Abstract
Guanine-rich DNA sequences that can adopt non-Watson-Crick structures in vitro are prevalent in the human genome. Whether such structures normally exist in mammalian cells has, however, been the subject of active research for decades. Here, we show that the G-quadruplex interacting drug pyridostatin promoted growth arrest in human cancer cells via inducing replication- and transcription-dependent DNA damage. Chromatin immunoprecipitation sequence (ChIP-Seq) analysis of the DNA damage marker γH2AX provided the genome-wide distribution of pyridostatin-induced sites of damage, and revealed that pyridostatin targets gene bodies containing clusters of sequences with a propensity for G-quadruplex formation. As a result, pyridostatin modulated the expression of these genes, including the proto-oncogene SRC. We observed that pyridostatin reduced SRC protein levels and SRC-dependent cellular motility in human breast cancer cells, validating SRC as a target. Our unbiased approach to define genomic sites of action for a drug establishes a framework for discovering functional DNA-drug interactions.
Chemical genetics seeks to create small molecule modulators of gene function to elucidate the complex cellular mechanisms that underlie human diseases1-3. The targeting of specific DNA loci to alter the expression of particular genes has been particularly challenging4 owing to the difficulty in identifying well-defined druggable sites. G-quadruplex nucleic acids may provide structural variations in the genome5,6 suitable for selective recognition by small molecules7-9 and have thus emerged as an attractive paradigm for chemical genetics despite the lack of functional evidence for their prevalence in human cells10. For example, drugs that induce dysfunctional telomeres11,12 or perturb ribosome biogenesis13 in human cells have been proposed to act via stabilizing clusters of G-quadruplex motifs at the ends of chromosomes and in regions containing clusters of ribosomal RNA genes, respectively. Furthermore, biologically relevant G-quadruplex motifs have been identified in several organisms14-16 suggesting that functional alternative DNA structures may also occur in human cells. However, a comprehensive genome-wide analysis of these motifs accessible to small molecules has not hitherto been described.
Here we provide an analysis of the genomic targets of the G-quadruplex binding small molecule pyridostatin (1). We found that cells treated with pyridostatin exhibit transcription and replication-dependent DNA damage, resulting in cell cycle arrest. High throughput sequencing and genome-wide analysis of the DNA associated to the DNA damage marker γH2AX demonstrated that pyridostatin targets gene bodies containing clusters of sequences able to adopt a G-quadruplex conformation. Cellular labelling of a pyridostatin analogue to visualize the localization of the small molecule revealed a striking overlap with a human DNA helicase known to exhibit G-quadruplex binding properties, lending support for the existence of functional alternative DNA structures in human cells. Finally, we determined that the proto-oncogene SRC is a target for pyridostatin, providing additional support for the druggability of certain cancer genes by G-quadruplex binding small molecules.
