Abstract
Background
Interferon (IFN)-alpha treatment for infectious disease and cancer causes high rates of depression and fatigue, and has been used to investigate the impact of inflammatory cytokines on brain and behavior. However, little is known about the transcriptional impact of chronic IFN-alpha on immune cells in vivo and its relationship to IFN-alpha-induced behavioral changes.
Methods
Genome-wide transcriptional profiling was performed on peripheral blood mononuclear cells from 21 patients with chronic hepatitis C either awaiting IFN-alpha therapy (n=10) or at 12 weeks of IFN-alpha treatment (n=11).
Results
Significance analysis of microarray data identified 252 up-regulated and 116 down-regulated gene transcripts. Of up-regulated genes, 2'-5'-oligoadenylate synthetase 2 (OAS2), a gene linked to chronic fatigue syndrome (CFS), was the only gene that was differentially expressed in patients with IFN-alpha-induced depression/fatigue, and correlated with depression and fatigue scores at 12 weeks (r=0.80, p=0.003 and r=0.70, p=0.017, respectively). Promoter-based bioinformatic analyses linked IFN-alpha-related transcriptional alterations to transcription factors involved in myeloid differentiation, IFN-alpha signaling, AP1 and CREB/ATF pathways, which were derived primarily from monocytes and plasmacytoid dendritic cells. IFN-alpha-treated patients with high depression/fatigue scores demonstrated up-regulation of genes bearing promoter motifs for transcription factors involved in myeloid differentiation, IFN-alpha and AP1 signaling, and reduced prevalence of motifs for CREB/ATF, which has been implicated in major depression.
Conclusions
Depression and fatigue during chronic IFN-alpha administration were associated with alterations in the expression (OAS2) and transcriptional control (CREB/ATF) of genes linked to behavioral disorders including CFS and major depression, further supporting an immune contribution to these diseases.
Keywords: interferon-alpha, gene array, RT-PCR, depression, fatigue, 2'-5'-oligoadenylate synthetase 2, CREB, ATF, TELiS, innate immunity
Introduction
Inflammatory cytokines are thought to contribute to depression, fatigue, and other neuropsychiatric disturbances in both medically ill and medically healthy individuals (Bower et al. 2002; Evans et al. 2005; Raison et al. 2006). For example, IFN-alpha, an inflammatory cytokine with antiviral and antiproliferative properties, used to treat malignant melanoma and chronic hepatitis C virus (HCV) infection, produces clinically significant depression and/or fatigue in up to 80% of patients depending on the dose (Lotrich 2009; Maddock et al. 2004; Miller 2009; Raison et al. 2005). Therefore, patients undergoing treatment with IFN-alpha have been studied to explore immune and neurobiological pathways through which peripheral inflammatory cytokines can influence the brain and behavior.
Although the physiological consequences of IFN-alpha and their relationship to IFN-alpha-induced behavioral changes have been well-studied, the molecular mechanisms involved have yet to be fully elucidated. Acutely, IFN-alpha binds to type-I-IFN receptors expressed on immune cells to activate Jak-STAT and tyrosine kinase signaling, which then increase the expression of cytokines and immunoregulatory genes (Darnell et al. 1994; Haque and Williams 1998; Pfeffer et al. 1998). Following chronic exposure to IFN-alpha, and cytokines induced by IFN-alpha, it is likely that multiple inflammatory as well as regulatory genes and signaling pathways are recruited. However, a careful analysis of gene regulation within peripheral immune cells during chronic IFN-alpha administration in relation to behavior has not been reported. Identification of genes and gene-regulating transcription factors associated with chronic IFN-alpha may help identify biomarkers or treatment targets for behavioral disorders associated with chronic inflammation, including major depression and chronic fatigue syndrome (CFS) (Klimas and Koneru 2007; Raison et al. 2009b; Yirmiya et al. 2000).
This study sought to map functional genomic changes resulting from chronic IFN-alpha exposure in a genome-wide transcriptional survey of peripheral blood mononuclear cells (PBMCs) from HCV patients treated for 12 weeks with IFN-alpha plus ribavirin compared to HCV patients awaiting IFN-alpha/ribavirin therapy. Patterns of differentially regulated genes were investigated, and individual gene transcripts that correlated with IFN-alpha-induced depression and fatigue were assessed. Bioinformatics analysis of differentially expressed genes sought to identify specific transcription factors that might mediate such effects, and define specific leukocyte subsets within which those transcriptional alterations occurred. Of note, transcription factors can bind to a number of closely related DNA promoter sequences, or transcription factor binding motifs (TFBMs), in the promoter regions of multiple genes to drive gene expression. Therefore, examination of promoter sequences of large sets of differentially expressed genes can identify TFBMs selectively over-represented among those genes, thus providing information about transcriptional control pathways potentially mediating observed differences in gene expression (Cole et al. 2005). It was hypothesized that TFBMs related to IFN-alpha signaling would be overrepresented in promoter regions of genes up-regulated in IFN-alpha-treated patients, and that the same TFBMs would be further overrepresented in promoter regions of genes that were specifically associated with IFN-alpha-induced depression and fatigue. Administration of IFN-alpha is associated with increases in monocyte chemotactic protein-1 (MCP-1/CCL2) and other innate immune, monocyte-related cytokines (e.g. interleukin (IL)-6, IL-1, and tumor necrosis factor-alpha) (Raison et al. 2009a; Taylor and Grossberg 1998), and therefore the signaling pathways related to monocyte/macrophage differentiation were also hypothesized to exhibit up-regulation. Finally, to examine TFBMs relevant to signaling pathways involved in depression and catecholamine control of immune function (Blendy 2006; Chen et al. 1999; Lutgendorf et al. 2009; Manji et al. 1999; Sanders and Straub 2002), CREB/ATF and the AP1/c-Fos family of TFBMs were analyzed.
Methods
Subjects
Twenty-one HCV-positive subjects (12 males, 9 females) were enrolled. Subjects were serum positive for anti-HCV antibodies or HCV-RNA positive by RT-PCR. Exclusion criteria included decompensated liver disease; liver disease from any cause other than HCV; infection with HIV (as reported by the subjects’ treating physician); unstable cardiovascular, endocrinologic, hematologic, renal, or neurologic disease (determined by physical examination and laboratory testing); history of schizophrenia or bipolar disorder or a diagnosis of major depression or substance abuse/dependence within 6 months of study entry (as determined by the Structured Clinical Interview for DSM-IV [SCID]) (First et al. 1997), and/or a score <28 on the Mini-Mental State Exam, indicating more than mild cognitive impairment (Folstein et al. 1975). Patients were required to be off all antidepressant, antipsychotic, or mood stabilizer medications for at least 4 months prior to blood sampling. Subjects were also required to discontinue other agents that might affect study results (i.e., narcotic analgesics, benzodiazepines, and anti-inflammatory agents) at least 2 weeks prior to sample collection. The subjects reported on herein represent a subsample of subjects included in previous studies on effects of IFN-alpha on cognitive performance, neuroendocrine function, and inflammatory responses (Felger et al. 2011; Majer et al. 2008; Raison et al. 2009a; Raison et al. 2010a; Raison et al. 2010b). All subjects provided written informed consent, and study procedures were approved by the Emory University Institutional Review Board.
Study Design
Study participants were enrolled in a longitudinal study examining immune, neuroendocrine, and neuropsychiatric variables at baseline and 12 weeks of either no treatment or treatment with IFN-alpha/ribavirin. For purposes of this study, PBMCs were obtained at 12 weeks from HCV patients treated with IFN-alpha plus ribavirin (n=11) and untreated HCV patients awaiting IFN-alpha/ribavirin therapy (control subjects, n=10). All subjects who underwent IFN-alpha treatment received either pegylated IFN-alfa-2b (Pegintron, Schering Plough, Kenilworth, NJ; 1.5µg/kg)(n=5) or pegylated IFN-alfa-2a (Pegasys, Roche-Genentech, San Francisco, CA; 180mg)(n=6) administered subcutaneously and ribavirin (800–1400 mg/day). Participation in treatment versus control group was determined by patients and their physician based on scheduling constraints and personal preferences and was not based on standardized criteria or controlled by study protocol. Blood was collected at 10 AM into EDTA-coated tubes via an indwelling catheter inserted at 8 AM. During blood sampling, subjects were asked to rest quietly for 30 minutes prior to blood withdrawal. PBMC were isolated by centrifugation (400×g, 15 min at 22°C) on lymphocyte separation medium (Mediatech, Manassas, VA). PBMC were stored in freezing serum (heat-inactivated FBS with 10% DMSO) at −80°C until RNA extraction. Urine drug screens were conducted at all visits to rule out substance abuse.
