Abstract
Tiling array and novel sequencing technologies have made available the transcription profile of the entire human genome. However, the extent of transcription and the function of genetic elements that occur outside of protein-coding genes, particularly those involved in disease, are still a matter of debate. In this review, we focus on long non-coding RNAs (lncRNAs) that are involved in cancer. We define lncRNAs and present a cancer-oriented list of lncRNAs, list some tools (for example, public databases) that classify lncRNAs or that scan genome spans of interest to find whether known lncRNAs reside there, and describe some of the functions of lncRNAs and the possible genetic mechanisms that underlie lncRNA expression changes in cancer, as well as current and potential future applications of lncRNA research in the treatment of cancer.
Keywords: long non-coding RNAs, cancer, online databases, function
INTRODUCTION
Non-protein-coding RNAs (ncRNAs) are gaining the attention of researchers in many fields, and the number of published articles is exponentially growing.1 MicroRNAs (miRNAs) belong to a small ncRNA group and are the most studied among ncRNAs; however, many more types of ncRNAs exist. In fact, tiling array and novel sequencing technologies have made available the transcription profile of the entire human genome, which showed a widespread transcription activity.2 However, the extent of transcription (that is, whether ncRNAs are mainly localized in close proximity to protein-coding genes (PCGs) or widespread throughout the genome) and the function of genetic elements that occur outside of PCGs are still a matter of debate.3–5 Moreover, by more traditional means, several researchers have cloned RNA transcripts whose nature is probably not to code proteins and that have a longer sequence than miRNAs do. These can be grouped under the classification of long ncRNAs (lncRNAs).
The human genome census reveals a striking predominance of non-coding regions (http://www.ncrna.org/statgenome/index.html?view=class&gid=hg18). In fact, PCG exons represent about 1.6% of the 3 × 109 base pairs of the human genome. Moreover, the number of PCGs is quite steady during evolution in metazoa (G value paradox), whereas the size of genomes tends to increase.6 Conservation among genomes also occurs within intergenic regions, suggesting that these regions are important in the fundamental processes involved in life. Finally, the largest part of the human genome, about 46%, is made up of repetitive elements (such as transposons) that probably have been one of the driving forces of evolution.7 It is worth mentioning that in most cases transposons do not code for proteins, and recently they have been found to be related to cancer processes.8,9
In this review, we focus our attention on lncRNAs that are involved in cancer. First, we will define lncRNA and present a cancer-oriented list of lncRNAs. Second, we will list some tools (for example, public databases) that classify lncRNAs or that scan genome spans of interest to find whether known lncRNAs reside there. Some of the databases can also be used to search for lncRNAs that are involved in a process or disease of interest (for example, cancer). Finally, we will describe some of the functions of lncRNAs, possible genetic mechanisms that underlie lncRNA expression changes in cancer, and current and potential future applications of lncRNA research in the treatment of cancer.
DEFINITION OF lncRNA
The most commonly used definition of lncRNA is an RNA molecule that is longer than 200 nucleotides and that is not translated into a protein. However, this definition may be too simple and does not take into account certain issues. First, the cutoff of 200 nucleotides was arbitrarily chosen and it was set more on the basis of RNA binding to silica columns during RNA purification rather than for its functional meaning.2 Second, a PCG is usually defined as a transcript that contains an open reading frame (ORF) longer than 100 amino acids.10 However, lncRNAs can contain ORFs longer than 100 amino acids and not necessarily synthesize polypeptides; plus, polypeptides shorter than 100 amino acids can be functional in organisms and are not by-products of canonical proteins.11 Finally, and even more confounding, the same RNA can contain both PCGs and non-coding functions.12–14 These issues demonstrate how little we currently know about ncRNAs (particularly lncRNAs) and how difficult it is to form a definition.
One updated definition that we agree with takes into account some of the aforementioned issues15 and defines lncRNAs as RNA molecules that may function as either primary or spliced transcripts and do not fit into known classes of small RNAs, such as miRNAs, piwi-interacting RNAs and small nucleolar RNAs, or into classes of structural RNAs (for example, transfer RNAs, small nuclear RNAs and spliceosomal RNAs). The strengths of this definition are the absence of ORF restriction, given the fact that a RNA molecule can possess both coding and non-coding activities, and the absence of length restriction that was arbitrarily set. Other investigators have proposed bioinformatic tools to clarify or adjust the 100-amino acid ORF length cutoff to determine whether an RNA molecule codes for a protein (reviewed in Dinger et al.).10 The strengths of Mercer’s definition are the absence of ORF restriction, as a matter of fact a RNA molecule can possess both coding and non-coding activities, and the absence of length restriction that was arbitrarily set.
Additionally, we must point out that in this review we use the abbreviation lncRNA, which should not be confused with long intergenic ncRNAs (lincRNAs)16,17, which are a subtype of lncRNAs.
CLASSIFICATION OF LNCRNAS AND PUBLIC DATABASES
Generating comprehensive classifications of lncRNAs is not an easy task. In fact, many lncRNA classifications are annotations from larger databases or projects (for example, GeneBank, Fantom3), and information about the real nature (protein-coding, non-protein-coding or mixed) and function of lncRNAs cannot be gleaned from these sources. Similarly, some lncRNAs have been described in only one published study and no further reports exist.18,19 Some lncRNAs have been grouped on the basis of their position relative to host PCG (for example, overlapping RNA, cis-antisense RNA, antisense RNA, bidirectional RNA, intronic RNA, promoter- or enhancer-correlated RNA).15 As it usually happens for all classifications, the same lncRNA may be listed under different groups. For example, lncRNAs predicted by computational models (for example, RNAz or Evofold) are often listed under different names in databases obtained from sequencing projects.
To facilitate the difficult task of organizing lncRNAs, we have listed the current online databases that include ncRNAs (Table 1). These databases collect lncRNAs originated from GenBank annotations or from published articles. Some of these databases list both ncRNAs that have been experimentally proven and those that are purely computational predictions (based on RNA Z or Evofold) or have been annotated as ncRNAs on the basis of the predicted size of their ORFs.
Table 1.
Public ncRNA databases
| Website (reference) | Name | Species | MicroRNA | Small nucleolar RNA | Infrastructural RNA (ribosomal RNA, transfer RNA, small nuclear RNA) | Notes |
|---|---|---|---|---|---|---|
| http://biobases.ibch.poznan.pl/ncRNA/ | Multiple kingdoms | Excluded | Excluded | Excluded | ||
| http://www.noncode.org/ 20 | Noncode | Multiple kingdoms | Included | Included | Small nuclear RNA excluded | Experiment-based, ncRNAs divided on the basis of function (pf classes) and disease association |
| http://research.imb.uq.edu.au/rnadb/ 21 | Rnadb | Multiple kingdoms | Included | Included | Excluded | |
| http://www.ncrna.org/ 22 | fRNA | Multiple kingdoms | Included | Functional ncRNA catalog, microarray info about ncRNA, dedicated UCSC page | ||
| http://escience.invitrogen.com/ncRNA/ | Human, mouse | Excluded | Excluded | Excluded | ||
| http://rnaqueen.sysu.edu.cn/ncRNAimprint/ 23 | ncRNA imprint | Mammals | Included | Included | Excluded | Focused only on imprinting ncRNAs |
| http://jsm-research.imb.uq.edu.au/nred/cgi-bin/ncrnadb.pl 24 | NRED | Human, mouse | Excluded | Excluded | Expression data | |
| http://www.lncrnadb.org/ 25 | lncRNAdb | Multiple kingdoms | Excluded | Excluded | Excluded | |
| http://rfam.sanger.ac.uk/ 26 | Rfam | Multiple kingdoms | Included | Included |
Abbreviations: fRNA, functional RNA; lncRNA, long non-coding RNA; ncRNA, non-protein-coding RNA; UCSC, University of California Santa Cruz. The website, reference and content are listed for each database, along with the most interesting feature of each website.