RESULTS
Pyridostatin induces DNA damage and cell cycle arrest
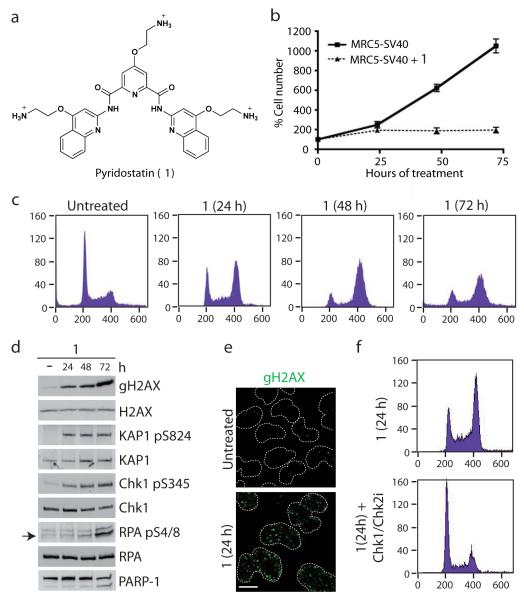
Pyridostatin is a highly selective G-quadruplex binding small molecule (Fig. 1a)17,18 that was designed to target polymorphic G-quadruplex structures regardless of sequence variability9. Consistent with our previous work indicating that pyridostatin exhibits anti-proliferative effects and induces dysfunctional telomeres17, we found that the drug decreased the proliferation of SV40-transformed human MRC5 fibroblasts (Fig. 1b) and various cancer cell lines (Supplementary Results, Supplementary Fig. 1). Moreover, we observed that, upon treatment with pyridostatin, cells predominantly accumulated in the G2 phase of the cell cycle (Fig. 1c) and exhibited markers indicating DNA damage response (DDR) activation19,20, including phosphorylation of histone H2AX on Ser-139 (termed γH2AX), the transcriptional repressor KAP1 (Ser-824), the checkpoint effector kinase Chk1 (Ser-345) and replication protein A (RPA Ser-4/8; Fig. 1d,e). Similar responses were observed in several cancer cell lines. Although treating cells with pyridostatin for 72 hours or longer caused apoptosis in some cells as evidenced by PARP-1 protein cleavage (Fig. 1d), most cells survived long-term pyridostatin incubation. Indeed, even after 10 days of treatment, cells still exhibited DDR signalling. However, a detectable proportion of long-term treated cells were arrested in G1, likely reflecting p21 protein induction at later time points (Supplementary Fig. 3a,b). Regardless of the duration of pyridostatin treatment, pharmacological inhibition of the DNA damage effector kinases Chk1 and Chk2 with AZD7762 (Chk1/Chk2i)21, or inhibition of the apical DNA double strand break (DSB)-sensing kinase ATM with KU55933 (ATMi)22, rapidly triggered the appearance of mitotic cells and the resumption of DNA replication (Fig. 1f and Supplementary Fig. 3c,d). Collectively, these results demonstrated that cell cycle arrest induced by pyridostatin arises primarily through DNA damage checkpoint activation.
Figure 1. Pyridostatin-induced DNA damage and checkpoint-dependent cell cycle arrest.
(a) Molecular structure of 1; trifluoromethanesulfonate counter anions are omitted for clarity. (b) 1 inhibited cell proliferation (N=3; error bars represent S.E.M.). (c) 1 caused cell accumulation in G2; cells were analyzed by FACS with cell count (y-axis) and DNA content (x-axis) as indicated. (d) 1 activated DDR signalling; western blots were performed with the indicated antibodies. Full gel images are displayed in Supplementary Fig. 2. (e) γH2AX foci induced by 1; cells were treated and analyzed by immunofluorescence (IF); dotted white lines indicate nuclear peripheries. (f) G2/M checkpoint-dependent arrest induced by 1; cells were analyzed as in Fig. 1c 2h after addition of Chk1/Chk2i. MRC5-SV40 cells were used throughout all Figures and were either untreated or treated with 2 μM 1 during 24 h unless otherwise stated. Scale bar, 10 μm.