Behavioral Assessments
Depression was evaluated using the Montgomery-Asberg Depression Rating Scale (MADRS) (Montgomery and Asberg 1979). The MADRS is a 10-item, clinician-administered scale that assesses severity of depressive symptoms. Fatigue was evaluated using the 20-item Multidimensional Fatigue Inventory (MFI-20) (Smets et al. 1995). The MFI assesses five dimensions of fatigue, including general fatigue, physical fatigue, mental fatigue, reduced activity and reduced motivation. In addition to scores for each subscale, a total score was derived by summing the 5-subscale scores (Wichers et al. 2005). Higher scores on the MADRS and MFI indicate greater symptom severity. The presence of IFN-alpha-induced depressive and fatigue symptoms after 12 weeks treatment were defined as a MADRS score ≥15 and MFI score ≥75, respectively. Of note, a MADRS score of 15 has been used as a cut-off for clinically significant depressive symptoms (Felger et al. 2011; Kearns et al. 1982; Potter et al. 2004), and a total MFI score ≥75 is similar to the mean score for patients diagnosed with CFS in a population-based study (Reeves et al. 2005).
Gene Expression Profiling
Total RNA was isolated from approximately 10 million PBMC using RNeasy kits (QIAGEN, Valencia, CA) according to manufacturer instructions. RNA sample concentrations and the A260/280 ratio were determined using the MBA 2000 System (Perkin-Elmer, Shelton, CT, USA). Each sample was linearly amplified by WT-Ovation RNA amplification system (NuGEN) and then submitted to the Emory Cancer Genomics Core for microarray analysis. After hybridization to Illumina Human HT-12 Expression BeadChips (Illumina, San Diego, CA) that target over 47,000 probes, BeadChips were scanned on the Illumina BeadArray Reader to determine probe fluorescence intensity. Raw probe intensities were normalized by the quantile normalization algorithm (Bolstad et al. 2003) using GenomeStudio software from Illumina, and data were deposited in NCBI Gene Expression Omnibus as series GSE31187.
RT-PCR
Total RNA was isolated as described above and reverse-transcribed into cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). Briefly, quantitative PCR was performed using Predesigned TaqMan Gene Expression Assay probes (Applied Biosystems) to target MCP-1/CCL2 (Hs00234140_m1), signal transducers and activators of transcription (STAT1) (Hs00234829_m1), STAT2 (Hs01013126_g1), protein kinase R (PKR/EIF2AK2) (Hs00169345_m1), and 2'-5'-oligoadenylate synthetase 2 (OAS2) (Hs00942643_m1) using TaqMan Universal PCR Master Mix (Applied Biosystems) and ABI 7900HT Sequence Detection System instrument and software (Applied Biosystems). All samples were run in duplicate. Expression levels were assessed in arbitrary units and normalized relative to housekeeping genes 18S and POL2RA. Preliminary experiments confirmed that expression of housekeeping genes was not affected by 12 weeks IFN-alpha (data not shown). Similar results were obtained using 18S or POL2RA, and 18S was employed for the analysis presented herein. Fold changes between control and IFN-alpha-treated patients were determined using the ∆∆Ct method.
Microarray Data Analysis and Bioinformatics
To derive a refined list of genes most affected by IFN-alpha, and to determine which of those genes were significantly different between subjects with low versus high MADRS (≥15) and MFI (≥75) scores at 12 weeks of IFN-alpha administration, the stringency of Significance Analysis of Microarray (SAM) (Tusher et al. 2001) was used. SAM is bioinformatic strategy that employs a variety of statistical methods to identify genes that are differentially expressed between groups while correcting for multiple comparisons (see Supplemental Methods). To identify pathways affected by IFN-alpha compared to control as well as genes for confirmation by RT-PCR, Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) was employed. Ingenuity Pathway Analysis evaluates changes in gene expression as they relate to known pathways associated with a variety of cellular functions and disease states. To determine the TFBMs and immune cell types associated with patterns of differential gene expression, the Transcription Element Listening System (TELiS; www.telis.ucla.edu) (Cole et al. 2005) and transcript origin analysis (TOA) (Cole et al. 2011) were used. TELiS searches promoters of differentially expressed genes to identify TFBMs that drive observed differences in gene expression. TOA examines specific patterns of gene expression that are associated with immune cell subtypes and generates a cell origin diagnosticity score, which provides information on which cell types contribute to changes in gene expression observed between groups. A comprehensive list of all genes with >50% expression difference (Bower et al. 2011; Cole et al. 2003; Miller et al. 2008) was entered into these analyses. See Supplementary Methods for details.
Statistical Analysis
Differences between groups were assessed using t-tests or chi-square or Fisher tests (as appropriate) for categorical clinical variables. Differences in gene expression (fold change) between control and IFN-alpha-treated patients as measured by RT-PCR were assessed using Wilcoxon rank-sum tests. Pearson correlations were computed to evaluate associations between gene expression and MADRS/MFI scores. Where indicated, multivariate analyses (backward and forward multiple linear regression) were performed to assess the potential contribution of age, sex, type of IFN-alpha administered, body mass index (BMI), history of major depression or substance abuse. SPSS software was used for statistical analyses.
Results
Subject Characteristics
As shown in Table 1, no significant differences between IFN-alpha/ribavirin-treated subjects and controls were observed for relevant clinical characteristics including age, race, gender, BMI, and past history of major depression or substance abuse.
Table 1.
Characteristics of Study Participants
| Characteristic | Control (n= 10) |
IFN-alpha (n= 11) |
P-value |
|---|---|---|---|
| Age (mean, SD) | 47.8 (3.7) | 48.5 (3.9) | 0.69 |
| Sex (n, %) Males | 5 (50) | 7 (63.6) | 0.67 |
| Race (n, %) | |||
| Caucasion | 4 (40) | 3 (27.3) | 0.49 |
| Black | 5 (50) | 7 (63.6) | |
| Hispanic | 0 (0) | 1 (9.1) | |
| American Indian | 1 (10) | 0 (0) | |
| Education (n, %) | |||
| Some high school | 1 (10) | 1 (9.1) | 0.09 |
| High school diploma | 1 (10) | 4 (36.4) | |
| Some college | 2 (20) | 5 (45.5) | |
| Standard college | 5 (50) | 0 (0) | |
| Post-graduate degree | 1 (10) | 1 (9.1) | |
| Past MD (n, %) | 0 (0) | 3 (27.3) | 0.21 |
| Past Substance Abuse (n, %) | 6 (60) | 7 (63.6) | 1.00 |
| Baseline MADRS (mean, SD) | 4.1 (5.6) | 4.7 (6.9) | 0.82 |
| Baseline MFI (mean, SD) | 40.1 (13.9) | 36.4 (8.4) | 0.88 |
| BMI (mean, SD) | 29.6 (6.0) | 30.0 (3.7) | 0.46 |
Differential Gene Expression after IFN-alpha Administration
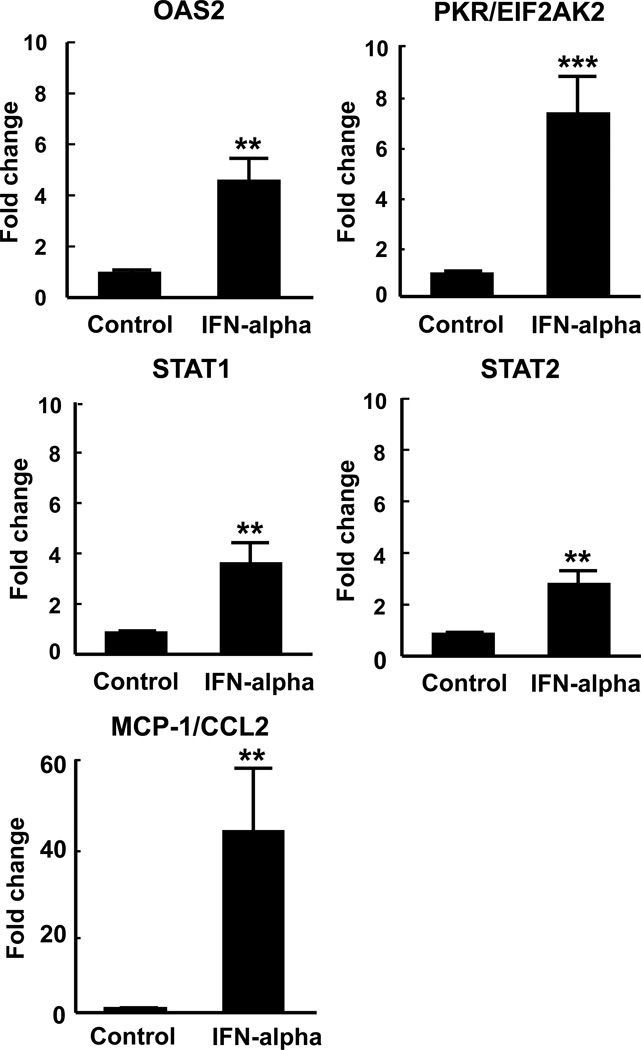
The SAM analysis revealed 252 up-regulated and 116 down-regulated gene transcripts out of 21,244 detected probes (Supplemental Table 1). Multiple probes representing a single gene were identified as significantly changed for some genes. Ingenuity Pathway Analysis demonstrated that 12 weeks of IFN-alpha administration significantly altered gene expression consistent with signaling involved in disease processes including systemic lupus erythematosus (SLE) and rheumatoid arthritis. As expected, it also identified that many genes increased by IFN-alpha were related to antiviral and inflammatory responses (see Supplemental Table 2). From these pathways, select genes were chosen for validation by RT-PCR, including OAS2, PKR/EIF2AK2, STAT1, STAT2, and MCP-1/CCL2. These gene were selected based on their role in IFN signaling (STAT1, STAT2), pattern recognition receptors in recognition of bacteria and viruses (OAS2, PKR/EIF2AK2), activation of interferon response factors (IRFs) by cytosolic pattern recognition receptors (STAT1, STAT2), dendritic cell maturation (STAT1, STAT2), role of PKR in interferon induction and antiviral response (PKR/EIF2AK2, STAT1), and IL-17 signaling (MCP-1/CCL2), and for their association with IFN-alpha-induced behavioral alterations. Expression of these genes was significantly increased in samples from IFN-alpha-treated patients compared to controls at 12 weeks (T=35.0,df=7,11,p<0.01 for OAS2), (T=28.0,df=7,11,p<0.001 for EIF2AK2/PKR), (T=31.0,df=7,11,p<0.01 for STAT1), (T=31.0,df=7,11,p<0.01 for STAT2), and (T=31.0,df=7,11,p<0.01 for MCP-1/CCL2) (Figure 1).