We found the functional RNA project database (fRNA) worth visiting. It uses a University of California Santa Cruz genome browser interface that contains many ncRNA tracks that have already been set up in a Genome Browser graphic interface, which allows the user to search for specific ncRNAs along with other features in the genomic context of interest. Although both fRNA and the Noncode project allow the user to search for functional classes or processes (for example, find all known ncRNAs that are involved in the cell cycle or in DNA replication or transcription), fRNA allows the user to search by disease (for example, cancer) as well. The ncRNA expression database, on the other hand, contains a large data set of ncRNA expression profiles that were obtained from three different experiment sets: Allen Brain Atlas (mouse), GNF Atlas (mouse and human) and V1.0 Compugen array (mouse). Although these expression data sets are not cancer-oriented, we foresee that eventually the ncRNA expression database, as well as others that are listed in Table 1, will be matched with other data sets that are more cancer-oriented (for example, Oncomine; https://www.oncomine.org). For now, the genomic positions of several lncRNAs can be matched to databases that collect lists of single-nucleotide polymorphisms (SNPs) associated with cancer (http://cistrome.dfci.harvard.edu/CaSNP/; Hindorff et al.)172 or cancer-associated genetic regions (for example, http://cancergenome.nih.gov, http://decipher.sanger.ac.uk, the Cancer Workbench at https://cgwb.nci.nih.gov/cgi-bin/heatmap and National Center for Biotechnology Information Gene Expression Omnibus at http://www.ncbi.nlm.nih.gov/geo/).14,27
CANCER-RELATED LNCRNAS
In this review, we focused our efforts on developing a list of lncRNAs that have been linked to cancer by various means. We mainly used three of the online databases to retrieve lncRNAs (that is, the LncRNA database, Noncode and the RNA Database), and we searched Pubmed for articles linking these lncRNAs to cancer. In Table 2, we report our findings.
Table 2.
lncRNAs that have been or might be (*) linked to cancer
| lncRNA | Molecular mechanism | Tumor | Reference | Genome position | Website |
|---|---|---|---|---|---|
| aHIF | Messenger RNA decay | Multiple cancers | 28–30 | hg19 chr14:61,283,843–61,285,036 | Noncode |
| Air | Epigenetic regulation | * | 31 | NA | lncRNAdb |
| ak023948 | Unknown | Papillary thyroid carcinoma | 32 | hg18 chr8:134136386-134139194 | lncRNAdb |
| alpha 250/alpha 280 | Antisense, transcription regulation | * | 33,34 | hg19 chr5:149,828,969-149,829,248 | lncRNAdb |
| anril | Antisense, transcription regulation | Prostate cancer | 35,36 | hg18 chr9:21,984,790-22,111,091 | lncRNAdb |
| anti-NOS2A | NA | Central nervous system tumors | 37 | hg18 chr17:57,692,139-57,696,081 | lncRNAdb |
| antisense tgf beta 3 | NA | * | 38,39 | NA | lncRNAdb |
| BA318C17.1 | NA | Colon cancer | 40 | hg19 chr20:14,864,899-14,910,132 | Rnadb |
| bc200 | Protein binding | Multiple cancers | 41,42 | hg19 chr2:47,562,454-47,562,653 | lncRNAdb |
| car intergenic 10 | Regulation of gene expression | * | 43 | hg18 chr10:127,690,946-127,693,326 | lncRNAdb |
| ccnd1-associated ncrnas | Regulation of gene expression | * | 44,45 | hg19 chr11:69,453,875-69,455,874 | lncRNAdb |
| dhfr upstream transcripts | Regulation of gene expression | * | 46 | hg18 chr5:79,985,935-79,986,521 | lncRNAdb |
| e2f4 antisense | NA | * | 47 | NA | lncRNAdb |
| emx2os | Antisense-sense pairing Dicer1 | * | 48–50 | hg18 chr10:119,233,794-119,294,569 | lncRNAdb |
| gas5 | Decoy of glucorticoid receptor | Breast cancer | 51 | hg18 chr1:172,099,662-172,103,748 | lncRNAdb |
| GNAS1-as RNA | NA | * | 52–56 | hg19 chr20:57,393,804-57,425,958 | Noncode |
| h19 | Transcription regulation (contains miR-675) | Multiple cancers | 57–59 | hg18 chr11:1,972,982-1,975,641 | lncRNAdb |
| h19 antisense | Regulation of gene expression | * | 60 | NA | lncRNAdb |
| h19 upstream conserved 1 and 2 | NA | * | 61 | NA | lncRNAdb |
| His-1 RNA | NA | * | 62,63 | hg19 chr2:145,456,944-145,465,439 | Noncode |
| HOTAIR | Epigenetic regulation | Multiple cancers | 64–66 | hg18 chr12:52,642,363-52,648,782 | lncRNAdb |
| hotairm1 | NA | * | 67–69 | hg18 chr7:27,102,268-27,106,109 | lncRNAdb |
| Hoxa11 antisense | NA | * | 70–73 | hg19 chr7:27,225,048-27,228,956 | Noncode |
| hoxd3as | NA | * | 74,75 | NA | lncRNAdb |
| HULC | Micro RNA decoy | Multiple cancers | 76–78 | hg18 chr6:8,597,441-8,599,080 | lncRNAdb |
| krasp1 | Micro RNA decoy | Prostate cancer | 14 | hg18 chr6:54,743,128-54,743,996 | lncRNAdb |
| KvlQT1-AS (Kcnq1ot1) | DNMT1 interaction and transcription gene silencing | Colon cancer | 79 | hg19 chr11:2,465,330-2,870,445 | Noncode |
| LEU2 | Pri-micro RNA, other | Chronic lymphocytic leukemia | 80 | hg19 chr13:50,556,688-50,699,677 | lncRNAdb |
| LOC285194 | NA | Osteosarcoma | 81 | hg18 chr3:117,911,325-117,918,575 | lncRNAdb |
| LUST | RNA–RNA interaction, RNA splicing | * | 19 | hg18 chr3:50,112,040-50,113,425 | lncRNAdb |
| MALAT-1 (NEAT2) | RNA splicing, small RNA production, protein interaction | Multiple cancers | 82–84 | hg18 chr11:65,021,809-65,030,513 | lncRNAdb |
| MEG3 | NA | Multiple cancers | 85,86 | hg18 chr14:100,362,198-100,397,121 | lncRNAdb |
| MER11C | RNA–protein interaction, regulation of gene expression | Cell lines | 83 | hg18 chr11:50,410,308-50,411,367 | lncRNAdb |
| Msx1 antisense | NA | * | 87–90 | NA | Noncode |
| ncR-uPAR | RNA–protein interaction | * | 18 | hg18 chr5:76,043,519-76,044,442 | lncRNAdb |
| NCRMS | NA | * | 91 | hg19 chr12:97,886,239-97,954,478 | Noncode |
| NDM29 | NA | Neuroblastoma | 92 | hg18 chr11:8,917,158-8,917,288 | lncRNAdb |
| NEAT1/TncRNA | RNA nuclear export, paraspeckle organization | * | 93–96 | hg18 chr11:64,946,845-64,950,577 | lncRNAdb |
| Nkx2.2AS | NA | * | 97,98 | NA | lncRNAdb |
| NRON | NFAT nuclear trafficking, RNA-protein binding | * | 99,100 | NA | lncRNAdb |
| NSCLC B2 | NA | * | 101,102 | hg19 chr6:11,192,694-11,205,944 | Rnadb |