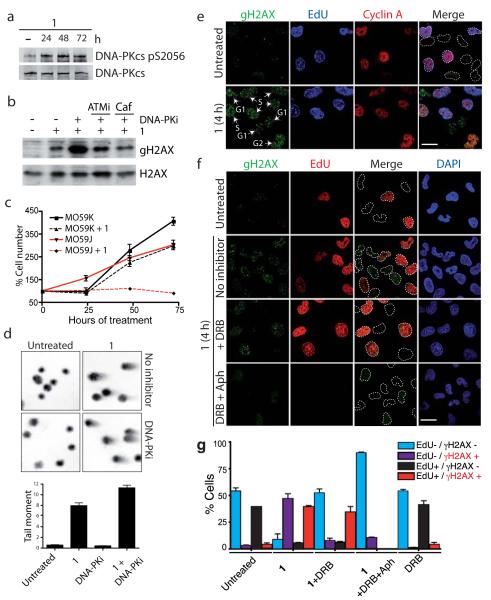
The production of γH2AX and other cellular markers of ATM activation after pyridostatin treatment suggested the induction of DSB. Consistent with this notion, pyridostatin activated the DSB repair protein kinase DNA-PKcs, as revealed by its auto-phosphorylation on Ser-2056 (Fig. 2a). Furthermore, incubating pyridostatin treated cells with the DNA-PKcs inhibitor NU7441 (DNA-PKi)23 markedly enhanced γH2AX production in a manner that was largely prevented when cells were additionally incubated with the ATMi or with caffeine (Caf), which inhibits ATM and the related DNA damage-responsive kinase ATR (Fig. 2b). It is noteworthy that DNA-PKcs inhibition triggered increased γH2AX production after short and long-term pyridostatin treatments (Fig. 2b and Supplementary Fig. 3e), suggesting that DNA-PKcs mediates ongoing DSB repair during exposure to pyridostatin. In agreement with this, DNA-PKcs deficient MO59J cells were considerably more sensitive to pyridostatin treatment than DNA-PKcs proficient MO59K cells (Fig. 2c). Neutral comet assays confirmed the presence of DSB in cells treated with pyridostatin and showed that these were enhanced upon DNA-PKcs inhibition (Fig. 2d).
Figure 2. Pyridostatin-induced transcription- and replication-dependent DNA damage.
(a) 1 activated DNA-PKcs; cells were analyzed as in Fig. 1d. (b) DNA-PKcs inhibition enhanced γH2AX formation in cells treated with 1 in an ATM-dependent manner; cells were pre-treated with the indicated inhibitors for 1 h before treatment with 1. Full gel images are displayed in Supplementary Fig. 4. (c) DNA-PKcs deficient MO59J cells were hypersensitive to 1; analysis was as in Fig. 1b. (d) DSB induced by 1; neutral comet assays were done in the presence or absence of 1 and DNA-PKi, tail moments were determined as described in Methods (N=3; error bars represent S.E.M.). (e) DNA damage in G1, S and G2 phases; cells were treated with 20 μM 1 for 4 h and analyzed as indicated, cell cycle phases are indicated with white arrows and labels, dotted white lines indicate nuclear peripheries; S phase cells are positive for EdU and Cyclin A, G2 cells are positive for Cyclin A only, while G1 cells are negative for EdU and Cyclin A. Scale bar, 20 μm. (f) DNA damage was prevented by transcription (DRB) and replication (Aph) inhibition; cells were pre-treated with inhibitors prior to addition of 20 μM 1 for 4 h; white arrows in cell treated with DRB and 1 indicate G1/G2 cells that are γH2AX-negative; dotted white lines indicate nuclear peripheries. Scale bar, 20 μm. (g) Quantification of cells in experiment Fig. 2f (N=3; >100 cells scored/condition/replica; error bars represent S.E.M; note that Aph prevents EdU incorporation).
Transcription- and replication-dependent DNA damage
To determine whether DSB formation induced by pyridostatin was affected by cell cycle status, we carried out immunofluorescence analyses of pyridostatin treated cells with anti-γH2AX antibodies to detect DNA damage, in conjunction with EdU (5-ethynyl-2′-deoxyuridine) staining to detect DNA replication in S phase, anti-Cyclin A antibodies to detect S and G2 cells, and DAPI (4′,6-diamidino-2-phenylindole) to stain double stranded DNA. We anticipated that this approach would enable a direct comparative analysis of all cell cycle phases simultaneously. Indeed, it revealed that the drug induced the appearance of DNA damage in G1, S and G2 cell cycle phases (Fig. 2e). Strikingly, pre-treating cells with the transcription inhibitor DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) prevented pyridostatin from inducing γH2AX foci only in EdU negative G1 and G2 cells (arrows in Fig. 2f; quantified in g). Moreover, pre-treatment with DRB plus the DNA replication inhibitor aphidicolin (Aph) markedly reduced both the number and intensity of γH2AX foci induced by pyridostatin in all cells (Fig. 2f; quantified in g). These data showed that pyridostatin induces DNA damage in G1 and G2 cells through transcription-dependent mechanisms, while damage in S phase cells also arises through ongoing DNA replication.