Fig. 1. Verification of gene expression for OAS2, PKR/EIF2AK2, STAT1, STAT2, and MCP-1/CCL2.
To verify altered mRNA expression in the context of IFN-alpha administration, changes in gene expression were compared in HCV control subjects and HCV patients treated with IFN-alpha/ribavirin for 12 weeks. Expression of IFN-alpha/antiviral genes, including OAS2, PKR/EIF2AK2, STAT1, and STAT2, and the inflammatory transcript, MCP-1/CCL2, were measured by RT-PCR. Expression of these genes was significantly increased after 12 weeks IFN-alpha administration (n=11) compared to control (n=7). IFN=interferon, MCP-1/CCL2=monocyte chemotractant protein-1, OAS2=2'-5'-oligoadenylate synthetase 2, PKR/EIF2AK2=protein kinase r, STAT1=signal transducers and activators of transcription. Data are summarized as mean+/−SE, **p<0.01, ***p<0.001
Genes Associated with Depressive Symptoms and Fatigue
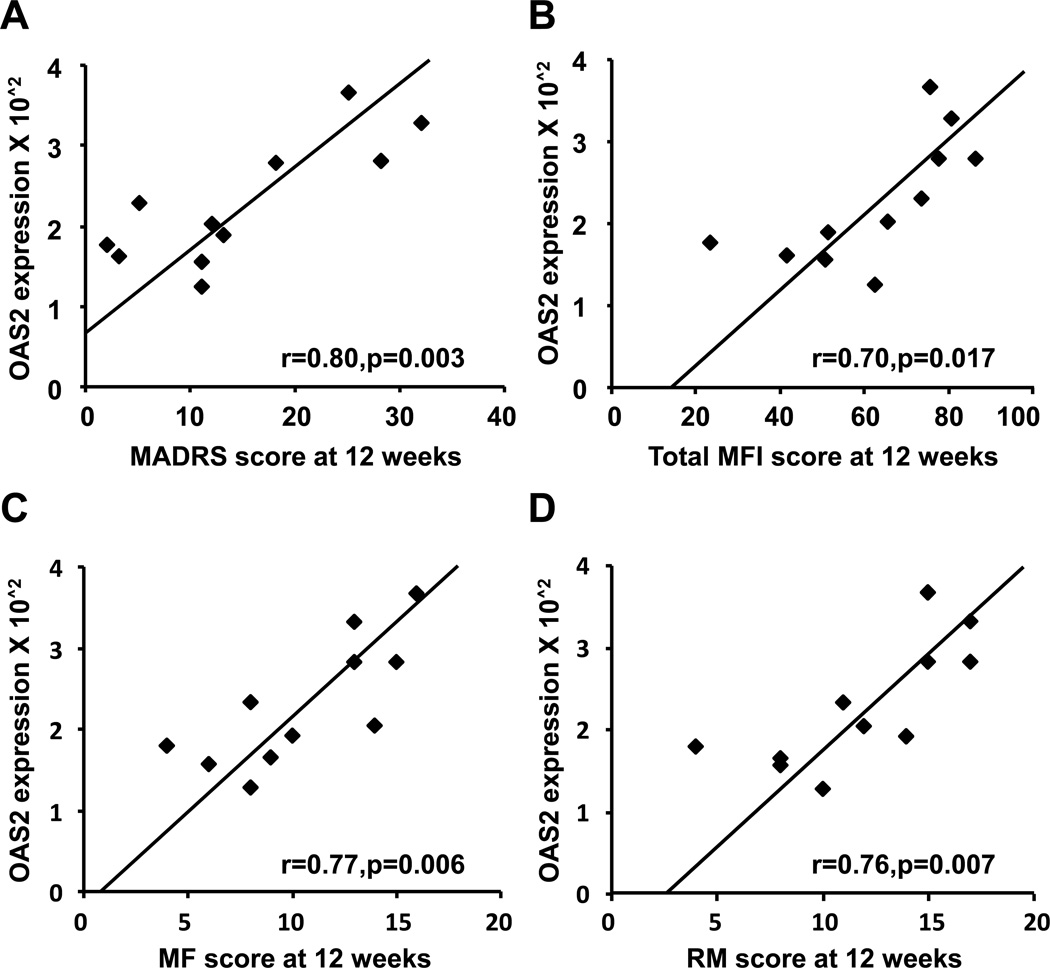
Of the 11 IFN-alpha-treated subjects, 4 subjects demonstrated both clinically significant depressive symptoms (MADRS score ≥15) and fatigue (total MFI ≥75) at 12 weeks of IFN-alpha administration. Three of these subjects also met symptom criteria for major depression according to DSM-IV. To identify genes that were increased in subjects with high MADRS and MFI scores, gene expression profiles of these individuals were compared to IFN-alpha-treated subjects with low depression (MADRS <15) and fatigue (MFI <75). SAM analysis of the 252 up-regulated and 116 down-regulated genes identified only 2 genes that were differentially expressed in high MADRS/MFI subjects; OAS2 (ILMN_2248970) was significantly up-regulated, and FCER1A (ILMN_1688423) was significantly down-regulated. Interestingly, OAS2 expression values significantly correlated with 12 week MADRS and MFI scores in IFN-alpha-treated subjects (r=0.80,df=9,p=0.003 and r=0.70,df=9,p=0.017, respectively)(Figure 2A,B). Significant correlations with OAS2 expression were also observed for mental fatigue (MF) (r=0.77,df=9,p=0.006) and reduced motivation (RM) (r=0.76,df=9,p=0.007)(Figure 1C,D). Similar results were found with OAS2 as measured by RT-PCR (data not shown). Multivariate analyses including age, sex, race, BMI, type of IFN-alpha, and past history of depression and substance abuse) as covariates revealed a final model including only OAS2 for MADRS (F[1,9]=14.25, p<0.01); MFI (F[1,9]=5.95, p<0.05); MF (F[1,9]=11.75, p<0.01); and RM (F[1,9]=14.68, p<0.01) in separate analyses. To evaluate the contribution of baseline depression and IFN-alpha-induced fatigue (week 12) to the association between OAS2 expression and high MADRS scores at week 12, partial correlations were employed. The analyses revealed that the relationship between OAS2 and week 12 MADRS scores persisted when controlling for baseline MADRS scores (r=0.80,df=8,p=0.005), as well as week 12 MFI scores (r=0.63,df=8,p=0.05). Decreased FCER1A was negatively correlated with MADRS scores at 12 weeks (r=−0.74,df=9,p=0.009), but this relationship was not significant when entered into multivariate analyses including clinical covariates.
