressing these fluorescent proteins and performed time-lapse imaging at a restrictive temperature of 25°C. Strikingly, when CdhA disappeared from SPBs in late G1 in normally cycling nuclei, CdhA remained at the SPBs of CB− nuclei in the same cell and was at a higher concentration in the nucleoplasm of CB− nuclei than in the CB+ nuclei (n = 21; Fig. 5 A). Thus, mipA-D159 causes failure of CdhA to dissociate from the SPB in a way that correlates with the failure of inactivation of APC/CCdhA. An obvious model is that mipA-D159 causes failure of dissociation of CdhA from the SPB at G1–S, which, in turn, causes constitutive activation of the APC/C.
Figure 5.
Projections of z-series stacks of CdhA and CB− nuclei in a strain carrying mipA-D159 at 25°C. (A) CdhA persists in CB− nuclei. The three nuclei are in the same cell. nup49-mCherry shows the periphery of each nucleus. The top nucleus is in S or early G2. CdhA-GFP is not detectable, but Cdk1-mCherry fills the nucleoplasm, except for a small region that corresponds to the nucleolus. The bottom two nuclei contain CdhA-GFP in the nucleoplasm and at their SPBs (arrows). They do not contain Cdk1 in the nucleoplasm, and they are, therefore, CB−. (B) CdhA disappears from the SPB (arrows) and nucleoplasm of CB− nuclei in mitosis. Images are z-stack projections from a time-lapse sequence, and the fluorescent proteins are the same as in A. The nucleus does not contain Cdk1-mCherry and is, thus, CB−. In mitosis (20 min), the CdhA-GFP disappears from the nucleoplasm and SPB, but, in the next interphase (220 min), it is again in the nucleoplasm and at the SPB, and the nucleus remains CB−.
Although CdhA-GFP did not disappear from the SPBs of CB− nuclei at G1–S, it did disappear in mitosis and reappear in G1 just as it did in CB+ nuclei (Fig. 5 B). We have previously found that once nuclei become CB−, they remain CB− (Nayak et al., 2010). Our data are consistent with the possibility that a stable, nuclear-autonomous change occurs to the SPB that precludes the destruction of CdhA at G1–S.
mipA-D159 is recessive (Jung et al., 2001; unpublished data) and is, thus, a loss-of-function rather than a gain-of-function mutation. The failure of dissociation of CdhA from the SPB is, thus, not a result of enhanced binding of CdhA to γ-tubulin, for example, and our data do not, in fact, provide any indication that CdhA interacts physically with γ-tubulin. Rather, they are more consistent with the possibility that γ-tubulin interacts, directly or indirectly, with a protein or protein complex required for destruction of CdhA or dislocation of CdhA from the SPB.
Materials and methods
Strains and media
The strains used for these experiments along with their genotypes are listed in Table S1. YAG (5 g/liter yeast extract, 20 g/liter d-glucose, and 15 g/liter agar supplemented with 400 µl/liter of a trace element solution; Vishniac and Santer, 1957) was used as a solid complete medium. Minimal medium consisted of 6 g/liter NaNO3, 0.52 g/liter KCl, 0.52 g/liter MgSO4 ⋅ 7H2O, 1.52 g/liter KH2PO4, 10 g/liter d-glucose, and 400 µl/liter of trace element solution (Vishniac and Santer, 1957) and appropriate nutrients to supplement nutritional markers carried by the strains. pH was adjusted to 6.0–6.5, and 15 g/liter of agar was added for solid minimal medium. For imaging, strains were cultured in liquid minimal medium in eight-chambered cover glasses (Lab-Tek; Thermo Fisher Scientific) with necessary supplements to complement nutritional markers.
