CYK4 activity as a GTPase-activating protein is required during anaphase to inhibit Rac1-dependent effector pathways associated with control of cell spreading and adhesion.
Abstract
In mitosis, animal cells lose their adhesion to the surrounding surfaces and become rounded. During mitotic exit, they reestablish these adhesions and at the same time physically contract and divide. How these competing processes are spatially segregated at the cell cortex remains mysterious. To address this question, we define the specific effector pathways used by RhoA and Rac1 in mitotic cells. We demonstrate that the MKlp1–CYK4 centralspindlin complex is a guanosine triphosphatase–activating protein (GAP) for Rac1 and not RhoA and that CYK4 negatively regulated Rac1 activity at the cell equator in anaphase. Cells expressing a CYK4 GAP mutant had defects in cytokinesis and showed elevated staining for the cell adhesion marker vinculin. These defects could be rescued by depletion of ARHGEF7 and p21-activated kinase, Rac1-specific effector proteins required for cell adhesion. Based on these findings, we propose that CYK4 GAP activity is required during anaphase to inhibit Rac1-dependent effector pathways associated with control of cell spreading and adhesion.
Introduction
Dividing cells undergo dramatic changes in shape during mitosis and cytokinesis (Glotzer, 2005; Barr and Gruneberg, 2007). Adherent animal cells transition from a flattened “leaflike” morphology to a sphere, which then contracts at the equator to give rise to two new cells that readhere to the surrounding growth surfaces and adjacent cells. These processes of cell adhesion and cytokinesis involve Rho family GTPase regulation of the actin cytoskeleton (Heasman and Ridley, 2008; Parsons et al., 2010). The GTPase RhoA is central to the process of cytokinesis in animal cells (Piekny et al., 2005). In anaphase, RhoA is activated at a tightly defined equatorial region of the cell cortex by the specific and conserved guanine nucleotide exchange factor (GEF) ECT2 (epithelial cell transforming 2; Tatsumoto et al., 1999; Kimura et al., 2000; Yüce et al., 2005). ECT2 targets to the midpoint of the dividing cells through interactions with the microtubule-bound centralspindlin complex and phosphatidylinositol phosphates in the inner leaflet of the plasma membrane (Su et al., 2011). Furthermore, a pool of Polo-like kinase 1 present on the central spindle locally activates ECT2 (Niiya et al., 2006; Petronczki et al., 2007; Wolfe et al., 2009). In this way, ECT2 drives the highly localized activation of RhoA and forms a signal for cytokinesis. Once activated, RhoA is then thought to recruit effector proteins, including the formin diaphanous and one or more members of the Rho kinase family (Castrillon and Wasserman, 1994; Madaule et al., 1998; Hickson et al., 2006; Bassi et al., 2011). Together, these effector proteins promote actin polymerization and myosin-dependent cell contractility in this region, thereby giving rise to a cleavage furrow and contractile ring (Glotzer, 2005).
Several studies have shown that RhoA actively cycles between active and inactive forms during this process, suggesting that a GTPase-activating protein (GAP) is needed in addition to the ECT2 GEF (Miller and Bement, 2009). The CYK4 subunit of the centralspindlin complex has a RhoGAP domain and has been viewed as the prime candidate for this Rho GAP activity (Jantsch-Plunger et al., 2000; Mishima et al., 2002; Somers and Saint, 2003; Zavortink et al., 2005). However, biochemical analysis of CYK4 has found that it has little or no activity toward RhoA and has greater activity toward Rac1 (Touré et al., 1998; Jantsch-Plunger et al., 2000), and genetic evidence in Drosophila melanogaster indicates that the fly CYK4 inhibits Rac activity (D’Avino et al., 2004). This conundrum was apparently resolved when it was found that phosphorylation of CYK4 by Aurora kinases at the central spindle in anaphase resulted in a switch in relative activity from Rac1 to RhoA (Minoshima et al., 2003). This simple picture is complicated by other studies that indicate that although CYK4 is an essential component of the centralspindlin complex (Mishima et al., 2002), the GAP activity of CYK4 can be dispensed with under some conditions (Goldstein et al., 2005; Yamada et al., 2006). Most strikingly, chicken B cell lines engineered to express a CYK4 GAP mutant grow and divide without obvious defect (Yamada et al., 2006). Furthermore, in Caenorhabditis elegans, mutations that reduce GAP activity, but have little effect on the localization or stability of CYK4, result in cytokinesis defects that can be suppressed by depletion of Rac (Canman et al., 2008). Thus, CYK4 may directly limit Rac rather than RhoA activity during anaphase.
Here, we define the specific Rac1 and RhoA effector pathways operating during mitosis and investigate the function and regulation of CYK4 GAP activity in human cell lines. The results support the view that CYK4 is a crucial regulator of Rac1 in adherent cells and that inhibition of Rac1-dependent effector pathways in anaphase is needed for cytokinesis.
Results
CYK4 is a Rac1 and Cdc42 GAP
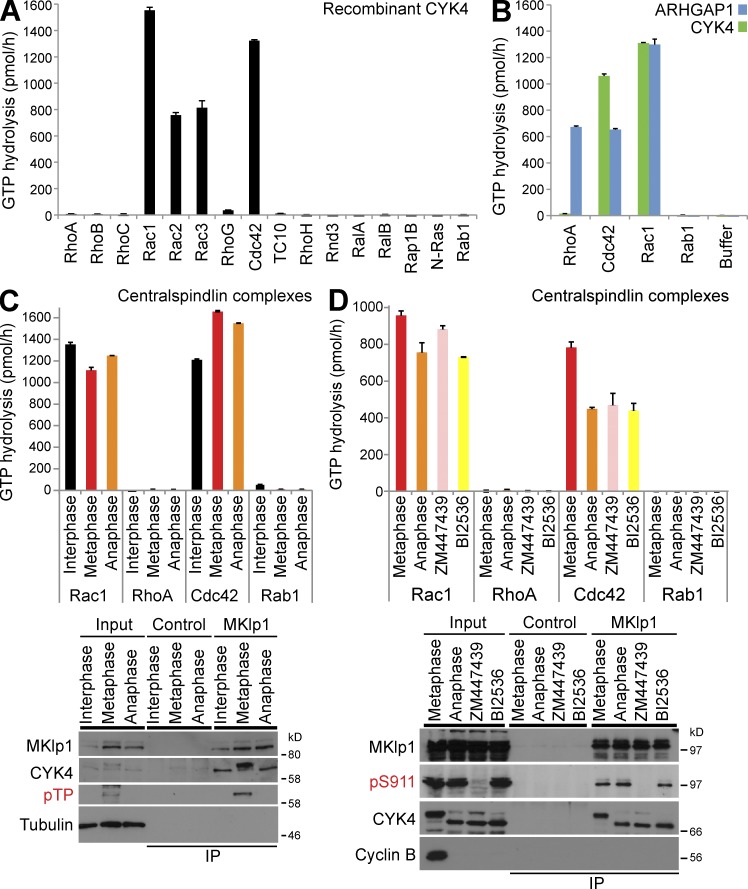
The CYK4 component of the centralspindlin complex has been reported to have both Rac1 and RhoA GAP activity (Jantsch-Plunger et al., 2000) and to be regulated by phosphorylation during progression through mitosis and cytokinesis (Minoshima et al., 2003). To test the specificity of CYK4, recombinant CYK4 purified from bacteria was screened against a panel of Rho family GTPases. RalA, RalB, Rap1B, N-Ras, and Rab1 are Ras superfamily GTPases outwith the Rho family and acted as negative controls. GTP hydrolysis assays revealed that CYK4 has GAP activity toward Rac1–3 and Cdc42 but not the other GTPases tested (Fig. 1 A). To eliminate the possibility that the RhoA preparations used were not competent for GTP hydrolysis, the broad specificity Rho family GAP ARHGAP1 was used (Rittinger et al., 1997a,b). ARHGAP1 was able to promote GTP hydrolysis by RhoA, Rac1, and Cdc42 but had no activity toward the non-Rho GTPase Rab1 (Fig. 1 B). These results suggest that CYK4 is not a RhoA GAP. However, in cells, CYK4 is bound to the kinesin motor MKlp1 as part of the centralspindlin complex and shows differential phosphorylation at different stages of mitosis, and it was therefore possible this might alter the specificity of the GAP activity.
Figure 1.
Human CYK4 is a GAP for Rac and Cdc42 but not RhoA. (A) Recombinant human CYK4 was tested against a representative panel of Rho family GTPases, RalA, RalB, Rap1B, Ras, and Rab1. In brief, 100 pmol of each GTPase was incubated in the presence or absence of 0.5 pmol hexahistidine-tagged CYK4 as described in the Materials and methods. The amount of CYK4-dependent GTP hydrolysis in picomoles per hour was calculated and plotted as a bar graph. (B) Recombinant human ARHGAP1 and CYK4 were tested against RhoA, Rac1, Cdc42, Rab1, or a GAP storage buffer blank. The amount of GAP-dependent GTP hydrolysis in picomoles per hour was calculated and plotted as a bar graph. (C) Endogenous MKlp1–CYK4 centralspindlin complexes were immune precipitated using MKlp1 antibodies from synchronized populations of HeLa cells in interphase, metaphase, or anaphase states. Control isolations were performed using GFP antibodies. These complexes were then used for GAP assays with RhoA, Rac1, Cdc42, and Rab1 as a negative control. The amount of centralspindlin-dependent GTP hydrolysis in picomoles per hour was calculated and plotted as a bar graph. Aliquots of the total cell lysate (input) and isolated complexes were Western blotted for MKlp1, CYK4, phosphothreonine Cdk1-phosphorylated CYK4 (pTP), and tubulin. (D) Endogenous MKlp1–CYK4 centralspindlin complexes were immune precipitated using MKlp1 antibodies from synchronized populations of HeLa cells in metaphase or anaphase cells treated with ZM447439 (Aurora B inhibitor) or BI2536 (Plk1 inhibitor). Control isolations were performed using GFP antibodies. These complexes were then used for GAP assays with RhoA, Rac1, Cdc42, and Rab1 as a negative control. The amount of centralspindlin-dependent GTP hydrolysis in picomoles per hour was calculated and plotted as a bar graph. Aliquots of the total cell lysate (input) and isolated complexes were Western blotted for MKlp1, phosphoserine 911 Aurora-phosphorylated MKlp1 (pS911), CYK4, and tubulin. Error bars indicate the standard deviations. IP, immunoprecipitation.