| NTT sense/antisense | NA | * | 103,104 | hg19 chr6:136,265,389-136,282,959 | Noncode |
| p53 mRNA | RNA protein binding | Multiple cancers | 12 | hg19 chr17:7571720-7590863 | lncRNAdb |
| p53int1 | NA | * | 105 | hg19 chr17:7,588,578-7,589,689 | Rnadb |
| PCA3/DD3 | NA | Prostate cancer | 106 | hg18 chr9:78,569,172-78,592,305 | Noncode |
| PCGEM1 | NA | Prostate cancer | 107 | hg18 chr2:193,322,816-193,349,870 | Noncode |
| PCNA-AS | NA | * | 108 | hg19 chr20:5,100,232-5,100,615 | Rnadb |
| PINC | NA | * | 109 | NA | lncRNAdb |
| PR Antisense | Regulation of gene expression | * | 110 | hg18 chr11:100,505,018-100,574,851 | lncRNAdb |
| PRINS | NA | * | 111,112 | hg18 chr10:24,576,060-24,584,981 | lncRNAdb |
| PTENP1 | Micro RNA decoy | Prostate cancer | 14 | hg18 chr9:33,663,502-33,667,418 | lncRNAdb |
| RMRP | Mitochondrial RNA processing endoribonuclease, hTERT-dependent small interfering RNA pathway | Leukemia and lymphoma | 113,114 | hg19 chr9:35,657,750-35,658,014 | Rnadb |
| RPS6KA2 antisense transcript | NA | Cell lines | 115 | NA | lncRNAdb |
| saf | NA | Cell lines | 116 | hg19 chr10:90,751,179-90,752,732 | Rnadb |
| SRA RNA | RNA-protein binding, transcription factor co-activator | Breast cancer | 117,118 | hg19 chr5:139,930,090-139,937,036 | Noncode |
| TERC | Telomere template | Multiple cancers | 119 | hg19 chr3:169,481,881-169,483,646 | Noncode |
| terra | Telomerase regulation | Multiple cancers | 120,121 | NA | lncRNAdb |
| tie-1AS | RNA–RNA interaction | * | 122–124 | NA | lncRNAdb |
| Tsix | Antisense of Xist | * | 125,126 | hg18 chrX:72,928,765-72,965,791 | lncRNAdb |
| UBE3A antisense | NA | * | 127,128 | hg19 chr15:25,264,182-25,299,063 | Noncode |
| uca1 | NA | Bladder cancer | 129,130 | hg19 chr19:15,939,757-15,946,226 | Rnadb |
| WT1-AS | NA | * | 131 | hg18 chr11:32,413,861-32,418,212 | lncRNAdb |
| xist | X inactivation | Multiple cancers | 125,132–134 | hg19 chrX:73,043,280-73,072,588 | Noncode |
| Zeb2NAT | NA | * | 135,136 | hg18 chr2:144,992,452-144,995,153 | lncRNAdb |
| Zfas1 | NA | Breast cancer | 137 | hg19 chr20:47,894,715-47,905,797 | lncRNAdb |
Abbreviations: lncRNA, long non-protein coding RNA; NA, not available. For each lncRNA, the name, function, tumor model in which it has been evaluated, and genomic position are listed, along with the public website where each lncRNA can be found.
In some cases the link between the lncRNA and cancer was obvious, and cancer was actually the model where these lncRNAs were first described for the first time (for example, MALAT-1, PCA3/DDR3 and HOTAIR). However, we also found some lncRNAs for which a link to cancer has not yet been fully elucidated, but preliminary findings indicate that it could be worthwhile to investigate the possible connection (these lncRNAs are marked with an asterisk in Table 2). For example, the lncRNA Air (antisense to Igf2r RNA) is involved in the imprinting of the Igfr2 locus.31 Despite the association of Igfr2 with cancer138 and the association of Air with Igfr2,139 we did not find any articles that directly examine the relationship between Air and cancer in humans. Moreover, although alpha 250/alpha 280 lncRNA regulates RPS14 transcription, which has been shown in short hairpin RNA screens to be a causal factor in 5q- syndrome,33 no studies have yet examined the direct involvement of alpha 250/alpha 280 lncRNA in 5q-syndrome.140
Some lncRNAs that we included contain small ncRNA (for example, miRNA and small nucleolar RNA). Although these lncRNAs host small RNAs, this may not be their exclusive function. For example, the knockout model of LEU2, which includes miR-15/16 as well, showed a more aggressive phenotype than did the miR-15/16 knockout model, which may indicate that LEU2 can participate in chronic lymphocytic leukemia development.80
Some lncRNAs have been associated with cancer but are not listed in the public data sets that we used to prepare Table 2. For example, regions that are extremely conserved among human, mouse and rat genomes141 are expressed in cancer tissue differently than in normal tissues and are regulated by methylation as well.142–146 The extremely high level of conservation among these lncRNAs, which are referred to as ultraconserved genes or transcribed UCRs, is their most peculiar feature.
LNCRNA FUNCTION
The function of ncRNAs is the most difficult and least understood aspect of ncRNA research. Better understanding ncRNA function will help clarify the real impact of genomic pervasive transcription on cell biology and evolution.147 As we gathered information about the lncRNAs involved in cancer, we also collected examples of lncRNA function (Figure 1).
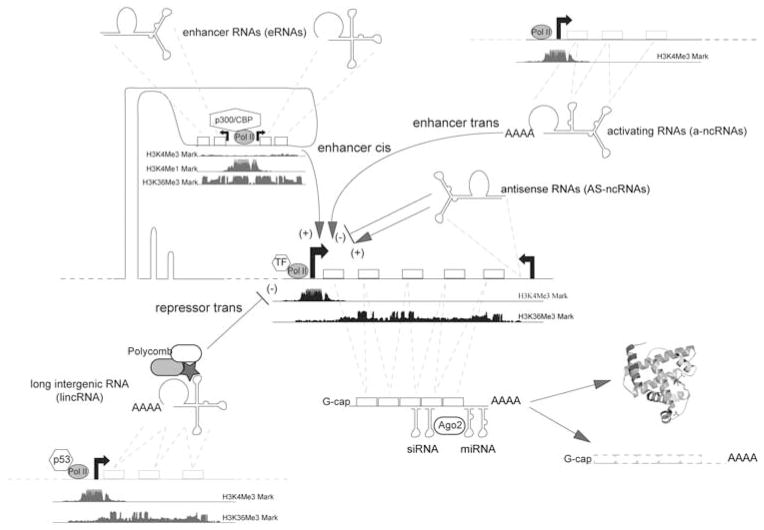
Figure 1.
LncRNA categories and functions. Several classes and functions of lncRNAs are depicted. The main function of lncRNA seems so far to regulate PCG transcription; indeed, lncRNA can either enhance or repress PCG transcription by changes in the chromatin state of the PCGs (for example, by histone methylating or acetylating). Enhancer RNAs derive from transcription of enhancer elements that can be located several kilobases upstream of target genes. Enhancer DNA can both regulate gene expression by DNA looping and direct DNA–DNA interaction with the target promoter, and they also transcribe non-polyA RNAs (that is, eRNA). The function and the role of eRNAs is at this moment unknown. Overall, both long ncRNAs (lincRNA, a-ncRNAs and AS-ncRNAs) and small ncRNAs (for example, siRNA and miRNA) regulate transcription and post transcription steps of protein synthesis, respectively. At the bottom of coding and non-coding transcription units that are shown in picture, the reader can find the peak diagram for CHIP-seq experiments concerning histone modifications: H3K4Me1, mono methylation at lysine 4 of histone 3 (often found near regulatory elements); H3K4Me3, tri methylation at lysine 4 of histone 3 (often found near promoters); H3K36Me3, tri methylation at lysine 36 of histone 3 (often found near active transcripts).