Gene targeting and transformation
Transforming molecules for the cdhA deletion and C-terminal fluorescent protein fusions were constructed by fusion PCR, as previously described (Nayak et al., 2006, 2010; Szewczyk et al., 2006). The cdhA deletion was created by transforming with a fragment carrying the selectable marker AfpyrG (Weidner et al., 1998) flanked by a 1,000-bp fragment upstream of cdhA and a 1,000-bp fragment downstream of cdhA. To create C-terminal fusion proteins, the transforming molecules consisted of ∼1,000 bp of the C-terminal coding sequence of the target protein fused in frame to a glycine–alanine linker (Yang et al., 2004), which was, in turn, fused in frame with the fluorescent protein coding sequence. The fluorescent protein coding sequence was followed by a selectable marker and an ∼1,000-bp sequence downstream of the target gene. The GFP variant we used was a plant-adapted GFP (Fernández-Abalos et al., 1998). The mCherry sequence was the original version described by Shaner et al. (2004), and the mRFP variant was as previously described (Campbell et al., 2002; Toews et al., 2004). The selectable markers used were AfpyrG, AfriboB, and AfpyroA from A. fumigatus (Weidner et al., 1998; Nayak et al., 2006). Recipient strains carried a deletion of the nkuA gene to minimize nonhomologous recombination (Nayak et al., 2006). A strain carrying nup49-mCherry was provided by C. De Souza and S. Osmani (The Ohio State University, Columbus, OH). Strains carrying nup49-mCherry along with other fluorescent protein fusions were constructed by crosses.
Diagnostic PCR
Transformants were verified by diagnostic PCR with multiple primer sets. A. nidulans genomic DNA was isolated as previously described (Lee and Taylor, 1990; Hervás-Aguilar et al., 2007), and diagnostic PCR was performed with AccuPrime Taq High Fidelity or Platinum Taq (both from Invitrogen). Alternatively, conidia were taken from the surface of a colony using a toothpick and suspended in 50 µl of TE buffer (10 mM Tris-HCl, pH 7.5, and 1 mM EDTA) in a microcentrifuge tube. 50 µl of 0.45–0.55-mm glass beads was added, and the tube was vortexed for 2 min. 2 µl of solution was immediately taken out and added to 18 µl of TE. 2 µl of this dilution was used for PCR using OneTaq Hot Start Quick-Load (New England Biolabs, Inc.) according to the manufacturer’s instructions.
Southern hybridizations
For Southern hybridizations, A. nidulans genomic DNA was isolated by either of two published protocols (Oakley et al., 1987; Lee and Taylor, 1990). Hybridizations were performed with a dried gel method (Oakley et al., 1987). Radioactively labeled full-length transforming DNA fragments were used as probes and were labeled using the Prime-It II Random Primer Labeling Kit (Agilent Technologies) and purified using spin-column chromatography through Sephadex G-50 (Sigma-Aldrich; Maniatis et al., 1982).
Microscopy
Imaging was performed with two systems. One was a spinning-disk confocal system (UltraVIEW VoX; PerkinElmer) mounted on an inverted microscope (IX71; Olympus). It was equipped with a constant-temperature chamber and a piezoelectric stage for rapid z-axis movement. Images were collected using a 60× 1.42 NA Plan Apo objective and an ORCA-ERAG camera (Hamamatsu Photonics). The system was controlled by Volocity software (PerkinElmer) running on a Power Mac computer (Apple). Solid-state 488- and 561-nm lasers were used for excitation. Fluorochrome-specific emission filters were used to prevent fluorochrome emission bleed-through. The second was an IX71 microscope equipped with a mercury illumination source along with Prior shutters, filter wheels, and z-axis drives and an ORCA-ERAG camera. For this system, we also collected images with a 60× 1.42 NA Plan Apo objective. A 459–481-nm bandpass excitation filter was used for GFP, and a 546–566-nm bandpass excitation filter was used for mRFP. We used a dual reflection band dichroic (457–480 and 542–565-nm reflection bands, 500–529 and 584–679-nm transmission bands) and a 499–529-nm emission filter for GFP and a 580–654-nm emission filter for mRFP. The system was controlled by Volocity software running on a Power Mac computer. Both systems were calibrated with a stage micrometer. Minimum and maximum intensity cutoffs (black level and white level) for each channel were chosen in Volocity before images were exported. No other adjustments were made to the images. Figures were prepared from exported images using CorelDRAW (Corel Corporation) with no further adjustments.
Online supplemental material
Fig. S1 shows the growth of CdhA-GFP fusion strains and control strains at temperatures ranging from 20 to 42°C. It also shows that CdhA and Cdk1 colocalize at the SPB. Fig. S2 shows the growth of cdhAΔ and control strains from 20 to 42°C, and it shows Southern hybridizations that verify that cdhA has been deleted. Table S1 shows the genotypes of strains used for this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201203115/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Tetsuya Horio for advice and assistance, C. Elizabeth Oakley for technical and editorial assistance, and Dr. Colin De Souza and Dr. Stephen Osmani for the nup49-mCherry construct.