Genomic localization of sites of DNA damage
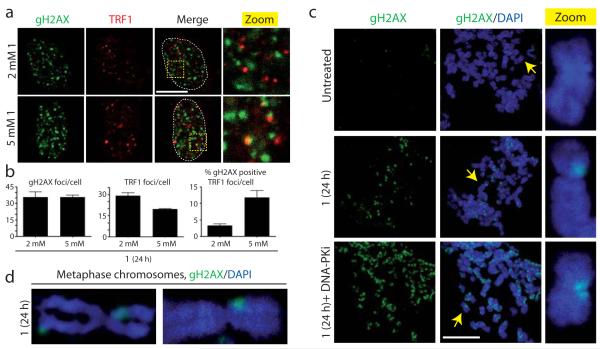
Previous studies have shown that treating cells with G-quadruplex interacting molecules can induce DNA damage signals at telomeres, suggesting the existence of such motifs at the ends of chromosomes11,12. However, we observed that, while relatively low concentrations of pyridostatin were able to inhibit proliferation and induce γH2AX foci in MRC5-SV40 cells, very few of these γH2AX foci co-localized with the telomere binding protein TRF1 (Fig. 3a; quantified in b). In contrast, higher concentrations increased the incidence of γH2AX-positive TRF1 foci and decreased the total numbers of TRF1 foci, thus indicating competition for binding at telomeres. We also found that the total numbers of γH2AX foci per cell did not increase proportionally with increasing concentrations of pyridostatin, suggesting that the drug targets defined DNA sites (Fig. 3b). Taken together, these data indicated that pyridostatin predominantly interacts with non-telomeric DNA loci at low concentrations, before targeting telomeres at higher doses. Indeed, immunofluorescence analyses of mitotic chromosomes following treatment revealed that most sites of γH2AX staining did not localize to chromosome ends (Fig. 3c,d). In line with our other data, DNA-PKcs inhibition increased the number of γH2AX domains on mitotic chromosomes following treatment with pyridostatin.
Figure 3. Visual analysis of pyridostatin targets.
(a) DNA damage signals induced by 1 were mainly non-telomeric; cells were analyzed as in Fig. 1e; areas within dotted lines denote nuclear DNA. Scale bar, 10 μm; zoomed images correspond to 4X magnifications of main images. (b) Quantification of experiment in Fig. 3a. N=3; >100 cells scored/condition/replica; error bars represent S.E.M. (c) γH2AX foci induced by 1 mainly marked non-telomeric sites on mitotic chromosomes; yellow arrows indicate chromosomes corresponding to the magnified images in the zoom column. Scale bar, 20 μm; zoomed images correspond to further 8X magnification (d) Zoomed images of damaged mitotic chromosomes from cells treated with 1 as in (c).