The first example that we describe is for lincRNAs. LincRNAs were first described using histone mark signatures, specifically trimethylation in lysine 4 and lysine 36 of histone 3 (H3K4m3, H3K36m3 or simply K4K36). The K4K36 mark detects active transcription units of both PCGs and ncRNAs. After excluding known genes (PCGs and ncRNAs), researchers have been able to retrieve novel transcriptional units. The first reports analyzed mouse and human cell lines, uncovering about 3000 lincRNAs.16,148 However, many more lincRNAs may remain to be discovered in other settings.149 Certain, lincRNAs were discovered before the use of the K4K36 signature such as MALAT-182 and HOTAIR, which was the first lncRNA ever described to interact with polycomb proteins and suppress gene transcription.64 Moreover, other histone signatures might reveal new lncRNAs.150
About 20% of lincRNAs bind to polycomb repressive complex 2, indicating that lincRNAs might regulate gene expression by directing the polycomb protein group to target DNA regions, inducing changes in histone marks and chromatin structure and ultimately suppressing transcription activity.148,151 The current model proposes that lincRNAs directly bind to the polycomb proteins and direct them to specific DNA segments in the human genome. However, how the lincRNA-polycomb complex recognizes the target DNA is not currently known.152 We do not currently know whether transcription factors bind lincRNAs as well, and whether RNA-binding proteins regulate lincRNAs as they do with miRNAs.153
Another class of lncRNAs that seems to regulate gene expression by changes in chromatin status includes antisense transcripts (reviewed in Morris and Vogt).154 Antisense ncRNA transcripts overlap PCG but are transcribed in the opposite direction. Although one would expect small interfering RNA (siRNA) machinery to degrade messenger RNA after the sense–antisense pairing, the mechanism in act instead seems to be the modifications of histone marks at the promoter region of the sense transcript (that is, PCGs). Apparently, antisense lncRNAs drive (cytosine-5)-methyltransferase 3A (DNMT3A) to the DNA of the host PCG to methylate histones at lysine 9 and 27 or CpG islands and ultimately silence transcription.
Several oncogenes or tumor-suppressor genes exhibit antisense transcription and consequent transcription gene silencing (for example, p21, c-Myc, p15, p53, TIE1 and PU.1).35 Interestingly, exogenous siRNAs that are in antisense orientation compared with PCG promoters are also effective at silencing transcription.155 However, how the antisense lncRNAs are regulated has not yet been explored.
LincRNAs, antisense lncRNAs, and other lncRNAs44,156 can be classified among the chromatin-associated RNAs (CARs) because their function apparently relies on the ability of the RNA to somehow bind to genomic DNA and consequently regulate chromatin states (euchromatin versus heterochromatin).44,156 Mondal et al.43 performed a thorough investigation of CARs throughout the genome of a human skin fibroblast cell line by deep sequencing of DNA-associated RNA after micrococcal nuclease treatment. They identified several CARs and reported that one CAR can activate transcription of neighboring genes.
Another class of lncRNAs that seems to regulate the transcription activity of host PCGs comprises the promoter upstream transcripts (PROMPTs). PROMPTs are localized upstream of promoters of some PCGs and they can be transcribed in both the sense and antisense orientations. PROMPTs seem to be a byproduct of RNA pol II activity; however, preliminary findings suggest that PROMPTs control promoter methylation.45 When Preker et al. first described PROMPT existence, they used a peculiar approach: they inhibited exosome key proteins by using siRNA to prolong the half-life of short-lived RNA transcripts. In this way, they were able to identify a plethora of PROMPTs. However, the function and impact of PROMPTs in cell biology have not yet been explored.
It is possible that lincRNAs, PROMPTs, and antisense RNAs, or CARs in general, have interdependent functions. For example, antigene RNAs are synthetic RNA molecules that when designed to be complementary to PCG promoters can either repress or activate gene expression. Antigene RNAs rely on RNA–RNA interaction with antisense transcripts that are generated nearby targeted promoters and on Ago proteins binding.157 It is possible that PROMPTs, lincRNAs and antisense lncRNAs interact and recapitulate antigene RNA mechanism; it is known that PROMPTs and antisense lncRNAs can interact with each other to trigger the antigene RNA pathway.158 In another example of lncRNA interdependent function, lincRNAs can interact with PROMPTs or antisense lncRNAs to ultimately direct polycomb protein complexes to targeted promoters of PCGs.36 Further examples of lncRNA function are discussed in other reviews.159,160
LNCRNA NETWORKS
Another interesting lncRNA function is target decoy or mimicry: lncRNAs can deceive another RNA or protein away from its natural target. For example, Poliseno et al.14 described pseudogenes as decoys for miRNAs. They reported that the PTEN gene and the PTEN pseudogenes (PTENP1) share a high degree of sequence homology and are targeted by the same miRNAs (that is, miR-17, -21, -214, -9 and -26 families). Thus, changes in PTENP1 expression levels indirectly affect PTEN expression levels by sequestering PTEN-targeting miRNAs. For instance, if PTENP1 expression levels decrease, miRNAs will be able to target PTEN and ultimately downregulate PTEN expression levels. Poliseno et al.14 also noted a similar mechanism for RAS pseudogenes. Another example of a lncRNA that acts as a miRNA decoy is the highly upregulated liver cancer transcript (HULC), which binds to and inhibits miR-372.76
Target decoys occur not only in cancer but also in infectious diseases: two studies reported that virus-encoded transcripts can act as miRNA decoys; in this case the net effect was to sequester and downregulate the miRNAs of the host organism.161,162 A similar example exists in plants for endogenous pseudogene transcripts that share a high degree of sequence homology with PCG transcripts, although in this case the pseudogenes contain point mutations within the miRNA-binding sites. Apparently, these pseudogenes not only sequester miRNAs from their PCG target, but also reduce miRNA expression levels.163
One particular type of lncRNA decoy involves proteins. PROMPTs, such as GAS5, can bind to transcription-factor proteins that would otherwise bind to the DNA promoters; thus, the RNA transcript decoy sequesters the transcription factor, which is no longer able to affect downstream target genes.164 GAS5 accomplishes this with a stem-loop structure in its sequence resembling the glucocorticoid receptor DNA-binding element. GAS5 seems to regulate other receptors (that is, androgen, mineralcorticoid and progesterone) by the same means. Interestingly, the interaction between GAS5 and the glucocorticoid receptor is modulated by dexamethasone, a glucocorticoid receptor agonist.164 At the same time, GAS5 has been shown to be regulated by mammalian target of rapamycin pathway and to mediate rapamycin effect on cell cycle in T cells (reviewed in Williams et al.).165
NcRNA decoys can target not only ncRNA–mRNA or DNA–protein interactions, but also interactions between ncRNAs. For example, miRNAs can target other ncRNAs as they do with messenger RNA; Calin et al.144 showed that miR-155 targets transcribed UCRs in both in vitro models and chronic lymphocytic leukemia patients. These findings support the existence of networks among ncRNAs and between ncRNAs and PCGs that are involved in cancer.
LNCRNA EXPRESSION IN CANCER
In cancer biology, one of the first evidences that researchers seek is gene expression differences between tumor and normal samples. The breadth of knowledge concerning lncRNA expression profiles in tumor and normal samples is quite modest at this time. It is likely that commercial gene expression arrays that have been used for PCGs contain probes that hybridize to lncRNAs, and it may be possible to retrieve cancer-related lncRNA expression profiles from public, tumor-specific gene-expression data sets (for example, Oncomine, Gene Expression Omnibus). However, to our knowledge this has not yet been done.