Fig. 2. Expression levels of the OAS2 gene correlated with MADRS and MFI scores at 12 weeks of IFN-alpha administration.
OAS2 expression was positively correlated with MADRS (A) and total MFI (B) scores at 12 weeks IFN-alpha administration. Assessment of subsets of the MFI score that were related to OAS2 expression revealed significant positive correlations for MF (C) and RM (D). IFN=interferon, MADRS=Montgomery Asberg Depression Rating Scale, MF=mental fatigue, MFI= Multidimensional Fatigue Inventory, OAS2=2'-5'-oligoadenylate synthetase 2, RM=reduced motivation.
Transcription Control Pathways
To examine transcriptional control pathways that might mediate observed differences in gene expression, a two-sample variant of TELiS bioinformatics analysis was used to assess the prevalence of TFBMs involved in IFN-alpha signaling, myeloid differentiation (MZF), CREB/ATF, and AP1 signaling pathways in promoter regions of differentially expressed genes (see Supplemental Table 3 for gene lists analyzed). The prevalence of TFBMs of interest is summarized in Table 2 as the mean ratio (fold difference), standard error, and p-value. We observed highly consistent results between the two databases that the TELiS system queries, TRANFAC and JASPAR, for the TFBMs investigated. As anticipated, IFN-alpha administration was associated with an increase in genes regulated by interferon-sensitive response element, V$ISRE_01, and IRFs (V$IRF1_01, V$IRF2_01, Jaspar Irf-1, and Jaspar Irf-2). The V$ISRE_01 and V$STAT1_01 were overrepresented in genes of high compared to low MADRS/MFI subjects. Genes with TFBMs associated with myeloid differentiation (V$MZF1_02 and Jaspar MZF_5-13) were increased in subjects treated with IFN-alpha compared to control. When comparing high and low MADRS/MFI groups, there was a significant increase in V$MZF1_01, Jaspar MZF_1-4, and Jaspar MZF_5-13 TFBM prevalence. Enrichment of CREB/ATF binding sites, V$CREB_01, V$CREB_Q2, and Jaspar CREB, was revealed in IFN-alpha compared to control subjects, yet a marked decrease in the prevalence of multiple CREB/ATF binding sites (V$CREB_02, V$CREB_Q2, V$CREB_Q4, V$ATF_01, and Jaspar CREB) was observed in differentially expressed genes from high compared to low MADRS/MFI subjects. The prevalence of AP1-related TFBMs was also found to be significantly increased in promoters of IFN-alpha-related genes compared to control (V$AP1_C, V$AP1_Q2, V$AP1_Q4, V$AP1_Q6, V$AP1FJ_Q2, and Jaspar c-Fos), as well as genes up-regulated in high versus low MADRS/MFI subjects (V$AP1_C, V$AP1_Q4, and Jaspar c-Fos).
Table 2.
Transcriptional activity of IFN-alpha, myeloid differentiation, CREB/ATF, and AP1 signaling pathways as assessed by TELiS bioinformatics analysis of the prevalence of response elements in the promoters of genes differentially expressed by IFN-alpha compared to control, and in patients with high versus low depression and fatigue scores.
| IFN-alpha vs. Control | High vs. Low MADRS/MFI |
|||||
|---|---|---|---|---|---|---|
| Transcription Factor | ||||||
| Binding Motif (TFBM) | Ratio‡ | SE | p-value* | Ratio‡ | SE | p-value* |
| IFN-alpha Signaling | ||||||
| V$ISRE_01 | 3.73 | 1.4 | 0.022 | 1.53 | 1.1 | 0.021 |
| V$IRF1_01 | 2.68 | 1.4 | 0.023 | −1.16 | 1.1 | 0.320 |
| V$IRF2_01 | 3.15 | 1.3 | 0.010 | 1.48 | 1.1 | 0.069 |
| V$STAT_01 | 0.78 | 1.4 | 0.529 | 1.60 | 1.1 | 0.011 |
| Jaspar Irf-1 | 2.88 | 1.3 | 0.003 | 1.10 | 1.1 | 0.429 |
| Jaspar Irf-2 | 2.47 | 1.1 | 0.000 | 1.34 | 1.2 | 0.250 |
| Myeloid Differentiation | ||||||
| V$MZF1_01 | 1.00 | 1.0 | 0.790 | 1.44 | 1.1 | 0.012 |
| V$MZF1_02 | 1.47 | 1.1 | 0.014 | 1.42 | 1.2 | 0.096 |
| Jaspar MZF_1–4 | 1.00 | 1.0 | 0.753 | 1.39 | 1.1 | 0.026 |
| Jaspar MZF_5–13 | 1.14 | 1.1 | 0.032 | 1.40 | 1.1 | 0.003 |
| CREB/ATF | ||||||
| V$CREB_01 | 1.23 | 1.1 | 0.007 | −1.10 | 1.1 | 0.399 |
| V$CREB_02 | 1.00 | 1.1 | 0.915 | −1.47 | 1.1 | 0.000 |
| V$CREB_Q2 | 1.25 | 1.1 | 0.013 | −2.01 | 1.3 | 0.029 |
| V$CREB_Q4 | 1.09 | 1.1 | 0.183 | −1.60 | 1.1 | 0.002 |
| V$ATF_01 | 1.23 | 1.1 | 0.070 | −1.72 | 1.1 | 0.004 |
| Jaspar CREB | 1.07 | 1.0 | 0.032 | −1.48 | 1.0 | 0.000 |
| AP1 | ||||||
| V$AP1_C | 1.57 | 1.1 | 0.002 | 2.02 | 1.2 | 0.010 |
| V$AP1_Q2 | 1.42 | 1.1 | 0.001 | −1.06 | 1.1 | 0.532 |
| V$AP1_Q4 | 1.56 | 1.1 | 0.004 | 1.73 | 1.2 | 0.019 |
| V$AP1_Q6 | 1.46 | 1.1 | 0.007 | 1.32 | 1.2 | 0.146 |
| V$AP1FJ_Q2 | 1.28 | 1.0 | 0.000 | −1.03 | 1.1 | 0.668 |
| Jaspar c-Fos | 1.44 | 1.1 | 0.001 | 2.25 | 1.3 | 0.007 |
Data represent the ratio (fold difference) and SE of response element prevalence in promoter regions of genes differentially expressed in subjects at 12 weeks IFN-alpha compared to controls, and in IFN-alpha-treated patients with high versus low MADRS and MFI scores.
p-values from single-sample t tests
Transcript Origin Analysis (TOA)
To investigate the cellular origin of differentially regulated transcripts, we conducted TOA (Cole et al. 2011) for differentially expressed genes in IFN-alpha compared to control, and high (n=4) versus low (n=7) MADRS/MFI subjects (Table 3). Transcripts that were up-regulated by IFN-alpha were derived primarily from monocytes and plasmacytoid dendritic cells. Whereas transcripts expressed by CD4+T lymphocytes, CD8+T lymphocytes, and NK cells appeared at approximately the same rate among IFN-alpha-increased transcripts as they did across a random sample of all human genes; transcripts originating from monocytes and B cells were significantly more prevalent in genes down-regulated by IFN-alpha. Interestingly, cell origin analysis revealed a significant presence of monocyte and plasmacytoid dendritic cell-related genes in the down-regulated transcripts from high versus low MADRS/MFI subjects. Conversely, the up-regulated genes of high MADRS/MFI subjects were characteristic of CD8+T lymphocytes, and NK cell-related genes were significantly present in both up- and down-regulated genes for these subjects. Similar to that of IFN-alpha compared to control, the down-regulated gene profile of high MADRS/MFI subjects was significantly represented by transcripts of B cell origin.
Table 3.