This work was supported by National Institutes of Health grant GM031837 and by the Irving S. Johnson fund of the University of Kansas Endowment.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- CB
- cyclin B
- mRFP
- monomeric RFP
- PMTOC
- polar microtubule–organizing center
- SPB
- spindle pole body
- WT
- wild type
References
- Acquaviva C., Pines J. 2006. The anaphase-promoting complex/cyclosome: APC/C. J. Cell Sci. 119:2401–2404 10.1242/jcs.02937 [DOI] [PubMed] [Google Scholar]
- Balczon R., Simerly C., Takahashi D., Schatten G. 2002. Arrest of cell cycle progression during first interphase in murine zygotes microinjected with anti-PCM-1 antibodies. Cell Motil. Cytoskeleton. 52:183–192 10.1002/cm.10043 [DOI] [PubMed] [Google Scholar]
- Bergen L.G., Morris N.R. 1983. Kinetics of the nuclear division cycle of Aspergillus nidulans. J. Bacteriol. 156:155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882 10.1073/pnas.082243699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Türkel N., Hemati N., Fuller M.T., Hunt A.J., Yamashita Y.M. 2008. Centrosome misorientation reduces stem cell division during ageing. Nature. 456:599–604 10.1038/nature07386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuschieri L., Nguyen T., Vogel J. 2007. Control at the cell center: The role of spindle poles in cytoskeletal organization and cell cycle regulation. Cell Cycle. 6:2788–2794 10.4161/cc.6.22.4941 [DOI] [PubMed] [Google Scholar]
- De Souza C.P., Hashmi S.B., Nayak T., Oakley B., Osmani S.A. 2009. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell. 20:2146–2159 10.1091/mbc.E08-08-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15:303–311 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Ferguson R.L., Maller J.L. 2010. Centrosomal localization of cyclin E-Cdk2 is required for initiation of DNA synthesis. Curr. Biol. 20:856–860 10.1016/j.cub.2010.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Abalos J.M., Fox H., Pitt C., Wells B., Doonan J.H. 1998. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol. Microbiol. 27:121–130 10.1046/j.1365-2958.1998.00664.x [DOI] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161:535–545 10.1083/jcb.200301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás-Aguilar A., Rodríguez J.M., Tilburn J., Arst H.N., Jr, Peñalva M.A. 2007. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J. Biol. Chem. 282:34735–34747 10.1074/jbc.M706723200 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Miller F.J., Cham M., Khodjakov A., Sluder G. 2001. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 291:1547–1550 10.1126/science.1056866 [DOI] [PubMed] [Google Scholar]
- Job D., Valiron O., Oakley B. 2003. Microtubule nucleation. Curr. Opin. Cell Biol. 15:111–117 10.1016/S0955-0674(02)00003-0 [DOI] [PubMed] [Google Scholar]
- Jung M.K., Prigozhina N., Oakley C.E., Nogales E., Oakley B.R. 2001. Alanine-scanning mutagenesis of Aspergillus γ-tubulin yields diverse and novel phenotypes. Mol. Biol. Cell. 12:2119–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B.M.H. 2002. Integration of the centrosome in cell cycle control, stress response and signal transduction pathways. Curr. Opin. Cell Biol. 14:35–43 10.1016/S0955-0674(01)00291-5 [DOI] [PubMed] [Google Scholar]
- Lee S.B., Taylor J.W. 1990. Isolation of DNA from fungal mycelia and single spores. PCR Protocols: A Guide to Methods and Applications. Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., Academic Press, Inc., San Diego/New York/Berkeley/Boston/London/Sydney/Tokyo/Toronto: 282–287 [Google Scholar]
- Li M., Zhang P. 2009. The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 4:2 10.1186/1747-1028-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., Sambrook J. 1982. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory, New York: 545 pp [Google Scholar]
- Maniotis A., Schliwa M. 1991. Microsurgical removal of centrosomes blocks cell reproduction and centriole generation in BSC-1 cells. Cell. 67:495–504 10.1016/0092-8674(91)90524-3 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Maller J.L. 2004. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 306:885–888 10.1126/science.1103544 [DOI] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. 2007. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 9:160–170 10.1038/ncb1529 [DOI] [PubMed] [Google Scholar]