To test this, CYK4 centralspindlin complexes were isolated from cells in G2 (interphase) or passing through mitosis (metaphase and anaphase). Analysis of these complexes using Western blotting confirmed the presence of both metaphase- and anaphase-specific phosphorylation events on both CYK4 on threonine residues and MKlp1 at S911 (Fig. 1, C and D; Neef et al., 2006). The mobility of CYK4 was reduced in metaphase compared with anaphase samples, and this could be reversed by treatment with a phosphatase (Fig. S1, A–C). Like the purified CYK4 subunit alone, these complexes showed activity toward Rac1 and Cdc42 but not RhoA or Rab1 (Fig. 1 C). This activity was not altered when CYK4 centralspindlin complexes were isolated from cells in which either Aurora B or Plk1 was inhibited during anaphase using ZM447439 or BI2536, respectively (Fig. 1 D).
A previous study has reported that CYK4 is phosphorylated by Aurora B in the GAP domain at serine 387 adjacent to the arginine finger catalytic residue at position 385 (Minoshima et al., 2003). To investigate this, the relative phosphorylation of centralspindlin complexes in metaphase and anaphase was analyzed using stable isotope labeling and cell culture (SILAC) and mass spectrometry (MS; Fig. S1 D). Although this analysis identified a series of metaphase- and anaphase-specific phosphorylations at candidate Cdk1, Aurora, and Polo kinase consensus motifs in the N- and C-terminal regions of CYK4, the putative regulatory phosphorylation at serine 387 was not identified (Fig. S1 D). In fact, no phosphorylation sites within the GAP domain were found. In contrast, the conserved Aurora B phosphorylation of MKlp1 at S911 was identified in anaphase samples by MS (Fig. S1 D) and Western blotting (Fig. 1 D), as expected (Neef et al., 2006). When recombinant CYK4 carrying S387A (nonphosphorylated) or S387D (phosphomimetic) mutations was compared with wild-type CYK4, the S387D mutant showed reduced Rac1 and Cdc42 GAP activity comparable with the R385A arginine finger catalytic site mutant (Fig. 2 A). However, no increase in activity toward RhoA was seen (Fig. 2 A). All of these mutant proteins can target to the central spindle region (Fig. 2 B) and interact with MKlp1 to form a centralspindlin complex (Fig. 2 C). Interestingly, the S387D mutation showed only a small decrease in GAP activity when CYK4 was incorporated into MKlp1 centralspindlin complexes isolated from stable cell lines (Fig. 2 C). This may be caused by the stabilizing effect of MKlp1 binding to CYK4 (Mishima et al., 2002). In contrast, the R385A arginine finger mutation still showed greatly reduced GAP activity toward Rac1 (Fig. 2 C).
Figure 2.
Arginine finger mutations reduce CYK4 GAP activity toward Rac1. (A) A schematic of CYK4 shows the conserved coiled coil, C1 putative membrane-binding region, and GAP domain with arginine finger residue at amino acid 385. Recombinant human ARHGAP1, CYK4, CYK4R385A, CYK4S387A, and CYK4S387D were tested against RhoA, Rac1, Cdc42, and Rab1. The amount of GAP-dependent GTP hydrolysis in picomoles per hour was calculated and plotted as a bar graph. (B). HeLa cells were transfected with Myc-tagged CYK4, CYK4R385A, CYK4S387A, and CYK4S387D for 24 h, fixed, and then stained with antibodies to the Myc epitope, MKlp1, or tubulin. DNA was stained with DAPI. Bar, 10 µm. (C) HeLa cells stably expressing inducible Myc-tagged copies of CYK4, CYK4R385A, CYK4S387A, and CYK4S387D were treated for 36 h with an siRNA directed to the 3′-UTR of CYK4 to deplete the endogenous protein or a control siRNA. At the same time, the cells were treated with doxycycline to induce the Myc-tagged transgenes. MKlp1–CYK4 centralspindlin complexes were then immune precipitated using Myc antibodies. Control isolations were performed using GFP antibodies. These complexes were then used for GAP assays with RhoA and Rac1. The amount of centralspindlin-dependent GTP hydrolysis in picomoles per hour was calculated and plotted as a bar graph. Aliquots of the total cell lysate (input) and isolated complexes were Western blotted for Myc, CYK4, MKlp1, and tubulin. Arrowheads indicate endogenous CYK4, and arrows show Myc-tagged CYK4. Error bars indicate the standard deviations. IP, immunoprecipitation; WT, wild type.
Collectively, these data support the view that CYK4 is a Rac1 and Cdc42 GAP rather than a Rho GAP and that this activity is not modulated by phosphorylation. Although the CYK4 S387D mutant had reduced GAP activity, endogenous CYK4 GAP activity was constant through mitosis, despite changes in the phosphorylation state of the protein.
CYK4 GAP activity is required for cytokinesis
The role of the CYK4 GAP activity toward Rac1 and Cdc42 was then tested using stable-inducible cell lines in which the endogenous copy of CYK4 can be replaced by a combination of siRNA to the CYK4 3′-untranslated region (UTR) and induction of a transgene comprising a myc-tagged CYK4 open reading frame lacking the 3′-UTR. Induction of wild-type CYK4 or the R385A GAP mutant in the presence of endogenous CYK4 had little effect on the outcome of cell division as measured by the number of binucleate cells (Fig. 3 A, siControl). Depletion of endogenous CYK4 without induction of the transgene resulted in a high frequency of binucleate cells (Fig. 3 A, siCYK4 3′-UTR uninduced). Induction of the wild-type CYK4 transgene, but not the R385A GAP mutant, rescued this effect (Fig. 3 A, siCYK4 3′-UTR induced). CYK4 GAP activity is therefore required for efficient cytokinesis.
Figure 3.
Rac1 inactivation by CYK4 is required for cytokinesis. (A) HeLa cells stably expressing inducible Myc-tagged copies of CYK4 or CYK4R385A were treated for 36 h with an siRNA directed to the 3′-UTR of CYK4 to deplete the endogenous protein or a control siRNA. At the same time, the cells were treated with doxycycline to induce the Myc-tagged transgenes (induced) or left untreated (uninduced). The cells were fixed and then stained with antibodies to tubulin, the Myc epitope, or DAPI to detect DNA. The number of binucleate cells was counted for each condition and plotted as a bar graph. Error bars show the standard deviation from the means (n = 3). Asterisks mark binucleate cells that have failed cytokinesis. (B) HeLa cells stably expressing inducible EGFP-tagged copies of Rac1 or activated Rac1Q61L were treated for the time indicated with doxycycline to induce the EGFP-tagged transgenes. The cells were fixed and then stained with antibodies to tubulin or DAPI to detect DNA. Rac1 was directly visualized by EGFP fluorescence. The number of binucleate cells was counted for each condition and plotted as in the line graph. (C) Alternatively, the cells were fixed using TCA and stained for RhoA and with antibodies to EGFP to detect EGFP-Rac1. DAPI was used to detect DNA. siControl, nonsilencing control; WT, wild type. Bars, 10 µm.
The GAP specificity data show that CYK4 can act on both Rac1 and Cdc42. It was therefore important to define which of these GTPases is expressed in the HeLa cells being used. First, antibodies to RhoA, Rac1, and Cdc42 were tested to show that they detect their target antigen equally well by Western blotting (Fig. S2 A). Western blot analysis of mitotic and asynchronous HeLa cell extract indicated that the cells express RhoA and Rac1 but little Cdc42 (Fig. S2 B). Furthermore, RhoA was the only one of these GTPases showing strong enrichment in the cleavage furrow region (Fig. S2, C–E). Because of these results, it was decided to focus on Rac1 as the most abundant target of CYK4 in HeLa cells.
The requirement for CYK4 GAP activity in cytokinesis is most simply explained if Rac1 has to hydrolyze GTP for this process to complete successfully. Experiments using cell lines expressing inducible copies of wild-type Rac1 or a hydrolysis-defective Rac1Q61L mutant support this idea. Induction of Rac1Q61L resulted in a time-dependent increase in binucleate cells (Fig. 3 B), and RhoA, which is normally restricted to the furrow region in anaphase, failed to concentrate properly despite apparently normal recruitment of the centralspindlin complex (Fig. 3 C). An excess of activated Rac1 therefore interferes with cytokinesis and normal cleavage furrow formation. Visual inspection of these cells revealed that the Rac1Q61L cells were highly flattened and spread when compared with the equivalent Rac1 wild type–expressing cells, suggesting that they are more adherent. This raises the obvious possibility that the normal function of CYK4 is to reduce Rac1-dependent signaling in the cleavage furrow, which would otherwise interfere with cytokinesis. To address this possibility, the Rac1 effector pathway in mitosis and the role of CYK4 GAP activity were investigated in more detail.