Transcript origin analysis (TOA) of IFN-alpha-induced transcriptional alterations in isolated leukocyte subsets compared to control, and in patients with high versus low depression and fatigue scores.
| IFN-alpha vs. Control | High vs. Low MADRS/MFI | |||||
|---|---|---|---|---|---|---|
| Cell Type | Up-Regulated | Up-Regulated | ||||
|
Mean TOA score |
Difference from genome (±SE)‡ |
p-value |
Mean TOA score |
Difference from genome (±SE)‡ |
p-value | |
| Monocytes | 1.44 | 1.23±0.18 | 0.000 | 0.5 | 0.29±0.27 | 0.136 |
| Plasmacytoid | ||||||
| dendritic cells | 1.10 | 0.71± 0.26 | 0.003 | −0.03 | −0.42±−0.37 | 0.868 |
| NK cells | 0.53 | −0.40±0.33 | 0.885 | 4.47 | 3.54±0.48 | 0.000 |
| CD4+ T cells | 0.10 | −0.31±0.10 | 0.998 | 0.41 | 0.00±0.15 | 0.492 |
| CD8+ T cells | −0.04 | −0.25±0.08 | 0.998 | 0.44 | 0.23±0.12 | 0.030 |
| CD19+B cells | −1.01 | −0.13±0.18 | 0.755 | −0.97 | −0.08± 0.27 | 0.621 |
| Cell Type | Down-Regulated | Down-Regulated | ||||
|
Mean TOA score |
Difference from genome (±SE)‡ |
p-value |
Mean TOA score |
Difference from genome (±SE)‡ |
p-value | |
| Monocytes | 0.67 | 0.46±0.21 | 0.014 | 1.55 | 1.34±0.27 | 0.000 |
| Plasmacytoid | ||||||
| dendritic cells | 0.71 | 0.32±0.29 | 0.133 | 1.08 | 0.69±0.37 | 0.034 |
| NK cells | 0.25 | −0.67± 0.37 | 0.964 | 5.32 | 4.40±0.48 | 0.000 |
| CD4+T cells | −0.02 | −0.42± 0.12 | 1.000 | −0.22 | −0.62± 0.15 | 1.000 |
| CD8+ T cells | −0.08 | −0.29±0.10 | 0.998 | −0.39 | −0.59±0.12 | 1.000 |
| CD19+ B cells | 0.46 | 1.34±0.21 | 0.000 | −0.22 | 0.66±0.27 | 0.008 |
Positive TOA diagnosticity scores indicate that differentially expressed genes originate predominately from the analyzed cell type. Negative values are uninformative, implying that transcripts originate from other cell types or from the analyzed cell type as well as other cell types.
Scores are presented as the difference from the genome mean and standard error (SE) of the difference.
Discussion
A systematic analysis of gene transcripts regulated by chronic IFN-alpha exposure using genome-wide profiling as it relates to IFN-alpha-induced depression and fatigue was performed. As expected, genes up-regulated at 12 weeks IFN-alpha administration were predominantly related to IFN-alpha/antiviral and inflammatory signaling pathways. However, there was a striking specificity of transcriptional alterations associated with the onset of IFN-alpha-induced depression and fatigue. OAS2 mRNA emerged as the primary transcript associated with the advent of depression and fatigue symptoms within the IFN-alpha-treated group. Subsequent bioinformatic analyses suggested that cytokine-induced depression and fatigue were also linked to a relative reduction in activity of the CREB/ATF signaling pathway, and a substantial transcriptional shift toward genes originating from NK and CD8+ T lymphocytes. These depression and fatigue-related transcriptional alterations were quite distinctive relative to the more general pattern of transcriptional (TFBMs) changes associated with IFN-alpha treatment compared to controls, which involved a shift to predominately myeloid lineage monocytes and plasmacytoid dendritic cells, and activation of IRF- and AP1-family transcription factors accompanied by a comparative increase in CREB/ATF activity. Thus, the present study identifies a distinctive pattern of leukocyte transcriptional alterations associated with the onset of clinically significant depression and fatigue during IFN-alpha administration that could serve as biomarkers and/or targets for diagnosis or intervention to reduce behavioral symptoms associated with chronic exposure to an inflammatory stimulus, as during medical illness or chronic stress.
Consistent with a role for IFN-alpha signaling in development of behavioral symptoms, functional clustering of genes using Ingenuity Pathways Analysis indicated patterns of differential gene expression consistent with systemic lupus erythematosis (SLE) and rheumatoid arthritis, both of which have well documented behavioral changes including depression and fatigue (Kojima et al. 2009; Kozora et al. 2006). This analysis further classified genes related to IFN-alpha/antiviral and inflammatory responses, and increased mRNA expression of common and behaviorally-relevant IFN-alpha-induced genes (STAT1, STAT2, PKR/EIF2AK2, and OAS2) and the inflammatory gene, MCP-1/CCL2 (confirmed by RT-PCR). STAT1 and PKR are activated in the brain following peripheral IFN-alpha administration, and are thought to be related to IFN-alpha-induced behavioral alterations in rodents (Wang et al. 2008; Wang et al. 2009). Furthermore, OAS2 and PKR are activated by IFN-alpha and have been associated with behavioral disturbances in patients with CFS (Suhadolnik et al. 1994b; Vojdani et al. 1997; Vojdani and Lapp 1999). Interestingly, we have previously reported increased MCP-1, an inflammatory chemokine released by monocytes, in the periphery and cerebrospinal fluid of IFN-alpha-treated patients (Raison et al. 2009a), and this mRNA transcript was expressed at high levels in IFN-alpha-treated patients.
Of the genes identified with SAM analysis as most affected by IFN-alpha, OAS2 was the only gene significantly increased in subjects that evinced high MADRS and MFI scores. Interestingly, OAS2 mRNA expression, as measured by both microarray and RT-PCR, was correlated with depression and fatigue scores in IFN-alpha-treated subjects at 12 weeks. Analysis of the MFI subscales indicated a significant association of OAS2 expression with subscales related to mental fatigue and reduced motivation. This relationship between OAS2 and IFN-alpha-induced fatigue is of special relevance in the context of CFS, in which OAS2 activity has been consistently found to be elevated and thought to indicate increased IFN-alpha-mediated antiviral activity in these subjects (Nijs and De Meirleir 2005; Suhadolnik et al. 1994a; Suhadolnik et al. 1994b; Vojdani and Lapp 1999). Although previous studies have reported increased OAS2 activity following IFN-alpha administration (Merritt et al. 1985; Okuno et al. 1991), this is the first study to directly link increased OAS2 expression with depression and fatigue in IFN-alpha-treated subjects. Further support for these findings were apparent when considering transcriptional control of differentially expressed genes. Not surprisingly, IFN-alpha signaling was increased in IFN-alpha patients compared to controls. However, IFN-alpha-related TFBMS were also overrepresented in genes of high versus low MADRS/MFI subjects, indicating that these innate antiviral signaling pathways are, in fact, directly related to depression and fatigue. The relationship between IFN-alpha signaling and peripheral OAS2 activity in IFN-alpha-induced depression and fatigue supports the hypothesis that chronic exposure to antiviral and/or inflammatory cytokines such as IFN-alpha may contribute to pathophysiology of diseases associated with chronic inflammation and/or viral infection including CFS (Suhadolnik et al. 1994b; Vojdani and Lapp 1999). In terms of inflammation, OAS2 is involved in innate immune responses to viral infection. Although the role of OAS2 in the brain has not been characterized in relation to behavioral alterations, OAS family transcripts are activated in rodent brains in response to viral infections (Saha and Rangarajan 2003; Sandberg et al. 1994) and to overexpression of IFN-alpha (Akwa et al. 1998). Whether activation of this enzyme in the brain is sufficient to induce behavioral change remains to be determined. Nevertheless, increased peripheral OAS2 observed in CFS, and in this study during IFN-alpha-induced depression/fatigue, indicates that OAS2 or downstream pathways activated by OAS2 may precipitate behavioral alterations associated with innate immune activation.
Interestingly, transcriptional differences related to IFN-alpha-induced depression and fatigue, were found in relation to CREB/ATF and AP1/c-Fos signaling pathways. Although genes related to CREB transcriptional-control were increased in IFN-alpha-treated subjects compared to controls, it was not surprising that patients with high MADRS/MFI scores displayed a marked decrease in genes controlled by CREB signaling. Unlike CREB, increased AP1/c-Fos transcriptional activity was apparent in genes differentially expressed by both IFN-alpha compared to control, as well as high compared to low MADRS/MFI scores. In relation to decreased CREB signaling and IFN-alpha-induced depression/fatigue, CREB activity has been implicated in major depression and increases in PBMC following response to antidepressant treatment (Blendy 2006; Koch et al. 2002; Lai et al. 2003; Yamada et al. 2003). Furthermore, CREB activity in PBMC is under control of catecholamines, which may be reduced in both the periphery and central nervous system during IFN-alpha-induced depression and fatigue (Miller 2009; Trask et al. 2000). Moreover, alterations in CREB and AP1/c-Fos activity in specific brain regions are related to exposure to chronic stress and the development of depressive and anxiety-like behaviors in rodents (Kuipers et al. 2006; Morinobu et al. 1995; Wallace et al. 2004). In the brain, these transcription factors are thought to act as gene expression regulators that produce long-term modifications at the neuronal level. Transcriptional activity of chronic IFN-alpha in the periphery may mirror regulation of gene expression in brain, and represent transcriptional modifications that potentially underlie chronic cytokine-induced behavioral alterations. In relation to cytokine-induced depression, AP1/c-Fos is a downstream target of MAPK/ERK signaling pathways (Kyriakis and Avruch 2001; Zarubin and Han 2005), and activation (phosphorylation) of p38-MAPK following the initial injection of IFN-alpha is associated with depression and fatigue during IFN-alpha treatment (Felger et al. 2011). Therefore, overrepresentation of AP1-regulated genes associated with IFN-alpha-induced depressed and fatigue may reflect over-activity of MAPK/ERK signaling pathways at 12 weeks IFN-alpha administration.