In cellulo chemical labelling of pyridostatin
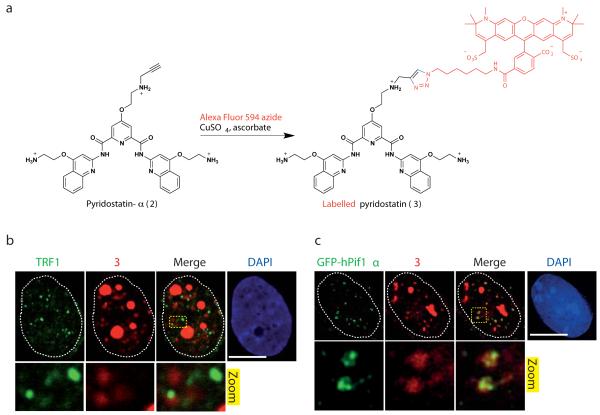
The inability to directly detect most small molecules in cells prompted us to develop a protocol enabling in cellulo covalent labelling of a pyridostatin analogue following treatment. Thus, we synthesized pyridostatin-α (2) that is structurally similar but contains an orthogonal alkyne fragment allowing selective chemical modification in cells via “click chemistry”, as depicted Fig. 4a. The copper-catalyzed alkyne-azide cycloaddition24 was chosen for its bio-compatibility and effectiveness in introducing the fluorophore. By using a well established Fluorescence Resonance Energy Transfer-melting protocol25, we observed that pyridostatin and pyridostatin-α promoted similar melting profiles to one another in vitro for a set of known G-quadruplex DNA motifs (Supplementary Fig. 5)9,18, demonstrating that the introduction of an alkyne fragment did not alter the recognition properties of the drug. Furthermore, pyridostatin-α exhibited growth inhibitory properties on cells and promoted DNA damage to extents that were comparable to those induced by the parent molecule, thus validating the suitability of this compound for this study. Chemical labelling performed on cells treated with 1 μM of pyridostatin-α for 12 h revealed small nuclear foci of fluorescently labelled pyridostatin (3) together with larger fluorescent patterns consistent with staining of nucleoli that contain abundant putative G-quadruplex forming sequences (PQS)6,13. Moreover, a similar staining pattern of labelled pyridostatin was observed when cells were first chemically fixed with formaldehyde to cross-link proteins with nucleic acids and freeze biochemical processes, then incubated with pyridostatin-α followed by chemical labelling (Fig. 4b). Only a few of these foci overlapped with TRF1 staining, which is in agreement with pyridostatin targeting non-telomeric genomic DNA sites. These data thereby pointed towards the existence of pre-folded G-quadruplex structures in human cells since cell fixation was performed prior to drug exposure.
Figure 4. Pyridostatin and hPif1 targeted overlapping sites in cells.
(a) Molecular structure of 2 and synthetic scheme for generating 3 in cells; a single isomer is shown for clarity, Alexa Fluor 594 is marked in red and newly formed chemical bonds are marked in blue. (b) 3 formed nuclear foci mainly at non-telomeric sites in MRC5-SV40 cells, fixed with formaldehyde prior to incubation with 2 followed by chemical labelling; dotted white lines indicate nuclear peripheries; zoomed images correspond to further 4X magnification. (c) GFP-hPif1α expressing U2OS cells display small nuclear foci of GFP-hPif1α that co-localizes with 3 in cells fixed with formaldehyde prior to incubation with 2 followed by chemical labelling; dotted white lines indicate nuclear peripheries; zoomed images correspond to further 5X magnification. Note that cells were first pre-extracted with CSK buffer as described in Supplementary Methods, then fixed with formaldehyde and stained with the indicated antibody. Scale bar, 10 μm.
The Saccharomyces cerevisiae DNA helicase Pif1 binds to and resolves G-quadruplexes during DNA replication26. Genome-wide analyses have revealed a correlation of Pif1 binding to genomic sequences containing PQS and to highly transcribed genes suggesting that Pif1 may also regulate transcription27. Furthermore, human Pif1 (hPif1) was recently shown to exhibit similar biochemical properties28. To establish whether hPif1 associates with pyridostatin at G-quadruplex-containing genomic loci, we developed a U2OS human osteosarcoma cell line that stably expresses the nuclear isoform of hPif1 (hPif1α) fused to a green-fluorescent protein (GFP; see Supplementary Methods) and studied the distribution of the protein as compared to the labelled small molecule by high-resolution microscopy. This revealed that, in the absence of drug treatment, GFP-hPif1α formed small nuclear foci whose pattern was comparable to that observed for the labelled small molecule (Supplementary Fig. 6). Moreover, in an independent experiment where cells were fixed prior to addition of pyridostatin-α, we observed a considerable overlap between the labelled small molecule and GFP-hPif1α foci (Fig. 4c). These data therefore demonstrated that the small molecule pyridostatin and the helicase hPif1 target overlapping genomic structures in human cells, and also indicated that such structures pre-exist prior to drug addition. These experiments thereby provided evidence for the existence of pre-folded G-quadruplex structures at non-telomeric locations within human genomic DNA, and suggested a role for hPif1 in the resolution of these structures in vivo.