Identification of RhoA and Rac1 effector pathways in mitotic cells
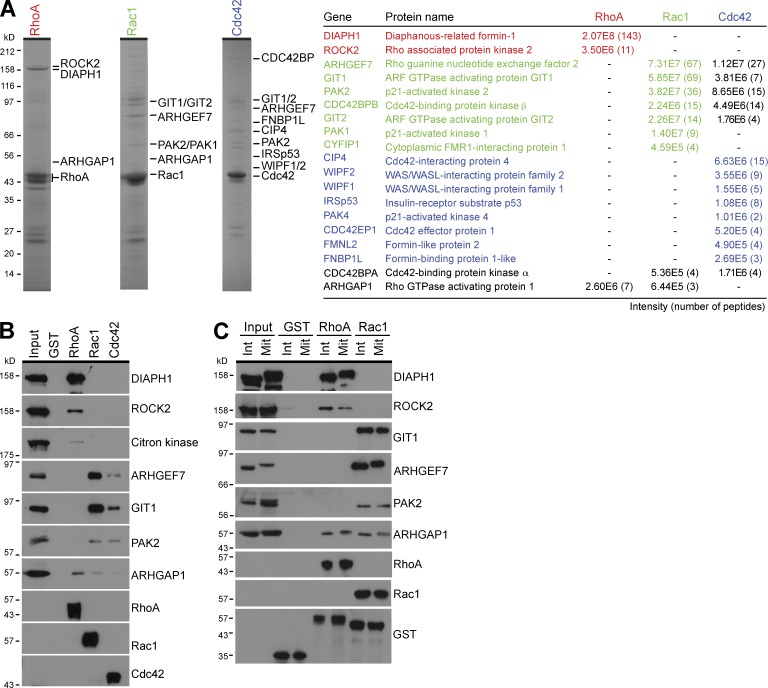
The different Rho family GTPases are known to couple to discrete effector pathways and thereby give rise to different modes of actin dynamics important for cell contraction, cell adhesion, and cell motility (Heasman and Ridley, 2008; Parsons et al., 2010). However, these studies have been mainly performed in interphase cells, and it is unclear whether mitosis-specific pathways exist. Pull-down experiments were therefore performed using recombinant RhoA, Rac1, and Cdc42 to identify which effector pathways they couple with during mitosis. This approach revealed that RhoA, Rac1, and Cdc42 bound to specific groups of effector proteins (Fig. 4 A). The formin DIAPH1 (diaphanous 1) and the ROCK2 (Rho kinase 2) bound to RhoA, whereas the p21-activated kinases (PAKs) PAK1 and PAK2, the Rac1 and Cdc42 GEF ARHGEF7/β-PIX (Manser et al., 1998), and the ARF6-GAPs (Vitale et al., 2000) GIT1 and GIT2 bound to Rac1 (Fig. 4, A and B). This pattern of Rac effectors is consistent with a previous study showing that PAK, ARHGEF7, and the GIT proteins form a modular signaling cassette essential for cell polarization and cell shape changes (Frank and Hansen, 2008). Some overlap between Rac1 and Cdc42 effectors was observed, but Cdc42 showed specific interactions with the Wiskott–Aldrich Syndrome protein (WASp)–interacting family, the inverse BAR (Bin/Amphiphysin/Rvs) domain protein IRSp53, and specific formins (Fig. 4 A and Table S1). ARHGAP1 was present in RhoA and Rac1 pull-downs (Fig. 4, A and B), fitting with its common GAP activity toward RhoA, Rac1, and Cdc42 (Fig. 1 B). These effector pathways are consistent with the idea that Rho, Rac, and Cdc42 have discrete cellular functions in cell contraction and adhesion during mitosis (Fig. S3) and are similar to those reported previously in interphase cells (Frank and Hansen, 2008; Heasman and Ridley, 2008; Parsons et al., 2010). Further experiments confirmed that RhoA and Rac1 have the same pattern of effector binding in both interphase and mitotic cells (Fig. 4 C). Because Rac1 does not act via mitosis-specific effectors, but may undergo a mitosis-specific form of regulation, the relationship between CYK4 Rac GAP activity and these specific Rac1 effector pathways was therefore investigated.
Figure 4.
RhoA and Rac1 couple to discrete effector pathways in mitotic cells. (A) Effector binding assays from mitotic HeLa cell lysate were performed using GST, RhoA, Rac1, and Cdc42 as described in the Materials and methods. Bound fractions were eluted using sample buffer and analyzed by SDS-PAGE and MS. Coomassie brilliant blue–stained gels of the RhoA, Rac1, and Cdc42 complexes are shown. Proteins identified by MS are listed in the table and marked by the side of the appropriate gel. Peptide number and intensity give a measure of the relative abundance. (B) Western blot analysis of the input and bound effector proteins was performed using the antibodies shown in the figure. (C) Effector binding assays from interphase (Int) and mitotic (Mit) HeLa cell lysate were performed using GST, RhoA, and Rac1 as described in the Materials and methods. Samples of the input material and bound fractions were analyzed by Western blotting using the antibodies shown in the figure.
CYK4 acts through Rac1-specific effector pathways
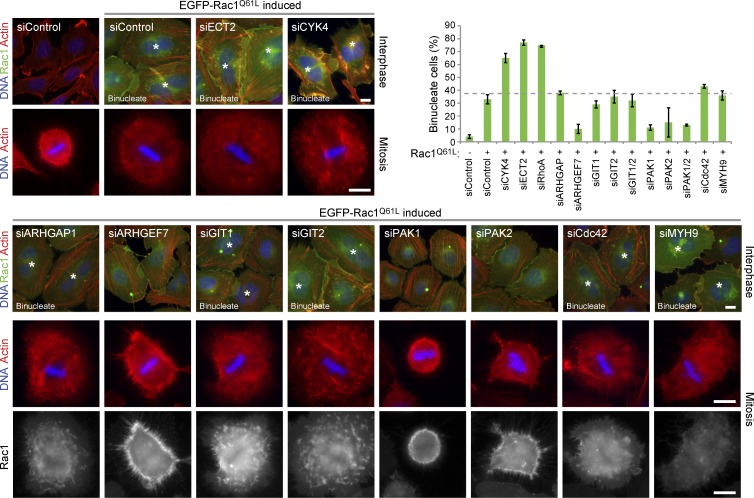
First, the effectors crucial for mediating the inhibitory effects of Rac1 on cytokinesis were defined. To do this, Rac1 effectors were depleted using specific siRNA duplexes (Fig. S4) from cells expressing an inducible copy of Rac1Q61L. Induction of Rac1Q61L expression resulted in increased cell spreading through mitosis, defective furrowing, and early cytokinesis failure in nearly 35–40% of cells (Fig. 5). Depletion of ECT2, the RhoA GEF required for furrow formation, RhoA itself, or the CYK4 component of centralspindlin required for central spindle integrity increased this to nearly 65–75% cytokinesis failure. In contrast, depletion of the Rac1 effectors ARHGEF7, PAK1, and PAK2 reduced cell spreading through mitosis and the extent of cytokinesis failure to 10% (Fig. 5). Furthermore, depletion of these effector proteins also rescue furrow formation (Fig. S5 A) defined by anillin, a marker for Rho-dependent organization of F-actin in the contractile ring (Gregory et al., 2008; Hickson and O’Farrell, 2008; Piekny and Glotzer, 2008; Kechad et al., 2012). No change was seen when ARHGAP1, GIT1, and GIT2 were depleted, showing that the inhibitory effect of activated Rac1 on cytokinesis did not require these proteins. Furthermore, depletion of Cdc42 had little effect on cell spreading and did not increase or decrease the rate of cytokinesis failure.
Figure 5.
Depletion of Rac1 effectors rescues the cytokinesis defect of cells expressing a hydrolysis-defective Rac1 mutant. HeLa cells expressing inducible EGFP-Rac1Q61L were transfected with siRNA duplexes targeting CYK4, ECT2, Rac1-specific effectors, Cdc42, a nonsilencing control (siControl), or Myh9, which was a protein binding nonspecifically to Sepharose beads in Rho GTPase pull-downs. Rac1Q61L expression was induced for 48 h, and the cells were fixed and then stained with phalloidin to detect actin or DAPI to detect DNA. Rac1 was directly visualized by EGFP fluorescence. Binucleate cells are marked by asterisks. The number of binucleate cells was counted for each condition and plotted as in the bar graph. Error bars indicate the standard deviation from the means (n = 3). The dotted line indicates the extent of cytokinesis failure. Bars, 10 µm.
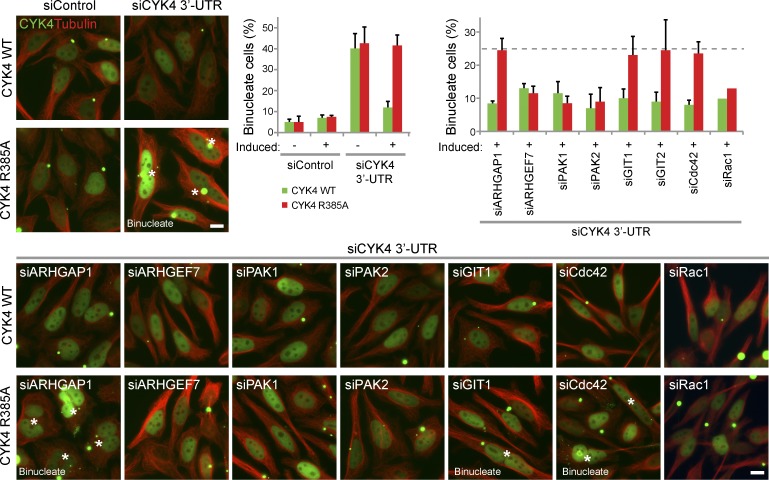
The relationship between Rac1 effectors and CYK4 GAP activity was then tested using stable-inducible cell lines in which the endogenous copy of CYK4 can be replaced by a combination of siRNA to the CYK4 3′-UTR and induction of a transgene comprising an EGFP-tagged CYK4 reading frame lacking the 3′-UTR. Replacement of wild-type CYK4 with the R385A GAP mutant resulted in 40% late stage cytokinesis failure after furrowing (Fig. 6) and perturbed anillin staining in early anaphase cells (Fig. S5 B). Similar to the results seen with the Rac1Q61L cells, depletion of ARHGEF7, PAK1, and PAK2 reduced the extent of cytokinesis failure to ∼10% (Fig. 6). Again, depletion of ARHGAP1, GIT1, GIT2, or Cdc42 had little effect on the rate of cytokinesis failure (Fig. 6). Importantly, depletion of Rac1 reduced the extent of cytokinesis failure, as expected if CYK4 acts via a Rac1-dependent pathway (Fig. 6). The inhibitory effects of Rac1 on cytokinesis in cells expressing the GAP mutant form of CYK4 or the activated Q61L mutant of Rac1 are therefore caused by specific effector pathways associated with cell spreading and adhesion.