Monocyte and plasmacytoid dendritic cell-related genes were significantly represented in the genes up-regulated by IFN-alpha as determined by TOA. Further support for IFN-alpha-mediated increases in monocyte signaling is the finding that MCP-1/CCL2 was the inflammatory transcript demonstrating the greatest fold-increase by IFN-alpha, and that genes differentially regulated by IFN-alpha demonstrated significant transcriptional control by MZF TFBMs related to monocyte/macrophage differentiation (Krishnaraju et al. 1995; Moeenrezakhanlou et al. 2008). Regarding plasmacytoid dendritic cells, these cells are a rich source of Type I IFNs, and are prominently activated in autoimmune disorders including SLE and Sjogren’s syndrome (Gottenberg et al. 2006; Ng and Bowman 2010; Ronnblom et al. 2003). Another interesting preliminary finding from subjects with high MADRS/MFI was that up-regulated genes were derived primarily from cytotoxic CD8+T cells and NK cells, and monocyte-related genes were more prevalent in the down-regulated genes. Nevertheless, a significant overrepresentation of genes under MZF transcriptional control was observed, indicating that increased myeloid-related signaling persisted in a background of cytotoxic lymphocyte transcriptional activity. Of note, NK cells contributed significantly to both up- and down-regulated genes associated with high MADRS/MFI, suggesting a shift in transcriptional activity within this cell type. Interestingly, alterations in NK cell activity have been frequently described in individuals with major depression (Evans et al. 1992; Irwin et al. 1990; Pariante and Miller 1995) and CFS (Klimas et al. 1990; Nijs and De Meirleir 2005). Future examination of PBMC by flow cytometry after 12 weeks IFN-alpha administration will determine whether transcriptional alterations in cytotoxic and monocytic cells translate into differences in cell number or activation state in patients with IFN-alpha-induced depression/fatigue.
In terms of limitations of this study, foremost is the small sample size. Thus, findings from genes differentially expressed in high compared to low MADRS/MFI subjects should be interpreted with caution. Furthermore, this study employed a cross sectional design that did not permit identification of gene expression at baseline that might predict development of IFN-alpha-induced depression/fatigue. It should also be noted that all IFN-alpha-treated patients were concomitantly treated with ribavirin, and it is possible that some transcriptional dynamics observed were induced by ribavirin. However, an in vitro study comparing transcriptional activity of ribavirin to that of pegylated IFN-alpha in PBMC, found that ribavirin induced differential expression of a negligible number of genes compared to IFN-alpha alone (Taylor et al. 2004). Additionally, the present study was not a randomized experiment in which patients were experimentally allocated to treatment and control groups. Although there does not appear to be any significant demographic or disease history-related confounded with IFN-alpha/ribavirin therapy in this sample, it remains possible that some other extraneous feature may differ between groups and potentially contribute to gene expression differences. However, the fact that IFN-alpha response genes dominated transcriptional signatures suggests that any confounding effects of ribavirn or other unmeasured variables likely contributed relatively little to the observed results. Furthermore, IFN-alpha mono-therapy for malignant melanoma has been associated with profound induction of depression and fatigue (Musselman et al. 2001; Raison et al. 2005). Thus, the transcriptional activity associated with depression and fatigue in this study is most likely attributed to specific effects of IFN-alpha.
In sum, this study affords a comprehensive examination of molecular alterations induced by chronic IFN-alpha exposure in a small sample of patients with HCV, and provides important clues and a theoretical framework for future studies examining changes in transcriptional activity related to cytokine-induced depression and fatigue.
Supplementary Material
Acknowledgements
This study was supported in part by grants from the National Institutes of Health to CLR (K23 MH064619, R01 MH070553), AHM (K05 MH069124, R01 HL073921, MHR01MH075102, T32 MH020018), and JCF (F32 MH093054) as well as the Emory Center for AIDS Research (P30 AI050409), and the Cancer Genomics Shared Resource of the Emory University School of Medicine. In addition, the study was supported by PHS Grant UL1 RR025008 from the Clinical and Translational Science Award program and PHS Grant M01 RR0039 from the General Clinical Research Center program, National Institutes of Health, National Center for Research Resources.
Footnotes
Financial Disclosures
All authors declare that there are no conflicts of interest, and all financial disclosures are listed for each author: Charles L. Raison serves as a consultant for Pamlab LLC and Biolex Therapeutics; Andrew H. Miller has served as a consultant for Abbott Laboratories, AstraZeneca, GlaxoSmithKline, Lundbeck Research USA, F. Hoffmann-La Roche Ltd., Schering-Plough Research Institute and Wyeth/Pfizer Inc. and has received research support from Centocor Inc., GlaxoSmithKline, and Schering-Plough Research Institute; Jennifer C. Felger, Steve W. Cole, Thaddeus W. W. Pace, Bobbi J Woolwine, Gregory H. Doho, and Fang Hu have nothing to declare.
References
- Akwa Y, Hassett DE, Eloranta ML, Sandberg K, Masliah E, Powell H, Whitton JL, Bloom FE, Campbell IL. Transgenic expression of IFN-alpha in the central nervous system of mice protects against lethal neurotropic viral infection but induces inflammation and neurodegeneration. J Immunol. 1998;161(9):5016–5026. [PubMed] [Google Scholar]
- Blendy JA. The role of CREB in depression and antidepressant treatment. Biological Psychiatry. 2006;59(12):1144–1150. doi: 10.1016/j.biopsych.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Aziz N, Fahey JL. Fatigue and proinflammatory cytokine activity in breast cancer survivors. Psychosomatic Medicine. 2002;64(4):604–611. doi: 10.1097/00006842-200207000-00010. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Irwin MR, Arevalo JM, Cole SW. Fatigue and gene expression in human leukocytes: increased NF-kappaB and decreased glucocorticoid signaling in breast cancer survivors with persistent fatigue. Brain, Behavior, and Immunity. 2011;25(1):147–150. doi: 10.1016/j.bbi.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Hasanat KA, Bebchuk JM, Moore GJ, Glitz D, Manji HK. Regulation of signal transduction pathways and gene expression by mood stabilizers and antidepressants. Psychosomatic Medicine. 1999;61(5):599–617. doi: 10.1097/00006842-199909000-00004. [DOI] [PubMed] [Google Scholar]
- Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: a PRIM approach. Bioinformatics. 2003;19(14):1808–1816. doi: 10.1093/bioinformatics/btg242. [DOI] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proceedings of the National Academy of Science U S A. 2011;108(7):3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005;21(6):803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Evans DL, Charney DS, Lewis L, Golden RN, Gorman JM, Krishnan KR, Nemeroff CB, Bremner JD, Carney RM, Coyne JC, Delong MR, Frasure-Smith N, Glassman AH, Gold PW, Grant I, Gwyther L, Ironson G, Johnson RL, Kanner AM, Katon WJ, Kaufmann PG, Keefe FJ, Ketter T, Laughren TP, Leserman J, Lyketsos CG, McDonald WM, McEwen BS, Miller AH, Musselman D, O'Connor C, Petitto JM, Pollock BG, Robinson RG, Roose SP, Rowland J, Sheline Y, Sheps DS, Simon G, Spiegel D, Stunkard A, Sunderland T, Tibbits P, Jr, Valvo WJ. Mood disorders in the medically ill: scientific review and recommendations. Biological Psychiatry. 2005;58(3):175–189. doi: 10.1016/j.biopsych.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Evans DL, Folds JD, Petitto JM, Golden RN, Pedersen CA, Corrigan M, Gilmore JH, Silva SG, Quade D, Ozer H. Circulating natural killer cell phenotypes in men and women with major depression. Relation to cytotoxic activity and severity of depression. Archives of General Psychiatry. 1992;49(5):388–395. doi: 10.1001/archpsyc.1992.01820050052009. [DOI] [PubMed] [Google Scholar]