- Nayak T., Szewczyk E., Oakley C.E., Osmani A., Ukil L., Murray S.L., Hynes M.J., Osmani S.A., Oakley B.R. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 172:1557–1566 10.1534/genetics.105.052563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T., Edgerton-Morgan H., Horio T., Xiong Y., De Souza C.P., Osmani S.A., Oakley B.R. 2010. γ-tubulin regulates the anaphase-promoting complex/cyclosome during interphase. J. Cell Biol. 190:317–330 10.1083/jcb.201002105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M.J., Osmani A.H., Morris N.R., Osmani S.A. 1992. An extra copy of nimEcyclinB elevates pre-MPF levels and partially suppresses mutation of nimTcdc25 in Aspergillus nidulans. EMBO J. 11:2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B.R. 2000. γ-Tubulin. Curr. Top. Dev. Biol. 49:27–54 10.1016/S0070-2153(99)49003-9 [DOI] [PubMed] [Google Scholar]
- Oakley C.E., Weil C.F., Kretz P.L., Oakley B.R. 1987. Cloning of the riboB locus of Aspergillus nidulans. Gene. 53:293–298 10.1016/0378-1119(87)90019-9 [DOI] [PubMed] [Google Scholar]
- Osmani A.H., van Peij N., Mischke M., O’Connell M.J., Osmani S.A. 1994. A single p34cdc2 protein kinase (encoded by nimXcdc2) is required at G1 and G2 in Aspergillus nidulans. J. Cell Sci. 107:1519–1528 [DOI] [PubMed] [Google Scholar]
- Raff J.W., Jeffers K., Huang J.Y. 2002. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J. Cell Biol. 157:1139–1149 10.1083/jcb.200203035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Faruki S., Khodjakov A. 2001. The centrosome in vertebrates: More than a microtubule-organizing center. Trends Cell Biol. 11:413–419 10.1016/S0962-8924(01)02085-2 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Sluder G. 2005. Two-way traffic: Centrosomes and the cell cycle. Nat. Rev. Mol. Cell Biol. 6:743–748 10.1038/nrm1712 [DOI] [PubMed] [Google Scholar]
- Srsen V., Gnadt N., Dammermann A., Merdes A. 2006. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J. Cell Biol. 174:625–630 10.1083/jcb.200606051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E., Nayak T., Oakley C.E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S.A., Oakley B.R. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 (published erratum appears in Nat. Protoc. 2006. 1:following 31120) 10.1038/nprot.2006.405 [DOI] [PubMed] [Google Scholar]
- Thornton B.R., Toczyski D.P. 2006. Precise destruction: An emerging picture of the APC. Genes Dev. 20:3069–3078 10.1101/gad.1478306 [DOI] [PubMed] [Google Scholar]
- Toews M.W., Warmbold J., Konzack S., Rischitor P., Veith D., Vienken K., Vinuesa C., Wei H., Fischer R. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45:383–389 10.1007/s00294-004-0495-7 [DOI] [PubMed] [Google Scholar]
- van Leuken R., Clijsters L., Wolthuis R. 2008. To cell cycle, swing the APC/C. Biochim. Biophys. Acta. 1786:49–59 [DOI] [PubMed] [Google Scholar]
- Vardy L., Toda T. 2000. The fission yeast γ-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19:6098–6111 10.1093/emboj/19.22.6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishniac W., Santer M. 1957. The thiobacilli. Bacteriol. Rev. 21:195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner G., d’Enfert C., Koch A., Mol P.C., Brakhage A.A. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378–385 10.1007/s002940050350 [DOI] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. 1999. γ-tubulin complexes and their interaction with microtubule-organizing centers. Curr. Opin. Struct. Biol. 9:250–259 10.1016/S0959-440X(99)80035-9 [DOI] [PubMed] [Google Scholar]
- Yang L., Ukil L., Osmani A., Nahm F., Davies J., De Souza C.P., Dou X., Perez-Balaguer A., Osmani S.A. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell. 3:1359–1362 10.1128/EC.3.5.1359-1362.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W., Schwab M., Nasmyth K., Seufert W. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 282:1721–1724 10.1126/science.282.5394.1721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.