Figure 6.
Depletion of Rac1 effectors rescues the cytokinesis defect of cells expressing the CYK4 arginine finger mutant. HeLa cells expressing inducible EGFP-CYK4 or GAP activity–defective CYK4R385A were transfected with control or CYK4 3′-UTR siRNA duplexes in combination with siRNA duplexes targeting Rac1-specific effectors and Cdc42. CYK4 expression was induced for 60 h, and then, the cells were fixed and stained with antibodies to tubulin and DAPI to detect DNA. CYK4 was directly visualized by EGFP fluorescence. Binucleate cells are marked with asterisks. The number of binucleate cells was counted for each condition and plotted as in the bar graph. Error bars indicate the standard deviation from the means (n = 3). The dotted line indicates the extent of cytokinesis failure. siControl, nonsilencing control; WT, wild type. Bars, 10 µm.
Increased cell spreading in anaphase in the absence of CYK4 GAP activity
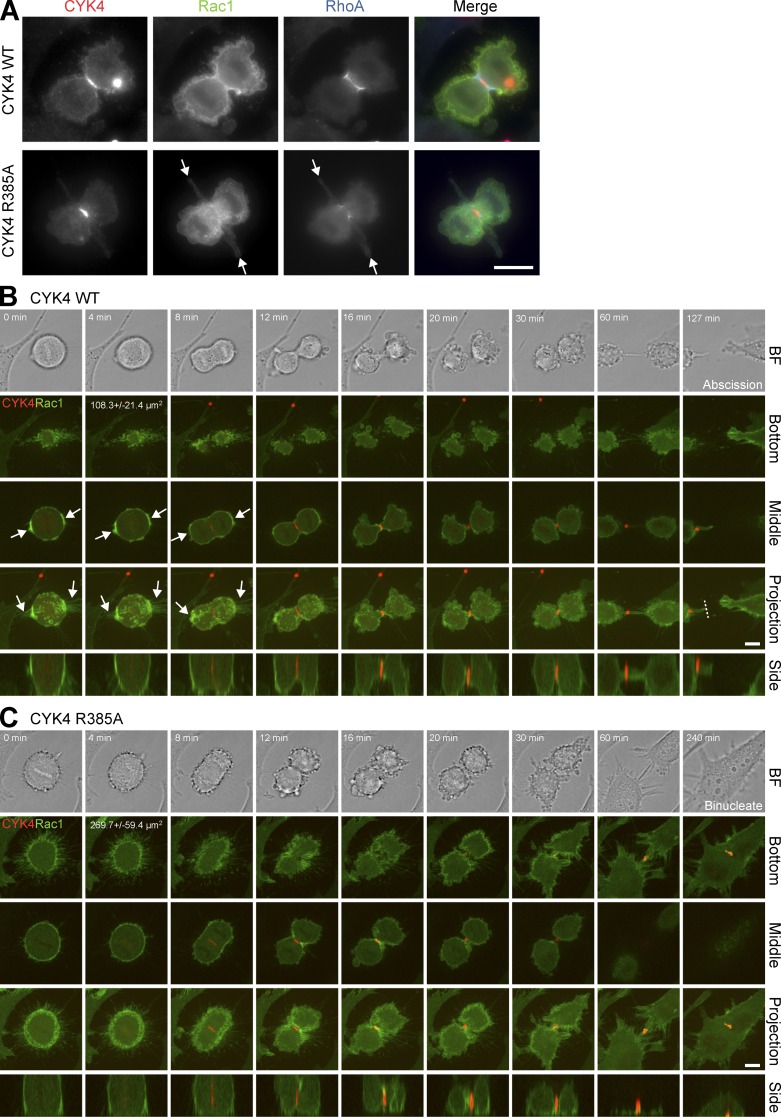
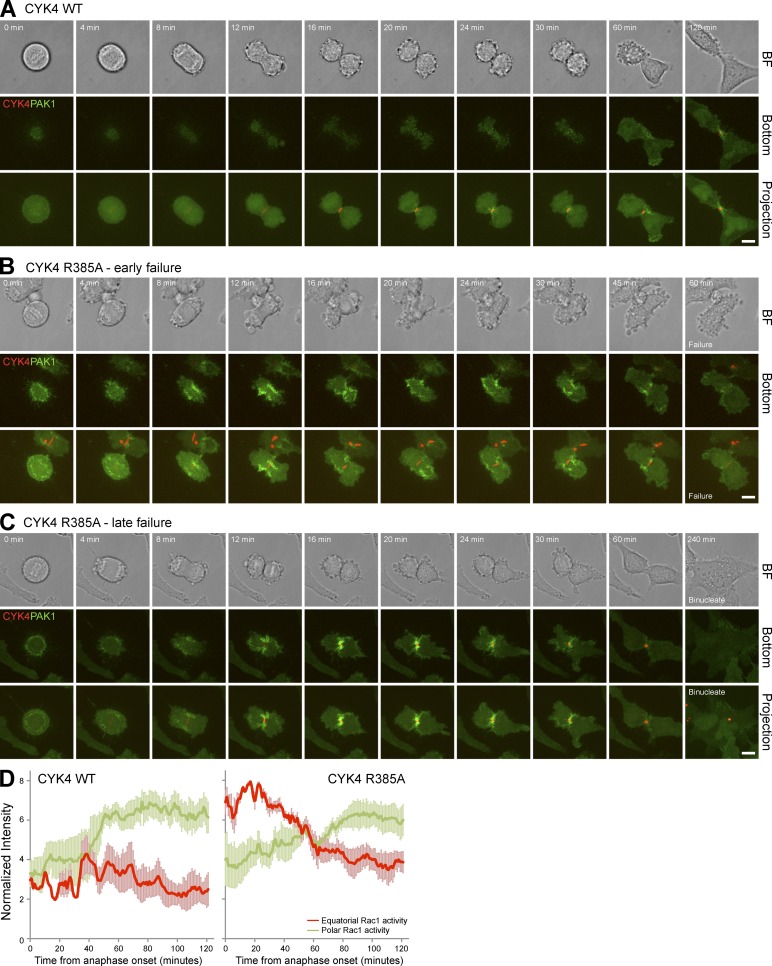
Careful inspection of CYK4 GAP mutant cells progressing through anaphase provided further support for the idea that altered cell spreading in the cleavage furrow region was perturbing RhoA function. These cells often displayed surface projections from the cleavage furrow region and reduced levels of RhoA in the cleavage furrow (Fig. 7 A and Fig. S5B). Live-cell imaging of Rac1 confirmed that the CYK4R385A cells have a larger contact with the growth surface than the equivalent CYK4 wild-type cells (Fig. 7, B and C, bottom). Measurements on 20 cells showed a 2.5-fold increase in attached surface area at the start of anaphase from 108.3 ± 21.4 µm2 in cells expressing wild-type CYK4 to 269.7 ± 59.4 µm2 in the CYK4R385A GAP mutant cells. Furthermore, retraction fibers and Rac1 that were enriched at the cell poles in cells expressing wild-type CYK4 (Fig. 7 B, CYK4 arrows) were spread evenly around the cell cortex when CYK4 GAP activity was compromised (Fig. 7 C, CYK4R385A). Examination of the side projection showed that cleavage furrow invagination was reduced in the CYK4R385A cells, and the midbody marked by CYK4 remained closer to the growth surface (Fig. 7 C), fitting with the idea that the cells are more adherent. The distribution of active Rac1 was then investigated using cells expressing a Cdc42 and Rac interactive binding (CRIB) domain reporter construct. Live-cell imaging revealed that in cells expressing the wild-type CYK4 GAP, active Rac1 accumulated at later time points at the spreading cell edges (Fig. 8 A and Video 1). This is consistent with previous work using fluorescence resonance energy transfer probes that also came to the conclusion that Rac1 activity is focused at the cell poles and away from the cleavage furrow region when cells are exiting mitosis (Yoshizaki et al., 2003, 2004). In contrast, in cells expressing the CYK4 R385A GAP mutant, active Rac1 accumulated in the central furrow region at the bottom of the cell, where it touched the growth surface (Fig. 8, B and C; and Videos 2 and 3). The CYK4 GAP mutant cells then underwent either an early (Fig. 8 B) or late stage failure in cytokinesis (Fig. 8 C). Although control cells have completed cytokinesis within 171.1 ± 29.8 min, the CYK4R385A cells do not form an extended midbody and fail cytokinesis by 81.0 ± 21.8 min or form an extended midbody and fail cytokinesis after 293.1 ± 53.6 min. Early failure was also associated with a near complete loss of furrowing, large movements of the cell cortex, and oscillation of the active pool of Rac1 (Fig. 8 B and Video 2). This suggests that CYK4 is needed to inactivate Rac1 from early stages of anaphase, even if cytokinesis failure may only become apparent at later time points. These observations are consistent with the idea that activated Rac1 in the equatorial furrow region is inhibitory to proper cell furrowing and that this can ultimately result in abscission defects and cytokinesis failure.
Figure 7.