To identify novel transcripts, some investigators have used the Affymetrix tiling array, which can test for lncRNA gene expression.166,167 Others have performed custom array profiling on large sample sets of a few lncRNAs.144,168 Most articles concerning lncRNA expression in cancer have shown a selected number of lncRNAs probed in tumor samples (Table 2 lists tumor types that have been tested for lncRNA expression). We also found a few articles (not included in Table 2) reporting the existence of transcriptionally active regions that are located outside known PCGs and are differentially expressed between normal and tumor tissues or are expressed under stress conditions.166,167 Gibb et al. used SAGE library generation to compare lncRNA expression in normal and dysplastic oral mucosa.169
Cancer biologists also seek to uncover genetic mutations (for example, amplifications, deletions and sequence mutations) in the lncRNA sequence. For example, sequence mutations in RNA component of mitochondrial RNA processing endoribonu-clease (RMRP) lncRNA are responsible for cartilage-hair hypoplasia syndrome, which is also known to increase the risk of developing several types of tumors.170,171 Some investigators have already sequenced select classes of lncRNAs to find mutations.166,172
In recent years, using SNP arrays to study very large populations (in the thousands), researchers have discovered several SNPs that are associated with certain traits or diseases, such as cancer (http://www.genome.gov/gwastudies contains a list of SNPs associated with several diseases).173 In some cases, disease-associated SNPs are in genomic spans outside of PCG transcripts;174,175 these genomic spans would be good candidates regions to search for novel transcripts. Some researchers have already found SNPs that are located within lncRNA transcripts and are associated with cancer. For example, Yang et al. showed that among six SNPs that are located within the boundaries of UCRs, two of them (that is, rs2056116 and rs9572903) were significantly associated with familial breast cancer.176 Cabili et al.,27 while reporting on a census of 8195 lincRNAs in 24 different human tissues, noted that the genomic positions of 414 lncRNAs were related to SNPs that have been associated with several diseases.173
DIAGNOSTIC AND THERAPEUTIC APPLICATIONS OF LNCRNAS
The relatively new field of lncRNA research is expanding quickly, but many gaps still need to be filled. Only recently has the number of lncRNAs in the human genome become clear.27 Moreover, researchers have not extensively investigated lncRNA expression in large and clinically controlled tumor data sets, nor is lncRNA function well understood.149 Few examples of transgenic models of lncRNA have been published to date.80,177
We foresee potential uses of lncRNAs in the clinical setting for oncology or for other fields. LncRNAs may be useful as novel biomarkers for diagnosis, prognosis and prediction of response to therapy. The lncRNA PCA3/DD3, for example, has already been assayed in controlled clinical settings. PCA3/DD3 was originally discovered in a differential display analysis comparing normal and tumor prostate samples.178 The features that make PCA3/DD3 a promising biomarker are its unique expression profile in prostate tumors compared with normal prostate and other tissues, its highly increased expression levels (that is, about 60 times) in prostate tumors compared with normal tissues, its expression in early-stage tumors and detectability in urine. PCA3/DD3 has been tested as a biomarker in clinical trials and compared with standard prostate markers (that is, prostate-specific antigen). However, the effectiveness of PC3/DD3 as a biomarker was about the same as that of prostate-specific antigen.106,179
The marked increase or decrease in lncRNA expression levels in tumors compared with normal tissues seems to be a feature shared among lncRNAs. Indeed, HOTAIR was found to be upregulated by hundreds or thousands of times in metastatic breast cancer tissue compared with normal breast tissue.64 Such a large difference in lncRNA expression levels in tumors compared with normal tissues is a topic for future clinical research, although lncRNAs must be assayed in larger clinical data sets. Other lncRNAs might be promising biomarkers as well.106,179
Another potential avenue of lncRNA research relates to the discovery of circulating miRNAs in serum, plasma and other body fluids, demonstrating that miRNAs may act not only within cells, but also at other sites within the body.180 It is highly probable that other types of ncRNAs, including lncRNAs, can be present in body fluids, as suggested by, for example, their presence in the secreted exosomes. LncRNAs found in numerous quantities in body fluids could be detected using simple quantitative reverse transcriptase polymerase chain reaction. This could represent an unexpected and yet unexplored gold mine of potential biomarkers predictive of survival or response to therapy.
LncRNAs might also be useful as therapeutic agents. The small size of miRNAs offers an intrinsic advantage in their use as therapeutic bullets by in vivo administration.181 However, because lincRNAs are much longer than miRNAs, they could not be used directly as therapeutic bullets but would require gene therapy delivery systems (for example, viruses), which would carry potential risks. On the other hand, lncRNAs could be targeted with synthetic siRNAs or miRNAs. Another way to target lncRNAs would be with drugs designed specifically to interact with lncRNAs, as vault RNAs naturally do. Vault RNAs belong to the largest ribonucleoprotein complex in eukaryotic cells (that is, vault), and they are involved in multidrug resistance.182 Gopinath et al. showed that vault RNAs directly bind to chemotherapeutic agents, indicating that it would also be possible to design small molecules that interact with lncRNAs. Of course, vault RNAs are technically short RNAs, ranging from 80 to 90 nucleotides; however, examples of longer RNAs involved with drug interaction exist, such as aptamers.183–186
Targeting transcripts the size of lncRNAs may seem like a daunting task, but there is a precedent for fragmenting large ribonucleoprotein complexes into more manageable sizes. This strategy has been applied in the design of ligands that can bind to expanded rCUG and rCAG repeat RNAs that are expressed in myotonic dystrophy type 1 and interact with the Muscleblind-like 1 protein.187 Moreover, systematic evolution of ligands by exponential enrichment (SELEX) approach can be used to identify chemicals that interact with lncRNAs.160
As well as being potential markers or therapeutic targets, lncRNAs could be used as models to develop novel strategies to target tumor cells. For example, synthetic RNA molecules that form hairpin structures simulating DNA transcription factor-binding elements can be generated to target and regulate transcription factor activity as GAS5 does.164 Synthetic lncRNAs that contain mutant miRNA-binding sites can sequester and reduce expression levels of miRNAs, as it happens in plants.163
Finally, small molecule compounds could be used to target lncRNAs. Indeed, small molecule compounds have already been tested for other uses in clinical trials to determine toxicity, body distribution and pharmacokinetics, and in some cases, their use in humans is already approved by the US Food and Drug Administration. Their use with lncRNAs requires only identifying, either by in silico predictions or by large library screens, the small molecules that target lncRNA or ribonucleoprotein complexes. If such compounds exist, the transition time from lab to clinic would be very short, which would be good news not only for scientists, but especially for patients with cancers and other diseases.