- Felger JC, Alagbe O, Pace TW, Woolwine BJ, Hu F, Raison CL, Miller AH. Early activation of p38 mitogen activated protein kinase is associated with interferon-alpha-induced depression and fatigue. Brain, Behavior, and Immunity. 2011;25(6):1094–1098. doi: 10.1016/j.bbi.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JB. Structured Clinical Interview for DSM-IV. Washington DC: American Psychiatric Press; 1997. [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Gottenberg JE, Cagnard N, Lucchesi C, Letourneur F, Mistou S, Lazure T, Jacques S, Ba N, Ittah M, Lepajolec C, Labetoulle M, Ardizzone M, Sibilia J, Fournier C, Chiocchia G, Mariette X. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjogren's syndrome. Proceedings of the National Academy of Science U S A. 2006;103(8):2770–2775. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque SJ, Williams BR. Signal transduction in the interferon system. Seminars in Oncology. 1998;25(1 Suppl 1):14–22. [PubMed] [Google Scholar]
- Irwin M, Patterson T, Smith TL, Caldwell C, Brown SA, Gillin JC, Grant I. Reduction of immune function in life stress and depression. Biological Psychiatry. 1990;27(1):22–30. doi: 10.1016/0006-3223(90)90016-u. [DOI] [PubMed] [Google Scholar]
- Kearns NP, Cruickshank CA, McGuigan KJ, Riley SA, Shaw SP, Snaith RP. A comparison of depression rating scales. British Journal of Psychiatry. 1982;141:45–49. doi: 10.1192/bjp.141.1.45. [DOI] [PubMed] [Google Scholar]
- Klimas NG, Koneru AO. Chronic fatigue syndrome: inflammation, immune function, and neuroendocrine interactions. Current Rheumatology Reports. 2007;9(6):482–487. doi: 10.1007/s11926-007-0078-y. [DOI] [PubMed] [Google Scholar]
- Klimas NG, Salvato FR, Morgan R, Fletcher MA. Immunologic abnormalities in chronic fatigue syndrome. Journal of Clinical Microbiology. 1990;28(6):1403–1410. doi: 10.1128/jcm.28.6.1403-1410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch JM, Kell S, Hinze-Selch D, Aldenhoff JB. Changes in CREB-phosphorylation during recovery from major depression. Journal of Psychiatric Research. 2002;36(6):369–375. doi: 10.1016/s0022-3956(02)00056-0. [DOI] [PubMed] [Google Scholar]
- Kojima M, Kojima T, Suzuki S, Oguchi T, Oba M, Tsuchiya H, Sugiura F, Kanayama Y, Furukawa TA, Tokudome S, Ishiguro N. Depression, inflammation, and pain in patients with rheumatoid arthritis. Arthritis and Rheumatism. 2009;61(8):1018–1024. doi: 10.1002/art.24647. [DOI] [PubMed] [Google Scholar]
- Kozora E, Ellison MC, West S. Depression, fatigue, and pain in systemic lupus erythematosus (SLE): relationship to the American College of Rheumatology SLE neuropsychological battery. Arthritis and Rheumatism. 2006;55(4):628–635. doi: 10.1002/art.22101. [DOI] [PubMed] [Google Scholar]
- Krishnaraju K, Nguyen HQ, Liebermann DA, Hoffman B. The zinc finger transcription factor Egr-1 potentiates macrophage differentiation of hematopoietic cells. Molecular and Cellular Biology. 1995;15(10):5499–5507. doi: 10.1128/mcb.15.10.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers SD, Trentani A, Westenbroek C, Bramham CR, Korf J, Kema IP, Ter Horst GJ, Den Boer JA. Unique patterns of FOS, phospho-CREB and BrdU immunoreactivity in the female rat brain following chronic stress and citalopram treatment. Neuropharmacology. 2006;50(4):428–440. doi: 10.1016/j.neuropharm.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81(2):807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- Lai IC, Hong CJ, Tsai SJ. Expression of cAMP response element-binding protein in major depression before and after antidepressant treatment. Neuropsychobiology. 2003;48(4):182–185. doi: 10.1159/000074635. [DOI] [PubMed] [Google Scholar]
- Lotrich FE. Major depression during interferon-alpha treatment: vulnerability and prevention. Dialogues in Clinical Neurosciences. 2009;11(4):417–425. doi: 10.31887/DCNS.2009.11.4/felotrich. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgendorf SK, DeGeest K, Sung CY, Arevalo JM, Penedo F, Lucci J, 3rd, Goodheart M, Lubaroff D, Farley DM, Sood AK, Cole SW. Depression, social support, and beta-adrenergic transcription control in human ovarian cancer. Brain, Behavior, and Immunity. 2009;23(2):176–183. doi: 10.1016/j.bbi.2008.04.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock C, Baita A, Orru MG, Sitzia R, Costa A, Muntoni E, Farci MG, Carpiniello B, Pariante CM. Psychopharmacological treatment of depression, anxiety, irritability and insomnia in patients receiving interferon-alpha: a prospective case series and a discussion of biological mechanisms. Journal of Psychopharmacology. 2004;18(1):41–46. doi: 10.1177/0269881104040230. [DOI] [PubMed] [Google Scholar]
- Majer M, Welberg LA, Capuron L, Pagnoni G, Raison CL, Miller AH. IFN-alpha-induced motor slowing is associated with increased depression and fatigue in patients with chronic hepatitis C. Brain, Behavior, and Immunity. 2008;22(6):870–880. doi: 10.1016/j.bbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji HK, Bebchuk JM, Moore GJ, Glitz D, Hasanat KA, Chen G. Modulation of CNS signal transduction pathways and gene expression by mood-stabilizing agents: therapeutic implications. Journal of Clinical Psychiatry. 1999;60(Suppl 2):27–39. discussion 40-21, 113-116. [PubMed] [Google Scholar]
- Merritt JA, Borden EC, Ball LA. Measurement of 2',5'-oligoadenylate synthetase in patients receiving interferon-alpha. Journal of Interferon Research. 1985;5(1):191–198. doi: 10.1089/jir.1985.5.191. [DOI] [PubMed] [Google Scholar]
- Miller AH. Norman Cousins Lecture. Mechanisms of cytokine-induced behavioral changes: psychoneuroimmunology at the translational interface. Brain, Behavior, and Immunity. 2009;23(2):149–158. doi: 10.1016/j.bbi.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Sze J, Marin T, Arevalo JM, Doll R, Ma R, Cole SW. A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biological Psychiatry. 2008;64(4):266–272. doi: 10.1016/j.biopsych.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeenrezakhanlou A, Shephard L, Lam L, Reiner NE. Myeloid cell differentiation in response to calcitriol for expression CD11b and CD14 is regulated by myeloid zinc finger-1 protein downstream of phosphatidylinositol 3-kinase. Journal of Leukocyte Biology. 2008;84(2):519–528. doi: 10.1189/jlb.1207833. [DOI] [PubMed] [Google Scholar]
- Montgomery SA, Asberg M. A new depression scale designed to be sensitive to change. Brtish Journal of Psychiatry. 1979;134:382–389. doi: 10.1192/bjp.134.4.382. [DOI] [PubMed] [Google Scholar]
- Morinobu S, Nibuya M, Duman RS. Chronic antidepressant treatment down-regulates the induction of c-fos mRNA in response to acute stress in rat frontal cortex. Neuropsychopharmacology. 1995;12(3):221–228. doi: 10.1016/0893-133X(94)00067-A. [DOI] [PubMed] [Google Scholar]
- Musselman DL, Lawson DH, Gumnick JF, Manatunga AK, Penna S, Goodkin RS, Greiner K, Nemeroff CB, Miller AH. Paroxetine for the prevention of depression induced by high-dose interferon alfa. New England Journal of Medicine. 2001;344(13):961–966. doi: 10.1056/NEJM200103293441303. [DOI] [PubMed] [Google Scholar]
- Ng WF, Bowman SJ. Primary Sjogren's syndrome: too dry and too tired. Rheumatology (Oxford) 2010;49(5):844–853. doi: 10.1093/rheumatology/keq009. [DOI] [PubMed] [Google Scholar]
- Nijs J, De Meirleir K. Impairments of the 2-5A synthetase/RNase L pathway in chronic fatigue syndrome. In Vivo. 2005;19(6):1013–1021. [PubMed] [Google Scholar]
- Okuno T, Shindo M, Arai K, Matsumoto M, Takeda M, Kashima K, Sokawa Y. 2',5' Oligoadenylate synthetase activity in peripheral blood mononuclear cells and serum during interferon treatment of chronic non-A, non-B hepatitis. Gastroenterology Japan. 1991;26(5):603–610. doi: 10.1007/BF02781676. [DOI] [PubMed] [Google Scholar]