ChIP-Seq analyses of sites of DNA damage
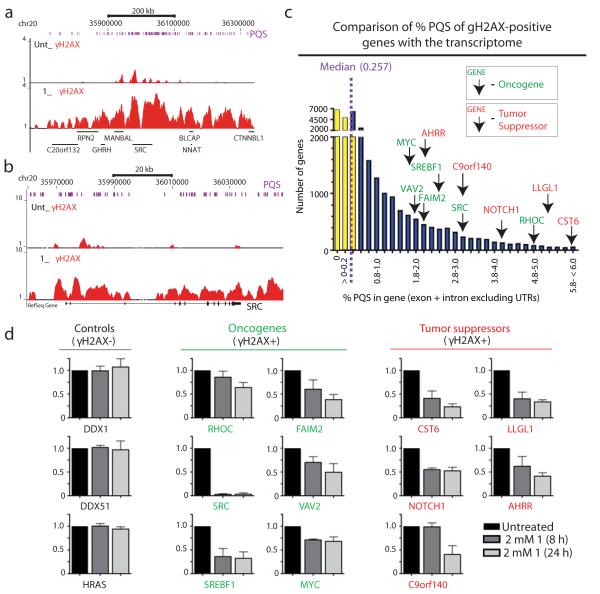
While PQS occur on average once per ten kilobases of the human genome6, with a propensity for them occurring in oncogenes29, structured G-rich sequences that are bona fide targets for pyridostatin are unknown. Our analyses suggested that the small molecule has relatively defined sites of interaction within the human genome; and furthermore, the transcription dependency of γH2AX foci implied that pyridostatin has a propensity for interacting with PQS within certain active genes. To explore these hypotheses, we carried out chromatin immunoprecipitation of γH2AX followed by high throughput sequencing (ChIP-Seq) analyses30,31 of samples obtained from cells treated with the compound and untreated control cells. Comparing the ensuing data sets across the human genome for each individual chromosome identified ~60 γH2AX domains induced by the small molecule. For example, Fig. 5a depicts a domain on chromosome 20, while γH2AX domain distribution across the whole genome is shown in Supplementary Fig. 7. Furthermore, consistent with our other data, while some of the γH2AX domains lay towards chromosomal ends (25%), 75% of them localized to interstitial chromosome regions. Additionally, although γH2AX domains occurred on most chromosomes, further analyses revealed that these domains were enriched on chromosomes that contained higher numbers of mapped PQS than would be expected for their respective sizes (Supplementary Fig. 8 and Supplementary Dataset 1). For example, chromosomes 16, 17, 19 and 20 have high PQS frequencies and displayed higher numbers of γH2AX domains than would have been predicted based on chromosome size.
Figure 5. ChIP-Seq analysis identified genomic targets of pyridostatin.