Acknowledgments
RS is supported as a fellow of the TALENTS Programme (7th R&D Framework Programme, Specific Programme: PEOPLE—Marie Curie Actions—COFUND). MIA is supported as a PhD fellow of the FCT (Fundação para a Ciência e Tecnologia), Portugal. GAC is supported as a fellow by The University of Texas MD Anderson Cancer Center Research Trust, as a research scholar by The University of Texas System Regents, and by the Chronic Lymphocytic Leukemia Global Research Foundation. Work in GAC’s laboratory is supported in part by the NIH/NCI (CA135444); a Department of Defense Breast Cancer Idea Award; Developmental Research Awards from the Breast Cancer, Ovarian Cancer, Brain Cancer, Multiple Myeloma and Leukemia Specialized Programs of Research Excellence (SPORE) grants from the National Institutes of Health; a 2009 Seena Magowitz–Pancreatic Cancer Action Network AACR Pilot Grant; the Laura and John Arnold Foundation and the RGK Foundation.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Bakel HNC, Blencowe BJ, Hughes TR. Response to “the reality of pervasive transcription”. PLoS Biol. 2011;9:e1001102. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 7.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washietl S, Findeiss S, Muller SA, Kalkhof S, von Bergen M, Hofacker IL, et al. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA. 2011;17:578–594. doi: 10.1261/rna.2536111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098–1105. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- 13.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 16.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shamovsky I, Nudler E. Gene control by large noncoding RNAs. Sci STKE. 2006;2006:pe40. doi: 10.1126/stke.3552006pe40. [DOI] [PubMed] [Google Scholar]
- 18.Madamanchi NR, Hu ZY, Li F, Horaist C, Moon SK, Patterson C, et al. A noncoding RNA regulates human protease-activated receptor-1 gene during embryogenesis. Biochim Biophys Acta. 2002;1576:237–245. doi: 10.1016/s0167-4781(02)00308-1. [DOI] [PubMed] [Google Scholar]
- 19.Rintala-Maki ND, Sutherland LC. Identification and characterisation of a novel antisense non-coding RNA from the RBM5 gene locus. Gene. 2009;445:7–16. doi: 10.1016/j.gene.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Bai B, Skogerbo G, Cai L, Deng W, Zhang Y, et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33 (Database issue):D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0–an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35(Supplement: Database issue):D178–D182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y, et al. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35(Supplement: Database issue):D145–D148. doi: 10.1093/nar/gkl837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Guan DG, Yang JH, Shao P, Zhou H, Qu LH. ncRNAimprint: a comprehensive database of mammalian imprinted noncoding RNAs. RNA. 2010;16 :1889–1901. doi: 10.1261/rna.2226910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37(Supplement: Database issue):D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39(Supplement: Database issue):D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37(Supplement: Database issue):D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cayre A, Rossignol F, Clottes E, Penault-Llorca F. aHIF but not HIF-1alpha transcript is a poor prognostic marker in human breast cancer. Breast Cancer Res. 2003;5:R223–R230. doi: 10.1186/bcr652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossignol F, Vache C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene. 2002;299:135–140. doi: 10.1016/s0378-1119(02)01049-1. [DOI] [PubMed] [Google Scholar]
- 30.Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript over-expressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 31.Yotova IY, Vlatkovic IM, Pauler FM, Warczok KE, Ambros PF, Oshimura M, et al. Identification of the human homolog of the imprinted mouse Air non-coding RNA. Genomics. 2008;92:464–473. doi: 10.1016/j.ygeno.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He H, Nagy R, Liyanarachchi S, Jiao H, Li W, Suster S, et al. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res. 2009;69:625–631. doi: 10.1158/0008-5472.CAN-08-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tasheva ES, Roufa DJ. Regulation of human RPS14 transcription by intronic antisense RNAs and ribosomal protein S14. Genes Dev. 1995;9:304–316. doi: 10.1101/gad.9.3.304. [DOI] [PubMed] [Google Scholar]
- 35.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korneev SA, Korneeva EI, Lagarkova MA, Kiselev SL, Critchley G, O’Shea M. Novel noncoding antisense RNA transcribed from human anti-NOS2A locus is differentially regulated during neuronal differentiation of embryonic stem cells. RNA. 2008;14:2030–2037. doi: 10.1261/rna.1084308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laverty HG, Wakefield LM, Occleston NL, O’Kane S, Ferguson MW. TGF-beta3 and cancer: a review. Cytokine Growth Factor Rev. 2009;20:305–317. doi: 10.1016/j.cytogfr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potts JD, Vincent EB, Runyan RB, Weeks DL. Sense and antisense TGF beta 3 mRNA levels correlate with cardiac valve induction. Dev Dyn. 1992;193:340–345. doi: 10.1002/aja.1001930407. [DOI] [PubMed] [Google Scholar]
- 40.Davison EJ, Tarpey PS, Fiegler H, Tomlinson IP, Carter NP. Deletion at chromosome band 20p12. 1 in colorectal cancer revealed by high resolution array comparative genomic hybridization. Genes Chromosomes Cancer. 2005;44:384–391. doi: 10.1002/gcc.20252. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Bocker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 42.Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J, et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25:2125–2133. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 43.Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Arai S, Song X, Reichart DD, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 46.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 47.Yochum GS, Cleland R, McWeeney S, Goodman RH. An antisense transcript induced by Wnt/beta-catenin signaling decreases E2F4. J Biol Chem. 2007;282:871–878. doi: 10.1074/jbc.M609391200. [DOI] [PubMed] [Google Scholar]
- 48.Noonan FC, Goodfellow PJ, Staloch LJ, Mutch DG, Simon TC. Antisense transcripts at the EMX2 locus in human and mouse. Genomics. 2003;81:58–66. doi: 10.1016/s0888-7543(02)00023-x. [DOI] [PubMed] [Google Scholar]
- 49.Okamoto J, Hirata T, Chen Z, Zhou HM, Mikami I, Li H, et al. EMX2 is epigenetically silenced and suppresses growth in human lung cancer. Oncogene. 2010;29:5969–5975. doi: 10.1038/onc.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spigoni G, Gedressi C, Mallamaci A. Regulation of Emx2 expression by antisense transcripts in murine cortico-cerebral precursors. PLoS One. 2010;5:e8658. doi: 10.1371/journal.pone.0008658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 52.Alakus H, Warnecke-Eberz U, Bollschweiler E, Monig SP, Vallbohmer D, Brabender J, et al. GNAS1 T393C polymorphism is associated with histopathological response to neoadjuvant radiochemotherapy in esophageal cancer. Pharmacogenomics J. 2009;9:202–207. doi: 10.1038/tpj.2009.5. [DOI] [PubMed] [Google Scholar]
- 53.Frey UH, Fritz A, Rotterdam S, Schmid KW, Potthoff A, Altmeyer P, et al. GNAS1 T393C polymorphism and disease progression in patients with malignant melanoma. Eur J Med Res. 2010;15:422–427. doi: 10.1186/2047-783X-15-10-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayward BE, Bonthron DT. An imprinted antisense transcript at the human GNAS1 locus. Hum Mol Genet. 2000;9:835–841. doi: 10.1093/hmg/9.5.835. [DOI] [PubMed] [Google Scholar]
- 55.Li T, Vu TH, Zeng ZL, Nguyen BT, Hayward BE, Bonthron DT, et al. Tissue-specific expression of antisense and sense transcripts at the imprinted Gnas locus. Genomics. 2000;69:295–304. doi: 10.1006/geno.2000.6337. [DOI] [PubMed] [Google Scholar]
- 56.Yoon AR, Gao R, Kaul Z, Choi IK, Ryu J, Noble JR, et al. MicroRNA-296 is enriched in cancer cells and downregulates p21WAF1 mRNA expression via interaction with its 3′ untranslated region. Nucleic Acids Res. 2011;39:8078–8091. doi: 10.1093/nar/gkr492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abulail R, Hochberg A, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2 :e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31 :350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 59.Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci USA. 2008;105:12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berteaux N, Aptel N, Cathala G, Genton C, Coll J, Daccache A, et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–6745. doi: 10.1128/MCB.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drewell RA, Arney KL, Arima T, Barton SC, Brenton JD, Surani MA. Novel conserved elements upstream of the H19 gene are transcribed and act as mesodermal enhancers. Development. 2002;129:1205–1213. doi: 10.1242/dev.129.5.1205. [DOI] [PubMed] [Google Scholar]