- Pariante CM, Miller AH. Natural killer cell activity in major depression: a prospective study of the in vivo effects of desmethylimipramine treatment. European Neuropsychopharmacology. 1995;5 Suppl:83–88. doi: 10.1016/0924-977x(95)00040-v. [DOI] [PubMed] [Google Scholar]
- Pfeffer LM, Dinarello CA, Herberman RB, Williams BR, Borden EC, Bordens R, Walter MR, Nagabhushan TL, Trotta PP, Pestka S. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Research. 1998;58(12):2489–2499. [PubMed] [Google Scholar]
- Potter GG, Kittinger JD, Wagner HR, Steffens DC, Krishnan KR. Prefrontal neuropsychological predictors of treatment remission in late-life depression. Neuropsychopharmacology. 2004;29(12):2266–2271. doi: 10.1038/sj.npp.1300551. [DOI] [PubMed] [Google Scholar]
- Raison CL, Borisov AS, Majer M, Drake DF, Pagnoni G, Woolwine BJ, Vogt GJ, Massung B, Miller AH. Activation of central nervous system inflammatory pathways by interferon-alpha: relationship to monoamines and depression. Biological Psychiatry. 2009a;65(4):296–303. doi: 10.1016/j.biopsych.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Borisov AS, Woolwine BJ, Massung B, Vogt G, Miller AH. Interferon-alpha effects on diurnal hypothalamic-pituitary-adrenal axis activity: relationship with proinflammatory cytokines and behavior. Molecular Psychiatry. 2010a;15(5):535–547. doi: 10.1038/mp.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Capuron L, Miller AH. Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends in Immunology. 2006;27(1):24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Dantzer R, Kelley KW, Lawson MA, Woolwine BJ, Vogt G, Spivey JR, Saito K, Miller AH. CSF concentrations of brain tryptophan and kynurenines during immune stimulation with IFN-alpha: relationship to CNS immune responses and depression. Molecular Psychiatry. 2010b;15(4):393–403. doi: 10.1038/mp.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Demetrashvili M, Capuron L, Miller AH. Neuropsychiatric adverse effects of interferon-alpha: recognition and management. CNS Drugs. 2005;19(2):105–123. doi: 10.2165/00023210-200519020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Lin JM, Reeves WC. Association of peripheral inflammatory markers with chronic fatigue in a population-based sample. Brain, Behavior, and Immunity. 2009b;23(3):327–337. doi: 10.1016/j.bbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Reeves WC, Wagner D, Nisenbaum R, Jones JF, Gurbaxani B, Solomon L, Papanicolaou DA, Unger ER, Vernon SD, Heim C. Chronic fatigue syndrome--a clinically empirical approach to its definition and study. BMC Medicine. 2005;3:19. doi: 10.1186/1741-7015-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnblom L, Eloranta ML, Alm GV. Role of natural interferon-alpha producing cells (plasmacytoid dendritic cells) in autoimmunity. Autoimmunity. 2003;36(8):463–472. doi: 10.1080/08916930310001602128. [DOI] [PubMed] [Google Scholar]
- Saha S, Rangarajan PN. Common host genes are activated in mouse brain by Japanese encephalitis and rabies viruses. J Gen Virol. 2003;84(Pt 7):1729–1735. doi: 10.1099/vir.0.18826-0. [DOI] [PubMed] [Google Scholar]
- Sandberg K, Eloranta ML, Campbell IL. Expression of alpha/beta interferons (IFN-alpha/beta) and their relationship to IFN-alpha/beta-induced genes in lymphocytic choriomeningitis. J Virol. 1994;68(11):7358–7366. doi: 10.1128/jvi.68.11.7358-7366.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders VM, Straub RH. Norepinephrine, the beta-adrenergic receptor, and immunity. Brain, Behavior, and Immunity. 2002;16(4):290–332. doi: 10.1006/brbi.2001.0639. [DOI] [PubMed] [Google Scholar]
- Smets EM, Garssen B, Bonke B, De Haes JC. The Multidimensional Fatigue Inventory (MFI) psychometric qualities of an instrument to assess fatigue. Journal of Psychosomatic Research. 1995;39(3):315–325. doi: 10.1016/0022-3999(94)00125-o. [DOI] [PubMed] [Google Scholar]
- Suhadolnik RJ, Reichenbach NL, Hitzges P, Adelson ME, Peterson DL, Cheney P, Salvato P, Thompson C, Loveless M, Muller WE, Schröder HC, Strayer DR, Carter WA. Changes in the 2-5A synthetase/RNase L antiviral pathway in a controlled clinical trial with poly(I)-poly(C12U) in chronic fatigue syndrome. In Vivo. 1994a;8(4):599–604. [PubMed] [Google Scholar]
- Suhadolnik RJ, Reichenbach NL, Hitzges P, Sobol RW, Peterson DL, Henry B, Ablashi DV, Muller WE, Schroder HC, Carter WA, Strayer DR. Upregulation of the 2-5A synthetase/RNase L antiviral pathway associated with chronic fatigue syndrome. Clinical Infectious Diseases. 1994b;18(Suppl 1):S96–S104. doi: 10.1093/clinids/18.supplement_1.s96. [DOI] [PubMed] [Google Scholar]
- Taylor JL, Grossberg SE. The effects of interferon-alpha on the production and action of other cytokines. Seminars in Oncology. 1998;25(1 Suppl 1):23–29. [PubMed] [Google Scholar]
- Taylor MW, Grosse WM, Schaley JE, Sanda C, Wu X, Chien SC, Smith F, Wu TG, Stephens M, Ferris MW, McClintick JN, Jerome RE, Edenberg HJ. Global effect of PEG-IFN-alpha and ribavirin on gene expression in PBMC in vitro. Journal of Interferon and Cytokine Research. 2004;24(2):107–118. doi: 10.1089/107999004322813354. [DOI] [PubMed] [Google Scholar]
- Trask PC, Esper P, Riba M, Redman B. Psychiatric side effects of interferon therapy: prevalence, proposed mechanisms, and future directions. Journal of Clinical Oncology. 2000;18(11):2316–2326. doi: 10.1200/JCO.2000.18.11.2316. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Science U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojdani A, Ghoneum M, Choppa PC, Magtoto L, Lapp CW. Elevated apoptotic cell population in patients with chronic fatigue syndrome: the pivotal role of protein kinase RNA. Journal of Internal Medicine. 1997;242(6):465–478. doi: 10.1111/j.1365-2796.1997.tb00019.x. [DOI] [PubMed] [Google Scholar]
- Vojdani A, Lapp CW. Interferon-induced proteins are elevated in blood samples of patients with chemically or virally induced chronic fatigue syndrome. Immunopharmacology and Immunotoxicology. 1999;21(2):175–202. doi: 10.3109/08923979909052757. [DOI] [PubMed] [Google Scholar]
- Wallace TL, Stellitano KE, Neve RL, Duman RS. Effects of cyclic adenosine monophosphate response element binding protein overexpression in the basolateral amygdala on behavioral models of depression and anxiety. Biological Psychiatry. 2004;56(3):151–160. doi: 10.1016/j.biopsych.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Wang J, Campbell IL, Zhang H. Systemic interferon-alpha regulates interferon-stimulated genes in the central nervous system. Molecular Psychiatry. 2008;13(3):293–301. doi: 10.1038/sj.mp.4002013. [DOI] [PubMed] [Google Scholar]
- Wang J, Dunn AJ, Roberts AJ, Zhang H. Decreased immobility in swimming test by homologous interferon-alpha in mice accompanied with increased cerebral tryptophan level and serotonin turnover. Neuroscience Letters. 2009;452(2):96–100. doi: 10.1016/j.neulet.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichers MC, Koek GH, Robaeys G, Praamstra AJ, Maes M. Early increase in vegetative symptoms predicts IFN-alpha-induced cognitive-depressive changes. Psychological Medicine. 2005;35(3):433–441. doi: 10.1017/s0033291704003526. [DOI] [PubMed] [Google Scholar]
- Yamada S, Yamamoto M, Ozawa H, Riederer P, Saito T. Reduced phosphorylation of cyclic AMP-responsive element binding protein in the postmortem orbitofrontal cortex of patients with major depressive disorder. Journal of Neural Transmission. 2003;110(6):671–680. doi: 10.1007/s00702-002-0810-8. [DOI] [PubMed] [Google Scholar]
- Yirmiya R, Pollak Y, Morag M, Reichenberg A, Barak O, Avitsur R, Shavit Y, Ovadia H, Weidenfeld J, Morag A, Newman ME, Pollmächer T. Illness, cytokines, and depression. Annals The New York Academy of Sciences. 2000;917:478–487. doi: 10.1111/j.1749-6632.2000.tb05412.x. [DOI] [PubMed] [Google Scholar]
- Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15(1):11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.