Cell surface projections emerge from the cleavage furrow in CYK4 GAP-defective cells. (A) HeLa cells expressing EGFP-Rac1 and inducible mCherry-CYK4 or GAP activity–defective CYK4R385A were transfected with control or CYK4 3′-UTR siRNA duplexes. CYK4 expression was induced for 60 h, and then, the cells were TCA fixed and stained with antibodies to mCherry to detect CYK4, RhoA, or EGFP to detect Rac1. Arrows mark the cell surface projections seen emerging from the cleavage furrow region in CYK4R385A cells. (B and C) Alternatively, these CYK4 (B) or CYK4R385A (C) cells were used for time-lapse imaging. A bright-field (BF) image is shown at the time points indicated together with the bottom section where the cells touch the glass growth surface, a middle section through the cell equator, a maximum intensity projection along the z axis of all sections, or a side projection along the y axis. The dotted line indicates where abscission has occurred. The 4-min time point, bottom section, includes a quantitation of the area of the cell in contact with the surface (n = 20). Arrows mark Rac1 accumulation at the cell poles and associated retraction fibers. WT, wild type. Bars, 10 µm.
Figure 8.
Increased Rac1 activity in the cleavage furrow of CYK4 GAP-defective cells. (A–C) HeLa cells expressing EGFP-PAK1 CRIB domain and inducible mCherry-CYK4 (A) or GAP activity–defective CYK4R385A (B and C) were transfected with CYK4 3′-UTR siRNA duplexes and then used for time-lapse imaging. A bright-field (BF) image is shown at the time points indicated together with the bottom section where the cells touch the glass growth surface and a maximum intensity projection along the z axis of all sections. Timings are relative to the onset of anaphase. Cells expressing only GAP activity–defective CYK4R385A showed either early (B) or late (C) stage failure of cytokinesis, and examples of both outcomes are therefore shown. CYK4 wild-type cells underwent abscission after 171.1 ± 29.8 min, whereas CYK4R385A cells showed early failure after 81.0 ± 21.8 min or late failure after 293.1 ± 53.6 min (n = 29). Bars, 10 µm. (D) Rac1 activity at the cell equator (red) and cell poles (green) was determined by measuring the intensity of the PAK1 CRIB domain probe every minute as cells exited mitosis using the Volocity 5 volume and quantitation tools. This was performed on cells expressing wild-type and R385A GAP mutant CYK4. Mean values are plotted in the graphs, and error bars indicate the standard deviation from the means (n = 5). WT, wild type.
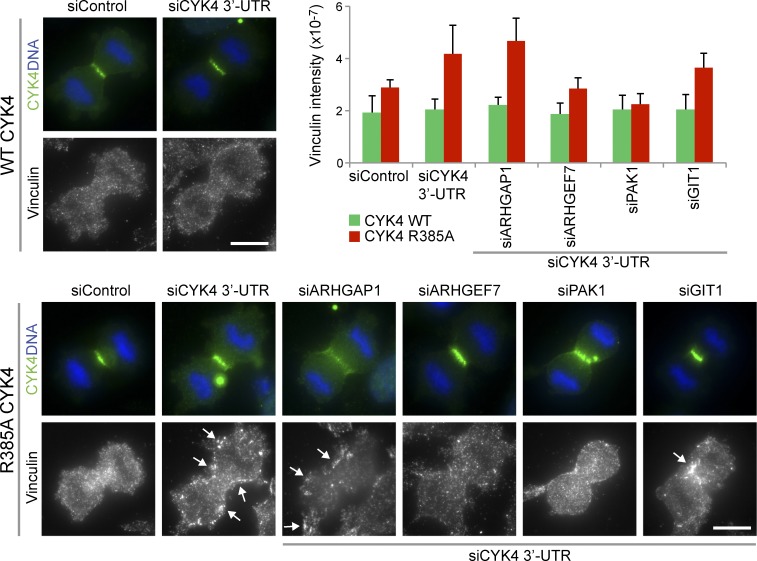
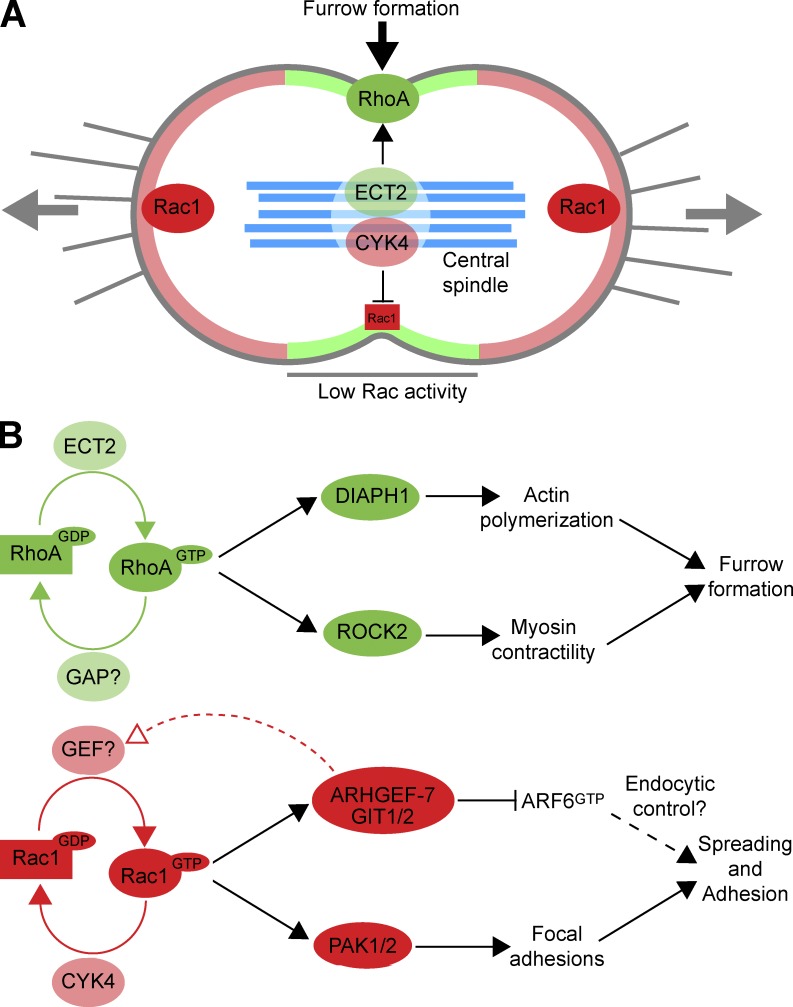
The role of Rac1 effectors in the formation of adhesion complexes was then investigated under conditions in which the CYK4 GAP was active or inactivate. Vinculin, a component of focal adhesion plaques, was used as a marker of these structures. In control cells expressing wild-type active CYK4, vinculin staining is diffuse in the cytoplasm during mitosis, consistent with a loss of adhesion, whereas CYK4 GAP mutant cells show defined punctate vinculin staining in the furrow region and at the cell cortex (Fig. 9). Depletion of PAK1 and ARHGEF7 caused a loss of this defined vinculin staining in the CYK4 GAP mutant cell lines (Fig. 9). In summary, these findings support the idea that the CYK4 GAP activity is required for the inhibition of Rac1 at cleavage furrow in anaphase (Fig. 10 A). In turn, this may reduce the activity of Rac1-dependent ARHGEF7 and PAK effector pathways associated with the regulation of cell spreading and adhesion (Fig. 10 B).
Figure 9.
Visualization of adhesion complexes using vinculin staining in CYK4 GAP-defective cells in anaphase. HeLa cells expressing inducible EGFP-CYK4 or GAP activity–defective CYK4R385A were transfected with control or CYK4 3′-UTR siRNA duplexes in combination with siRNA duplexes targeting Rac1-specific effectors. CYK4 expression was induced for 60 h, and the cells were fixed and then stained with antibodies to GFP to detect CYK4 and vinculin and DAPI to detect DNA. Arrows indicate especially bright vinculin spots. The vinculin staining intensity was measured using ImageJ and is plotted in the bar graph. Error bars indicate the standard deviation from the means (n = 7). siControl, nonsilencing control; WT, wild type. Bars, 10 µm.
Figure 10.
A model for CYK4 regulation of Rac1 activity in anaphase. (A) At the onset of anaphase, cells adhere at the cell poles and contract in the equatorial region, driven by Rac1 and RhoA, respectively. The RhoA exchange factor ECT2 is enriched at the cell equator because of interactions with the MKlp1–CYK4 centralspindlin complex. This leads to local activation of RhoA in the furrow region (shaded pale green). Rac1 is inactivated in this region by the CYK4 GAP component of centralspindlin, creating a zone of low Rac1 activity. This does not occur at the cell poles, so Rac1 activity is higher in these regions (shaded pale red). Central spindle microtubules are shown in pale blue. (B) Regulatory and effector pathways for RhoA and Rac1 are summarized, together with the events they are known to control. The identity of the mitotic RhoA GAP remains to be assigned. GIT1/2 have been previously assigned as ARF6 GAPs and are therefore shown as inhibitors of ARF6 function. ARF6 has a complex function in cytokinesis together with Rab35 and modulates endocytic recycling (Chesneau et al., 2012).
Discussion
Regulation of CYK4 activity by phosphorylation
Earlier work using the CYK4 GAP domain has suggested that CYK4 GAP activity is regulated by Aurora B phosphorylation (Minoshima et al., 2003), yet we find no evidence for this. Purified centralspindlin complexes containing full-length CYK4 protein were purified in metaphase and anaphase phosphorylated forms but showed equivalent activity toward Rac1 and no detectable activity toward RhoA. Furthermore, phosphomimetic mutation of the putative Aurora phosphorylation site at serine 387 did not cause a switch in specificity from Rac1 to RhoA (Minoshima et al., 2003) but rather partial inactivation of Rac1 and Cdc42 GAP activity. This serine residue is conserved in other Rho family GAPs, including ARHGAP1, and the published crystal structures show that it lies at the interface between the GAP and the substrate GTPase (Rittinger et al., 1997a,b). ARHGAP1 binds to RhoA without phosphorylation at this site, so this is not an obligatory modification required for GAP activity, and there is no mechanism explaining why it would select for RhoA above Rac1.