- 62.Li J, Rhodes JC, Askew DS. Evolutionary conservation of putative functional domains in the human homolog of the murine His-1 gene. Gene. 1997;184:169–176. doi: 10.1016/s0378-1119(96)00591-4. [DOI] [PubMed] [Google Scholar]
- 63.Li J, Witte DP, Van Dyke T, Askew DS. Expression of the putative proto-oncogene His-1 in normal and neoplastic tissues. Am J Pathol. 1997;150:1297–1305. [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long non-coding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 66.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 67.Owens BM, Hawley RG. HOX and non-HOX homeobox genes in leukemic hematopoiesis. Stem Cells. 2002;20:364–379. doi: 10.1634/stemcells.20-5-364. [DOI] [PubMed] [Google Scholar]
- 68.Rice KL, Licht JD. HOX deregulation in acute myeloid leukemia. J Clin Invest. 2007;117:865–868. doi: 10.1172/JCI31861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chau YM, Pando S, Taylor HS. HOXA11 silencing and endogenous HOXA11 antisense ribonucleic acid in the uterine endometrium. J Clin Endocrinol Metab. 2002;87:2674–2680. doi: 10.1210/jcem.87.6.8527. [DOI] [PubMed] [Google Scholar]
- 71.Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. 2005;11:531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- 72.Fiegl H, Windbichler G, Mueller-Holzner E, Goebel G, Lechner M, Jacobs IJ, et al. HOXA11 DNA methylation–a novel prognostic biomarker in ovarian cancer. Int J Cancer. 2008;123:725–729. doi: 10.1002/ijc.23563. [DOI] [PubMed] [Google Scholar]
- 73.Potter SS, Branford WW. Evolutionary conservation and tissue-specific processing of Hoxa 11 antisense transcripts. Mamm Genome. 1998;9:799–806. doi: 10.1007/s003359900870. [DOI] [PubMed] [Google Scholar]
- 74.Bedford M, Arman E, Orr-Urtreger A, Lonai P. Analysis of the Hoxd-3 gene: structure and localization of its sense and natural antisense transcripts. DNA Cell Biol. 1995;14:295–304. doi: 10.1089/dna.1995.14.295. [DOI] [PubMed] [Google Scholar]
- 75.Okubo Y, Hamada J, Takahashi Y, Tada M, Tsutsumida A, Furuuchi K, et al. Transduction of HOXD3-antisense into human melanoma cells results in decreased invasive and motile activities. Clin Exp Metastasis. 2002;19:503–511. doi: 10.1023/a:1020346211686. [DOI] [PubMed] [Google Scholar]
- 76.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688–692. doi: 10.1097/meg.0b013e328306a3a2. [DOI] [PubMed] [Google Scholar]
- 78.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 79.Tanaka K, Shiota G, Meguro M, Mitsuya K, Oshimura M, Kawasaki H. Loss of imprinting of long QT intronic transcript 1 in colorectal cancer. Oncology. 2001;60 :268–273. doi: 10.1159/000055328. [DOI] [PubMed] [Google Scholar]
- 80.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 81.Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13. 31 in osteosarcoma. Cancer Res. 2010;70:160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- 82.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 83.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci USA. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 86.Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blin-Wakkach C, Lezot F, Ghoul-Mazgar S, Hotton D, Monteiro S, Teillaud C, et al. Endogenous Msx1 antisense transcript: in vivo and in vitro evidences, structure, and potential involvement in skeleton development in mammals. Proc Natl Acad Sci USA. 2001;98:7336–7341. doi: 10.1073/pnas.131497098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chetcuti A, Aktas S, Mackie N, Ulger C, Toruner G, Alkan M, et al. Expression profiling reveals MSX1 and EphB2 expression correlates with the invasion capacity of Wilms tumors. Pediatr Blood Cancer. 2011;57:950–957. doi: 10.1002/pbc.23003. [DOI] [PubMed] [Google Scholar]
- 89.Revet I, Huizenga G, Chan A, Koster J, Volckmann R, van Sluis P, et al. The MSX1 homeobox transcription factor is a downstream target of PHOX2B and activates the Delta-Notch pathway in neuroblastoma. Exp Cell Res. 2008;314:707–719. doi: 10.1016/j.yexcr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 90.Sliwinski T, Synowiec E, Czarny P, Gomulak P, Forma E, Morawiec Z, et al. The c.469+46_56del mutation in the homeobox MSX1 gene–a novel risk factor in breast cancer? Cancer Epidemiol. 2010;34:652–655. doi: 10.1016/j.canep.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 91.Chan AS, Thorner PS, Squire JA, Zielenska M. Identification of a novel gene NCRMS on chromosome 12q21 with differential expression between rhabdo-myosarcoma subtypes. Oncogene. 2002;21:3029–3037. doi: 10.1038/sj.onc.1205460. [DOI] [PubMed] [Google Scholar]
- 92.Castelnuovo M, Massone S, Tasso R, Fiorino G, Gatti M, Robello M, et al. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. FASEB J. 2010;24:4033–4046. doi: 10.1096/fj.10-157032. [DOI] [PubMed] [Google Scholar]
- 93.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim YS, Hwan JD, Bae S, Bae DH, Shick WA. Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer. 2010;10:576. doi: 10.1186/1471-2407-10-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rivera MN, Kim WJ, Wells J, Stone A, Burger A, Coffman EJ, et al. The tumor suppressor WTX shuttles to the nucleus and modulates WT1 activity. Proc Natl Acad Sci USA. 2009;106:8338–8343. doi: 10.1073/pnas.0811349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR, et al. Expression profiling of EWS/FLI identifies NKX2. 2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9:405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 98.Tochitani S, Hayashizaki Y. Nkx2. 2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun. 2008;372:691–696. doi: 10.1016/j.bbrc.2008.05.127. [DOI] [PubMed] [Google Scholar]
- 99.Mancini M, Toker A. NFAT proteins: emerging roles in cancer progression. Nat Rev Cancer. 2009;9:810–820. doi: 10.1038/nrc2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 101.Jacquot C, Carbonnelle D, Tomasoni C, Papaconstadinou A, Roussis V, Roussakis C. Identification of a novel putative non-coding RNA involved in proliferation arrest of a non-small cell lung carcinoma cell line treated with an original chemical substance, methyl-4-methoxy-3-(3-methyl-2-butanoyl) benzoate. Int J Oncol. 2004;25:519–527. [PubMed] [Google Scholar]
- 102.Malleter M, Jacquot C, Moreau D, Tomasoni C, Tsvetanova M, Chinou I, et al. A novel large regulator RNA, B2, partially overlaps the HEF1/NEDD9/Cas-L gene. Int J Mol Med. 2010;25:897–903. doi: 10.3892/ijmm_00000420. [DOI] [PubMed] [Google Scholar]
- 103.Delgado Andre N, De Lucca FL. Non-coding transcript in T cells (NTT): antisense transcript activates PKR and NF-kappaB in human lymphocytes. Blood Cells Mol Dis. 2008;40:227–232. doi: 10.1016/j.bcmd.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 104.Liu AY, Torchia BS, Migeon BR, Siliciano RF. The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics. 1997;39:171–184. doi: 10.1006/geno.1996.4463. [DOI] [PubMed] [Google Scholar]
- 105.Reisman D, Balint e, Loging WT, Rotter V, Almon E. A novel transcript encoded within the 10-kb first intron of the human p53 tumor suppressor gene (D17S2179E) is induced during differentiation of myeloid leukemia cells. Genomics. 1996;38:364–370. doi: 10.1006/geno.1996.0639. [DOI] [PubMed] [Google Scholar]
- 106.Lee GL, Dobi A, Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nat Rev Urol. 2011;8:123–124. doi: 10.1038/nrurol.2011.10. [DOI] [PubMed] [Google Scholar]
- 107.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tommasi S, Pfeifer GP. In vivo structure of two divergent promoters at the human PCNA locus. Synthesis of antisense RNA and S phase-dependent binding of E2F complexes in intron 1. J Biol Chem. 1999;274:27829–27838. doi: 10.1074/jbc.274.39.27829. [DOI] [PubMed] [Google Scholar]
- 109.Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, et al. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci USA. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38 :7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- 112.Szegedi K, Sonkoly E, Nagy N, Nemeth IB, Bata-Csorgo Z, Kemeny L, et al. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol. 2010;19:269–278. doi: 10.1111/j.1600-0625.2010.01066.x. [DOI] [PubMed] [Google Scholar]
- 113.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Makitie O, Kaitila I. Cartilage-hair hypoplasia–clinical manifestations in 108 Finnish patients. Eur J Pediatr. 1993;152:211–217. doi: 10.1007/BF01956147. [DOI] [PubMed] [Google Scholar]
- 115.Monti L, Cinquetti R, Guffanti A, Nicassio F, Cremona M, Lavorgna G, et al. In silico prediction and experimental validation of natural antisense transcripts in two cancer-associated regions of human chromosome 6. Int J Oncol. 2009;34:1099–1108. doi: 10.3892/ijo_00000237. [DOI] [PubMed] [Google Scholar]