Although phosphorylation at serine 387 was not detected and does not appear to be important for CYK4 activity in our hands, other phosphorylations were identified. These might be relevant for controlling CYK4 localization, which undergoes a switch from the cytosol in metaphase to the central spindle and equatorial cell cortex in anaphase. In this context, the Cdk1 consensus sites at T342 adjacent to the C1 domain and at T580 near to the C terminus may be significant. This might provide a way to control the access of CYK4 to its substrate GTPases at the cell cortex, and further studies will be required to address this issue.
CYK4 GAP activity and adherent cell divisions
Previous studies have painted a somewhat confused picture on the role of the CYK4 GAP activity in cytokinesis (Glotzer, 2009). Chicken B cell lines expressing only a GAP mutant form of CYK4 are able to carry out the process of cytokinesis without any detectable defects (Yamada et al., 2006). Similarly, developing Drosophila neuroblasts do not appear to have cell division defects when CYK4 GAP activity is compromised (Goldstein et al., 2005). However, other studies indicate that CYK4 GAP activity is needed for assembly of a functional contractile ring and successful cell division in Drosophila (Somers and Saint, 2003; Zavortink et al., 2005) and Xenopus laevis oocytes (Miller and Bement, 2009). This has resulted in complex arguments postulating noncatalytic functions for the CYK4 GAP domain that in some way impact on RhoA function (Loria et al., 2012) and claims that phosphorylation can switch the specific activity of CYK4 from Rac1 to RhoA (Minoshima et al., 2003). We resolve some of these arguments by a careful analysis of CYK4 GAP activity in mitotic cells and by defining the specific effector pathways to which RhoA and Rac1 couple. Cells lacking CYK4 GAP activity are more adherent during anaphase, and this interferes with the formation of the cleavage furrow and hence cytokinesis. Depletion of Rac1 or Rac1 effectors involved in cell spreading and adhesion rescues this defect. This provides a simple explanation for the observation that chicken B cells do not require CYK4 GAP activity to divide because they are grown in suspension and therefore nonadherent (Yamada et al., 2006). This may also be relevant in fly neuroblast divisions, in which the cells delaminate from the neuroectoderm as they undergo asymmetric cell divisions and therefore don’t display focal adhesions or junctions to neighboring cells (Wodarz, 2005).
Our findings on adherent mammalian cell lines are broadly in agreement with those made in early embryonic development in C. elegans in which it was also reported that depletion of Rac could suppress the phenotype of CYK-4 E448K mutations (Canman et al., 2008). In that case, Rac function was linked to the Arp2/3 complex because depletion of WASp and WAVE could also recover the CYK-4 E448K mutant (Canman et al., 2008). In adherent human cells, we have identified a specific role for Rac1 and the PAK–ARHGEF7 effector pathway required for cell spreading and adhesion. Other data suggest that Cdc42 couples to the WASp pathway through WIPF1 and WIPF2 in these cells. However, because depletion of Cdc42 did not rescue the CYK4 GAP mutant phenotype, this pathway could not explain the division defect in these cells, so it was not investigated further.
Additional indirect support for the idea that CYK-4 is a Rac GAP comes from studies in C. elegans that showed CYK-4 is important for polarization of the early embryo and the targeting of the atypical protein kinase C and Par-3 complex (Portereiko et al., 2004; Jenkins et al., 2006). Because Par-3 has been directly linked to Rac and Cdc42 rather than Rho (Lin et al., 2000), these findings fit well with the idea that CYK4 is a Rac and Cdc42 GAP.
Collectively, these results obtained in highly different systems support the view that the crucial function of CYK4 in cytokinesis is the inactivation of Rac and thereby ensuring the correct balance of Rac and Rho activity (Mandato et al., 2000; Yoshizaki et al., 2004; D’Avino et al., 2005). In adherent somatic cells, this may be most crucial for regulation of cell spreading adhesion, whereas in early embryonic divisions, it may be more important for the regulation of Rac and Cdc42 during anteroposterior polarization events.
Materials and methods
Reagents and antibodies
Standard laboratory reagents were obtained from Thermo Fisher Scientific and Sigma-Aldrich. Antibodies to MKlp1 (1–444 aa), CYK4 (1–633 aa), and anillin (614–960 aa) were raised in sheep and rabbit using hexahistidine-tagged human proteins expressed in and purified from bacteria. Specific antibodies were purified using the antigens conjugated to Affigel-15, eluted with 0.2 M glycine, pH 2.8, and then dialyzed against PBS before storage at −80°C. Rabbit antibodies to full-length recombinant PRC1 and an MKlp1 pS911 peptide, CRKRR(phospho-S)STVA, have been described previously (Neef et al., 2003, 2006, 2007). Commercially available antibodies were used to ARHGAP1 (rabbit Cdc42 GAP; Bethyl Laboratories, Inc.), ARHGEF7 (rabbit Cool1/β-Pix; Cell Signaling Technology), Cdc42 (mouse; Cytoskeleton), Citron kinase (mouse Citron Rho-interacting kinase; BD), Cyclin B1 (mouse GNS3; EMD Millipore), DIAPH1 (rabbit; Bethyl Laboratories, Inc.), GIT1 (rabbit; Bethyl Laboratories, Inc.), GIT2 (rabbit; Bethyl Laboratories, Inc.), PAK1 (rabbit; Bethyl Laboratories, Inc.), PAK2 (rabbit; Bethyl Laboratories, Inc.), pTP (mouse; Cell Signaling Technology), Rac1 (mouse; Cytoskeleton), RhoA (mouse; Cytoskeleton), ROCK2 (rabbit; Bethyl Laboratories, Inc.), tubulin (mouse DM1A [Sigma-Aldrich] or sheep [Cytoskeleton]), Vinculin (rabbit; Abcam), and WASF1 (rabbit; Sigma-Aldrich). Secondary antibodies, raised in donkey, to mouse, rabbit, and sheep/goat conjugated to HRP, Alexa Fluor 488, Alexa Fluor 555, Alexa Fluor 568, and Alexa Fluor 647 were obtained from Molecular Probes. Affinity-purified primary and secondary antibodies were used at 1 µg/ml, and sera were used at a 1:1,000 dilution in PBS. A benchtop microfuge (5417R; Eppendorf) was used for all centrifugations unless otherwise indicated.
Molecular biology and protein expression
Rho family GTPases were amplified from cDNA by PCR and cloned into a hexahistidine-GST tag bacterial expression vector pFAT2. The proteins were expressed in BL21(DE3) cells and purified using nickel–nitrilotriacetic acid agarose (Fuchs et al., 2007). ARHGAP1 and CYK4 were amplified from cDNA by PCR and cloned in the hexahistidine tag bacterial expression vector pQE32. CYK4 and ARHGAP1 were expressed in JM109 cells and purified using nickel–nitrilotriacetic acid agarose. For mammalian expression, inserts were amplified by PCR from cDNA and then cloned into the pcDNA4 and pcDNA5 vectors containing EGFP, mCherry, FLAG, or Myc tags. The siRNA duplexes used in this study are listed in Table S1.
Cell culture media preparation and cell culture conditions
HeLa were cultured in growth medium (DME containing 10% fetal bovine serum) at 37°C and 5% CO2. For plasmid transfection and siRNA transfection, TransIT LT1 (Mirus Bio LLC) and Oligofectamine (Invitrogen), respectively, were used according to the manufacturers’ instructions. HeLa Flp-In cells (Invitrogen) expressing single copies of epitope-tagged transgenes were generated by transfecting pcDNA5-FLAG–, EGFP-, or mCherry vector–encoded transgenes and Flp-recombinase at a ratio of 1:9 into HeLa S3 Flp-In cells. Transfected cells were expanded onto 15-cm dishes 24 h after transfection, and stable clones were selected using 0.4 µg/ml blasticidin and 100 µg/ml hygromycin. Single colonies were picked after a 14-d selection and amplified in 24-well plates, and expression was checked by Western blotting and microscopy.
Isolation of CYK4 and CYK4R385A centralspindlin complexes
Wild-type and catalytically inactive CYK4 complexes were isolated from 3 × 15–cm dishes of Myc-CYK4 and Myc-CYK4R385A stable-inducible HeLa cell lines, or untransfected control cells were grown to 60% confluence. The cells were treated with CYK4 3′-UTR siRNA to deplete the endogenous CYK4 for 36 h. The cells were then arrested in mitosis with 100 ng/ml nocodazole for 16 h. Mitotic cells were harvested by shake off, washed three times with warm PBS, and then resuspended in warm growth medium. The cells were incubated in a 5% CO2 incubator at 37°C for 20 min and then washed twice with cold PBS. For the anaphase condition, cells were replated in growth medium and harvested when the peak of anaphase cells became visible under the microscope, typically after 45–60 min. Cells were harvested and washed in ice-cold PBS. The cells were lysed in 1 ml cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 1% [vol/vol] Triton X-100 with protease and phosphatase inhibitor cocktails [Sigma-Aldrich]), left on ice for 20 min, and then centrifuged at 20,000 gav for 20 min at 4°C to remove insoluble debris. To isolate CYK4 complexes, 3 µg of sheep anti-MKlp1 or anti-Myc protein G–Sepharose beads were added into 900 µl cell lysate and rotated at 4°C for 3 h. The beads were washed three times with cell lysis buffer and then used for GAP assays.
SILAC analysis of centralspindlin complexes
For SILAC experiments, DME formulated to lack l-arginine and l-lysine was used. Before use, l-arginine was added to a final concentration of 84 mg/ml, and l-lysine was added to 146 mg/ml, with additional proline at 200 mg/ml to prevent metabolic conversion of arginine to proline. For “light,” Arg0Lys0 medium standard amino acids were used (Sigma-Aldrich), and for “heavy,” Arg6Lys6 medium [13C6]-l-arginine-HCl and [13C6]-l-lysine-HCl were used (Cambridge Isotope Laboratories). Bovine serum (Invitrogen) was dialyzed overnight at 4°C against PBS using a 10-kD molecular mass cutoff membrane to remove any amino acids. HeLa cells were grown for 3 wk in this medium to reach steady-state labeling with “light” and “heavy” amino acids. For the metaphase condition, 8 × 15–cm confluent dishes of “light” cells were treated with 100 ng/ml nocodazole for 16 h. Mitotic cells were harvested by shake off, washed three times with 37°C PBS, and then resuspended in warm growth medium. The cells were incubated in a 5% CO2 incubator at 37°C for 20 min and then washed twice with ice-cold PBS. For the anaphase condition, 8 × 15–cm confluent dishes of “heavy” cells were treated with 100 ng/ml nocodazole for 16 h, washed three times in 37°C PBS, and then replated in “heavy” growth medium and harvested when the peak of anaphase cells became visible under the microscope, typically after 45–60 min. Cells were harvested and washed in ice-cold PBS. Cell pellets were lysed by addition of 1.2 ml mitotic cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 40 mM β-glycerophosphate, and 1% [vol/vol] IGEPAL with protease and phosphatase inhibitor cocktails [Sigma-Aldrich]). After 15 min on ice, lysates were centrifuged at 20,000 gav for 20 min at 4°C to remove insoluble debris. Lysates were adjusted to 10 mg/ml, and then 3 mg of each lysate was combined into a single tube together with 5 µg of sheep anti-GFP (control) or sheep anti-MKlp1 and 40 µl of packed protein G–Sepharose. After 4-h incubation at 4°C, immune complexes were recovered by centrifugation at 10,000 gav for 1 min and then washed once in lysis buffer and three times with 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% (vol/vol) IGEPAL. Samples were then resuspended in sample buffer and analyzed by MS.
MS
Protein samples for MS were separated on 4–12% gradient NuPAGE gels (Invitrogen) and then stained using a colloidal Coomassie blue stain. Gel lanes were typically cut into 12 slices and then digested with trypsin. The resulting tryptic peptide mixtures in 0.05% trifluoracetic acid were then analyzed by online liquid chromatography–MS/MS with a chromatography column (nanoAcquity UPLC; Waters) and mass spectrometer (Orbitrap XL ETD; Thermo Fisher Scientific) fitted with a nanoelectrospray source (Proxeon). Peptides were loaded on to a 5-cm × 180-µm symmetry trap column (part no. 186003514; BEH-C18; Waters) in 0.1% formic acid at 15 µl/min and then resolved using a 25-cm × 75-µm column (part no. 186003815; BEH-C18) in 99–37.5% acetonitrile in 0.1% formic acid at a flow rate of 400 nl/min. The mass spectrometer was set to acquire an MS survey scan in the Orbitrap (R = 30,000) and then perform MS/MS on the top five ions in the linear quadrupole ion trap after fragmentation using collision ionization (30 ms with 35% energy). A 90-s rolling exclusion list with n = 3 was used to prevent redundant analysis of the same ions. MaxQuant and Mascot (Matrix Science) were then used to compile and search the raw data against the human International Protein Index database (Cox and Mann, 2008; Cox et al., 2009). Protein group and peptide lists were sorted and analyzed in Excel (Microsoft) and MaxQuant. MS and MS/MS spectra were manually inspected using Xcalibur Qual Browser (Thermo Fisher Scientific).
GAP assays
For GTP-loading reactions, 10 µl assay buffer, 73 µl H2O, 10 µl of 10-mM EDTA, pH 8.0, 5 µl of 1-mM GTP, 2 µl γ-[32P]GTP (10 mCi/ml; 5,000 Ci/mmol; GE Healthcare), and 100 pmol Rho and other Ras superfamily GTPases were mixed on ice. After 15 min of incubation at 30°C, loaded GTPases were stored on ice. GAP reactions were started by the addition of 0.5 pmol GAP or purified CYK4 centralspindlin complexes as specified in the figures. A 2.5-µl aliquot of the assay mix was scintillation counted to measure the specific activity in counts per minute per picomoles of GTP. Reactions were then incubated at 30°C for 20 min and then split into two equal aliquots. Each aliquot was immediately added to 795 µl of ice-cold 5% (wt/vol) activated charcoal slurry in 50 mM NaH2PO4, left for 1 h on ice, and centrifuged at 16,100 gav to pellet the charcoal. A 400-µl aliquot of the supernatant was scintillation counted, and the amount of GTP hydrolyzed was calculated from the specific activity of the reaction mixture.
Rho family GTPase effector pull-downs
For binding assays, 25 µg GST-tagged Rho family GTPase was bound to 10 µl of packed glutathione–Sepharose (GE Healthcare) in a 1-ml total volume of nucleotide loading buffer (NL100; 20 mM Hepes-NaOH, pH 7.5, 100 mM NaOAc, 5 mM MgCl2, and 0.1% [vol/vol] Triton X-100) for 120 min at 4°C. The beads were first washed three times in 500 µl with NL100. 1 ml of either 10 mg/ml interphase or mitotic cell extract was added to the beads together with 100 µl of 10-mM GTP. Binding was then allowed to proceed for 120 min at 4°C, rotating to mix. The beads were then washed three times with 1 ml NL100, and bound proteins were eluted by the addition of 30 µl of 3× loading buffer and incubated for 5 min at 95°C. Eluted proteins were then analyzed on by SDS-PAGE and Western blotting or MS.
Microscopy and live-cell imaging
For PFA fixation, coverslips were washed twice with 2 ml PBS and fixed with 2 ml of 3% (wt/vol) PFA in PBS for 15 min. Coverslips were quenched with 2 ml of 50-mM NH4Cl in PBS and then incubated in a further 2 ml of quench solution for 10 min. Coverslips were washed three times in 2 ml PBS before permeabilization in 0.2% (vol/vol) Triton X-100 for 5 min. For TCA fixation, coverslips were washed twice with 2 ml PBS and fixed with 2 ml of ice-cold 10% (wt/vol) TCA solution for 15 min. Coverslips were quenched three times with 2 ml of 30-mM glycine in PBS for 10 min each and then washed three times in 2 ml PBS. In all cases, primary and secondary antibody staining was performed in PBS for 60 min at room temperature. Affinity-purified antibodies were used at 1 µg/ml, whereas commercial antibodies were used as directed by the manufacturers.
Fixed samples on glass slides were imaged using a 60×, 1.35 NA oil immersion objective on an upright microscope system (BX61; Olympus) with filter sets for DAPI, EGFP/Alexa Fluor 488, Alexa Fluor 555, Alexa Fluor 568, and Alexa Fluor 647 (Chroma Technology Corp.), a camera (CoolSNAP HQ2; Roper Scientific), and imaging software (MetaMorph 7.5; Molecular Devices). Illumination was provided by a 200-W metal halide light source (Lumen; Prior Scientific). For live-cell imaging, cells were plated in 2-cm dishes with a coverglass window in the bottom. Imaging was performed at 37°C in 5% CO2 using an inverted microscope (IX81; Olympus) with a 60×, 1.42 NA oil immersion objective coupled to a spinning-disk confocal system (UltraVIEW VoX; PerkinElmer) fitted with an EM charge-coupled device camera (C9100-13; Hamamatsu Photonics). Image stacks of 25–35 planes spaced 0.5–0.7 µm were taken at one to four stage positions every minute for 2–12 h. Exposure times were 10–33 ms with an EM gain setting of 200 for EGFP- or mCherry-tagged proteins using 3% power on the 488- and 561-nm lasers, respectively. A bright-field reference image stack was collected using 10-ms exposures. Maximum intensity projection images of the fluorescent channels without gamma adjustment were cropped in ImageJ (National Institutes of Health) and placed into Illustrator CS3 (Adobe) to produce the figures. Intensity measurements were performed on the full 3D dataset using the volume quantitation and object tracking tools of Volocity 5 (PerkinElmer).
Online supplemental material
Fig. S1 is a SILAC analysis of centralspindlin complex phosphorylation in metaphase and anaphase cells. The Rho GTPases expressed in HeLa cells were defined in Fig. S2, and a schematic of their effector pathways is shown in Fig. S3. Western blot controls for antibody specificity and siRNA efficiency for RhoA, Rac1, Cdc42, and their effector proteins are shown in Fig. S4. Staining for the cleavage furrow protein anillin in cells expressing hydrolysis-defective mutants of Rac1 is shown in Fig. S5. Table S1 contains a list of the siRNA duplexes directed toward cell cycle regulators, GTPases, and effector proteins investigated in the course of this study. Video 1 shows localization of activated Rac1 in anaphase cells expressing wild-type CYK4. Video 2 shows localization of activated Rac1 and early cytokinesis failure in cells expressing the GAP mutant CYK4. Video 3 shows localization of activated Rac1 and late cytokinesis failure in cells expressing the GAP mutant CYK4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201204107/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Ulrike Grüneberg for much discussion and practical advice.
This work was supported by a Cancer Research UK program grant to F.A. Barr (C20079/A9473).