- 116.Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet. 2005;14:1465–1474. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- 117.Colley SM, Leedman PJ. SRA and its binding partners: an expanding role for RNA-binding coregulators in nuclear receptor-mediated gene regulation. Crit Rev Biochem Mol Biol. 2009;44:25–33. doi: 10.1080/10409230802661719. [DOI] [PubMed] [Google Scholar]
- 118.Foulds CE, Tsimelzon A, Long W, Le A, Tsai SY, Tsai MJ, et al. Research resource: expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Mol Endocrinol. 2010;24:1090–1105. doi: 10.1210/me.2009-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cao Y, Bryan TM, Reddel RR. Increased copy number of the TERT and TERC telomerase subunit genes in cancer cells. Cancer Sci. 2008;99:1092–1099. doi: 10.1111/j.1349-7006.2008.00815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 121.Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37:1152–1159. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115:133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nakayama T, Inaba M, Naito S, Mihara Y, Miura S, Taba M, et al. Expression of angiopoietin-1, 2 and 4 and Tie-1 and 2 in gastrointestinal stromal tumor, leiomyoma and schwannoma. World J Gastroenterol. 2007;13:4473–4479. doi: 10.3748/wjg.v13.i33.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rees KA, Singh H, Brindle NP. The receptor tyrosine kinase Tie1 is expressed and activated in epithelial tumour cell lines. Int J Oncol. 2007;31:893–897. [PubMed] [Google Scholar]
- 125.Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, et al. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111:393–405. doi: 10.1016/s0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- 126.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 127.Mishra A, Godavarthi SK, Jana NR. UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol Dis. 2009;36 :26–34. doi: 10.1016/j.nbd.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 128.Numata K, Kohama C, Abe K, Kiyosawa H. Highly parallel SNP genotyping reveals high-resolution landscape of mono-allelic Ube3a expression associated with locus-wide antisense transcription. Nucleic Acids Res. 2011;39:2649–2657. doi: 10.1093/nar/gkq1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 130.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 131.Hancock AL, Brown KW, Moorwood K, Moon H, Holmgren C, Mardikar SH, et al. A CTCF-binding silencer regulates the imprinted genes AWT1 and WT1-AS and exhibits sequential epigenetic defects during Wilms’ tumourigenesis. Hum Mol Genet. 2007;16:343–354. doi: 10.1093/hmg/ddl478. [DOI] [PubMed] [Google Scholar]
- 132.Agrelo R, Wutz A. ConteXt of change–X inactivation and disease. EMBO Mol Med. 2010;2:6–15. doi: 10.1002/emmm.200900053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop–a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nelles L, Van de Putte T, van Grunsven L, Huylebroeck D, Verschueren K. Organization of the mouse Zfhx1b gene encoding the two-handed zinc finger repressor Smad-interacting protein-1. Genomics. 2003;82:460–469. doi: 10.1016/s0888-7543(03)00169-1. [DOI] [PubMed] [Google Scholar]
- 137.Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.De Souza AT, Hankins GR, Washington MK, Orton TC, Jirtle RL. M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nat Genet. 1995;11:447–449. doi: 10.1038/ng1295-447. [DOI] [PubMed] [Google Scholar]
- 139.Oliva J, Bardag-Gorce F, French BA, Li J, French SW. The regulation of non-coding RNA expression in the liver of mice fed DDC. Exp Mol Pathol. 2009;87:12–19. doi: 10.1016/j.yexmp.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kumar MS, Narla A, Nonami A, Mullally A, Dimitrova N, Ball B, et al. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q-syndrome. Blood. 2011;118:4666–4673. doi: 10.1182/blood-2010-12-324715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 142.Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J, et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene. 2010;29:3583–3592. doi: 10.1038/onc.2010.106. [DOI] [PubMed] [Google Scholar]
- 143.Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2011;108:786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 145.Shen H, Lu C, Jiang Y, Tang J, Chen W, Zhang H, et al. Genetic variants in ultraconserved elements and risk of breast cancer in Chinese population. Breast Cancer Res Treat. 2011;128:855–861. doi: 10.1007/s10549-011-1395-4. [DOI] [PubMed] [Google Scholar]
- 146.Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.van Bakel H, Hughes TR. Establishing legitimacy and function in the new transcriptome. Brief Funct Genomic Proteomic. 2009;8:424–436. doi: 10.1093/bfgp/elp037. [DOI] [PubMed] [Google Scholar]
- 148.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 152.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2011;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Vrielink JO, et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Morris KV, Vogt PK. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle. 2010;9:2544–2547. doi: 10.4161/cc.9.13.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–217. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- 156.Kanduri C. Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol. 2011;22:343–350. doi: 10.1016/j.semcdb.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 157.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19 (R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Buck AH, Perot J, Chisholm MA, Kumar DS, Tuddenham L, Cognat V, et al. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010;16:307–315. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 164.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Williams GT, Mourtada-Maarabouni M, Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem Soc Trans. 2011;39:482–486. doi: 10.1042/BST0390482. [DOI] [PubMed] [Google Scholar]
- 166.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 167.Silva JM, Perez DS, Pritchett JR, Halling ML, Tang H, Smith DI. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics. 2010;95:355–362. doi: 10.1016/j.ygeno.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 168.Scaruffi P, Stigliani S, Moretti S, Coco S, De Vecchi C, Valdora F, et al. Transcribed-ultra conserved region expression is associated with outcome in high-risk neuroblastoma. BMC Cancer. 2009;9:441. doi: 10.1186/1471-2407-9-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gibb EA, Enfield KS, Stewart GL, Lonergan KM, Chari R, Ng RT, et al. Long non-coding RNAs are expressed in oral mucosa and altered in oral premalignant lesions. Oral Oncol. 2011 doi: 10.1016/j.oraloncology.2011.07.008. (e-pub ahead of print 9 August 2011) [DOI] [PubMed] [Google Scholar]
- 170.Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 171.Taskinen M, Ranki A, Pukkala E, Jeskanen L, Kaitila I, Makitie O. Extended follow-up of the Finnish cartilage-hair hypoplasia cohort confirms high incidence of non-Hodgkin lymphoma and basal cell carcinoma. Am J Med Genet A. 2008;146A:2370–2375. doi: 10.1002/ajmg.a.32478. [DOI] [PubMed] [Google Scholar]
- 172.Wojcik SE, Rossi S, Shimizu M, Nicoloso MS, Cimmino A, Alder H, et al. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 2010;31:208–215. doi: 10.1093/carcin/bgp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 176.Yang R, Frank B, Hemminki K, Bartram CR, Wappenschmidt B, Sutter C, et al. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 2008;29 :351–355. doi: 10.1093/carcin/bgm290. [DOI] [PubMed] [Google Scholar]
- 177.Lanz RB, Chua SS, Barron N, Soder BM, DeMayo F, O’Malley BW. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol Cell Biol. 2003;23:7163–7176. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 179.Day JR, Jost M, Reynolds MA, Groskopf J, Rittenhouse H. PCA3: from basic molecular science to the clinical lab. Cancer Lett. 2011;301:1–6. doi: 10.1016/j.canlet.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 180.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 182.Gopinath SC, Matsugami A, Katahira M, Kumar PK. Human vault-associated non-coding RNAs bind to mitoxantrone, a chemotherapeutic compound. Nucleic Acids Res. 2005;33:4874–4881. doi: 10.1093/nar/gki809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hanson S, Berthelot K, Fink B, McCarthy JE, Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- 184.Hermann T. Chemical and functional diversity of small molecule ligands for RNA. Biopolymers. 2003;70:4–18. doi: 10.1002/bip.10410. [DOI] [PubMed] [Google Scholar]
- 185.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 186.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 187.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, et al. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]