Footnotes
Abbreviations used in this paper:
- CRIB
- Cdc42 and Rac interactive binding
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- MS
- mass spectrometry
- PAK
- p21-activated kinase
- SILAC
- stable isotope labeling and cell culture
- UTR
- untranslated region
- WASp
- Wiskott–Aldrich syndrome protein
References
- Barr F.A., Gruneberg U. 2007. Cytokinesis: placing and making the final cut. Cell. 131:847–860 10.1016/j.cell.2007.11.011 [DOI] [PubMed] [Google Scholar]
- Bassi Z.I., Verbrugghe K.J., Capalbo L., Gregory S., Montembault E., Glover D.M., D’Avino P.P. 2011. Sticky/Citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis. J. Cell Biol. 195:595–603 10.1083/jcb.201105136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman J.C., Lewellyn L., Laband K., Smerdon S.J., Desai A., Bowerman B., Oegema K. 2008. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science. 322:1543–1546 10.1126/science.1163086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D.H., Wasserman S.A. 1994. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 120:3367–3377 [DOI] [PubMed] [Google Scholar]
- Chesneau L., Dambournet D., Machicoane M., Kouranti I., Fukuda M., Goud B., Echard A. 2012. An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr. Biol. 22:147–153 10.1016/j.cub.2011.11.058 [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J.V., Mann M. 2009. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4:698–705 10.1038/nprot.2009.36 [DOI] [PubMed] [Google Scholar]
- D’Avino P.P., Savoian M.S., Glover D.M. 2004. Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J. Cell Biol. 166:61–71 10.1083/jcb.200402157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino P.P., Savoian M.S., Glover D.M. 2005. Cleavage furrow formation and ingression during animal cytokinesis: a microtubule legacy. J. Cell Sci. 118:1549–1558 10.1242/jcs.02335 [DOI] [PubMed] [Google Scholar]
- Frank S.R., Hansen S.H. 2008. The PIX-GIT complex: a G protein signaling cassette in control of cell shape. Semin. Cell Dev. Biol. 19:234–244 10.1016/j.semcdb.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Haas A.K., Spooner R.A., Yoshimura S., Lord J.M., Barr F.A. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177:1133–1143 10.1083/jcb.200612068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. 2005. The molecular requirements for cytokinesis. Science. 307:1735–1739 10.1126/science.1096896 [DOI] [PubMed] [Google Scholar]
- Glotzer M. 2009. Cytokinesis: GAP gap. Curr. Biol. 19:R162–R165 10.1016/j.cub.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.Y., Jan Y.N., Luo L. 2005. Function and regulation of Tumbleweed (RacGAP50C) in neuroblast proliferation and neuronal morphogenesis. Proc. Natl. Acad. Sci. USA. 102:3834–3839 10.1073/pnas.0500748102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S.L., Ebrahimi S., Milverton J., Jones W.M., Bejsovec A., Saint R. 2008. Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Curr. Biol. 18:25–29 10.1016/j.cub.2007.11.050 [DOI] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Hickson G.R., O’Farrell P.H. 2008. Rho-dependent control of anillin behavior during cytokinesis. J. Cell Biol. 180:285–294 10.1083/jcb.200709005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson G.R., Echard A., O’Farrell P.H. 2006. Rho-kinase controls cell shape changes during cytokinesis. Curr. Biol. 16:359–370 10.1016/j.cub.2005.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V., Gönczy P., Romano A., Schnabel H., Hamill D., Schnabel R., Hyman A.A., Glotzer M. 2000. CYK-4: A Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149:1391–1404 10.1083/jcb.149.7.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N., Saam J.R., Mango S.E. 2006. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 313:1298–1301 10.1126/science.1130291 [DOI] [PubMed] [Google Scholar]
- Kechad A., Jananji S., Ruella Y., Hickson G.R. 2012. Anillin acts as a bifunctional linker coordinating midbody ring biogenesis during cytokinesis. Curr. Biol. 22:197–203 10.1016/j.cub.2011.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Tsuji T., Takada Y., Miki T., Narumiya S. 2000. Accumulation of GTP-bound RhoA during cytokinesis and a critical role of ECT2 in this accumulation. J. Biol. Chem. 275:17233–17236 10.1074/jbc.C000212200 [DOI] [PubMed] [Google Scholar]
- Lin D., Edwards A.S., Fawcett J.P., Mbamalu G., Scott J.D., Pawson T. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:549–547 10.1038/35019582 [DOI] [PubMed] [Google Scholar]
- Loria A., Longhini K.M., Glotzer M. 2012. The RhoGAP domain of CYK-4 has an essential role in RhoA activation. Curr. Biol. 22:213–219 10.1016/j.cub.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P., Eda M., Watanabe N., Fujisawa K., Matsuoka T., Bito H., Ishizaki T., Narumiya S. 1998. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 394:491–494 10.1038/28873 [DOI] [PubMed] [Google Scholar]
- Mandato C.A., Benink H.A., Bement W.M. 2000. Microtubule-actomyosin interactions in cortical flow and cytokinesis. Cell Motil. Cytoskeleton. 45:87–92 [DOI] [PubMed] [Google Scholar]
- Manser E., Loo T.H., Koh C.G., Zhao Z.S., Chen X.Q., Tan L., Tan I., Leung T., Lim L. 1998. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell. 1:183–192 10.1016/S1097-2765(00)80019-2 [DOI] [PubMed] [Google Scholar]
- Miller A.L., Bement W.M. 2009. Regulation of cytokinesis by Rho GTPase flux. Nat. Cell Biol. 11:71–77 10.1038/ncb1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y., Kawashima T., Hirose K., Tonozuka Y., Kawajiri A., Bao Y.C., Deng X., Tatsuka M., Narumiya S., May W.S., Jr, et al. 2003. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell. 4:549–560 10.1016/S1534-5807(03)00089-3 [DOI] [PubMed] [Google Scholar]
- Mishima M., Kaitna S., Glotzer M. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2:41–54 10.1016/S1534-5807(01)00110-1 [DOI] [PubMed] [Google Scholar]
- Neef R., Preisinger C., Sutcliffe J., Kopajtich R., Nigg E.A., Mayer T.U., Barr F.A. 2003. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162:863–875 10.1083/jcb.200306009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R., Klein U.R., Kopajtich R., Barr F.A. 2006. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr. Biol. 16:301–307 10.1016/j.cub.2005.12.030 [DOI] [PubMed] [Google Scholar]
- Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E.A., Sillje H., Barr F.A. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444 10.1038/ncb1557 [DOI] [PubMed] [Google Scholar]
- Niiya F., Tatsumoto T., Lee K.S., Miki T. 2006. Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene. 25:827–837 10.1038/sj.onc.1209124 [DOI] [PubMed] [Google Scholar]
- Parsons J.T., Horwitz A.R., Schwartz M.A. 2010. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11:633–643 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Glotzer M., Kraut N., Peters J.M. 2007. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell. 12:713–725 10.1016/j.devcel.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Piekny A.J., Glotzer M. 2008. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr. Biol. 18:30–36 10.1016/j.cub.2007.11.068 [DOI] [PubMed] [Google Scholar]
- Piekny A., Werner M., Glotzer M. 2005. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 15:651–658 10.1016/j.tcb.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Portereiko M.F., Saam J., Mango S.E. 2004. ZEN-4/MKLP1 is required to polarize the foregut epithelium. Curr. Biol. 14:932–941 10.1016/j.cub.2004.05.052 [DOI] [PubMed] [Google Scholar]
- Rittinger K., Walker P.A., Eccleston J.F., Nurmahomed K., Owen D., Laue E., Gamblin S.J., Smerdon S.J. 1997a. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 388:693–697 10.1038/41805 [DOI] [PubMed] [Google Scholar]
- Rittinger K., Walker P.A., Eccleston J.F., Smerdon S.J., Gamblin S.J. 1997b. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 389:758–762 10.1038/39651 [DOI] [PubMed] [Google Scholar]
- Somers W.G., Saint R. 2003. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell. 4:29–39 10.1016/S1534-5807(02)00402-1 [DOI] [PubMed] [Google Scholar]
- Su K.C., Takaki T., Petronczki M. 2011. Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev. Cell. 21:1104–1115 10.1016/j.devcel.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Tatsumoto T., Xie X., Blumenthal R., Okamoto I., Miki T. 1999. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell Biol. 147:921–928 10.1083/jcb.147.5.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré A., Dorseuil O., Morin L., Timmons P., Jégou B., Reibel L., Gacon G. 1998. MgcRacGAP, a new human GTPase-activating protein for Rac and Cdc42 similar to Drosophila rotundRacGAP gene product, is expressed in male germ cells. J. Biol. Chem. 273:6019–6023 10.1074/jbc.273.11.6019 [DOI] [PubMed] [Google Scholar]
- Vitale N., Patton W.A., Moss J., Vaughan M., Lefkowitz R.J., Premont R.T. 2000. GIT proteins, A novel family of phosphatidylinositol 3,4, 5-trisphosphate-stimulated GTPase-activating proteins for ARF6. J. Biol. Chem. 275:13901–13906 10.1074/jbc.275.18.13901 [DOI] [PubMed] [Google Scholar]
- Wodarz A. 2005. Molecular control of cell polarity and asymmetric cell division in Drosophila neuroblasts. Curr. Opin. Cell Biol. 17:475–481 10.1016/j.ceb.2005.08.005 [DOI] [PubMed] [Google Scholar]
- Wolfe B.A., Takaki T., Petronczki M., Glotzer M. 2009. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7:e1000110 10.1371/journal.pbio.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Hikida M., Kurosaki T. 2006. Regulation of cytokinesis by mgcRacGAP in B lymphocytes is independent of GAP activity. Exp. Cell Res. 312:3517–3525 10.1016/j.yexcr.2006.07.026 [DOI] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Kurokawa K., Itoh R.E., Nakamura T., Mochizuki N., Nagashima K., Matsuda M. 2003. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 162:223–232 10.1083/jcb.200212049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Parrini M.C., Dulyaninova N.G., Bresnick A.R., Mochizuki N., Matsuda M. 2004. Cell type-specific regulation of RhoA activity during cytokinesis. J. Biol. Chem. 279:44756–44762 10.1074/jbc.M402292200 [DOI] [PubMed] [Google Scholar]
- Yüce O., Piekny A., Glotzer M. 2005. An ECT2–centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170:571–582 10.1083/jcb.200501097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavortink M., Contreras N., Addy T., Bejsovec A., Saint R. 2005. Tum/RacGAP50C provides a critical link between anaphase microtubules and the assembly of the contractile ring in Drosophila melanogaster. J. Cell Sci. 118:5381–5392 10.1242/jcs.02652 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.