(a) ChIP-Seq (Supplementary Methods) identified γH2AX regions in oncogenes and tumor suppressors containing PQS clusters; cells were treated with 2 μM 1 during 24h; γH2AX ChIP-Seq of chromosomal region containing SRC is shown (zoomed view in b); chromosome locations and transcripts are shown; purple bars represent mapped PQS (Supplementary Methods). (c) γH2AX-positive oncogenes and tumor suppressors; transcriptome was plotted; purple-dashed line indicates the median % PQS value; yellow bars represent genes below median value and blue bars the genes above median value. Note that all γH2AX-positive genes exhibit higher PQS contents compared to the median value. (d) 1 down-regulated the mRNAs of genes in which it induces γH2AX. Total RNA was purified then reversed transcribed and analyzed by quantitative qRT-PCR for the indicated genes. Expression data for each gene are graphed, with the y-axis indicating expression data for each gene normalized to the housekeeping genes ALAS1 and B2M, and then normalized to untreated samples (N=3; error bars represent S.E.M.). Data from untreated, 8 h and 24 h treated samples are shown for each gene.
To assess the effects of pyridostatin at specific genomic loci, we focused on a gene set comprising 385 designated oncogenes and 763 tumor suppressors32. As anticipated, γH2AX domains were not identifiable with standard peak finding protocols due to the broad coverage of γH2AX signatures33. Consequently, we manually scored the gene set to identify those that displayed γH2AX enrichment in treated versus untreated ChIP-Seq libraries across the entire gene length (Supplementary Dataset 2). For example, Fig. 5b depicts γH2AX enrichment for the proto-oncogene SRC. This analysis identified 25 γH2AX-positive genes from our chosen gene set (Supplementary Fig. 9,10). As shown in Supplementary Fig. 11, γH2AX induction by pyridostatin was further validated by ChIP-qPCR analyses on some genes. We next calculated the percentage of bases that were located within PQS (% PQS in gene) for each individual gene of the human transcriptome (Fig. 5c and Supplementary Dataset 3), which yielded a median value of 0.257%. Markedly, all 25 genes of the gene set that we identified as γH2AX-positive after treatment with pyridostatin exhibited PQS contents that were higher than this median value and contained PQS on both coding and non-coding strands of each target. For example, the SRC and MYC genes contained 12-fold and 7-fold increased PQS levels across their lengths as compared to the median PQS value. It is also noteworthy that in the absence of treatment, the basal level of DNA damage for some of the genes studied showed a good correlation with PQS clustering compared to γH2AX-negative control genes that contained no PQS (Fig 5a,b and Supplementary Fig. 7,9). For these genes, we found that pyridostatin treatment enhanced the pre-existing γH2AX-enrichment at these loci. These results were therefore in agreement with there being G-quadruplex clusters that promote DNA damage in untreated cells, with this effect being amplified upon treatment with the G-quadruplex targeting drug. These data were also in line with our observation that hPif1 and pyridostatin target overlapping genomic sites that contain structured PQS clusters. It is noteworthy that our analyses also identified genes containing PQS clusters that were γH2AX-negative. For example, the HRAS gene exhibited high PQS content with a % PQS value of 9.484, but did not display detectable γH2AX-enrichment in cells treated with pyridostatin. Thus, while there was a good correlation between PQS density and γH2AX formation for particular genes, PQS density alone was not an accurate predictor of DNA damage induction through pyridostatin targeting. This revealed that additional local features of individual loci must contribute to rendering them responsive to pyridostatin.
Pyridostatin alters mRNA levels of damaged genes
Since local DNA damage within a genomic locus can trigger transcriptional inhibition in cis34, we explored whether pyridostatin affected the mRNA levels for MYC and the top ten γH2AX-positive genes that contained the highest PQS densities identified in the above analyses. We also analyzed the housekeeping genes ALAS1 and B2M as controls to normalize gene expression levels since these genes contain low levels of PQS clusters and were γH2AX-negative (Supplementary Fig. 12). Additional γH2AX-negative controls we used were HRAS, DDX1 that contains moderate PQS content, and DDX51 that exhibits a contiguous PQS of over 1400 nucleotides (Supplementary Fig. 12). We found that while the expression levels of control genes were mostly unaffected by pyridostatin, all the γH2AX-positive target genes analyzed were down-regulated after 8 hours of drug treatment (Fig. 5d). Of these, the proto-oncogene SRC was most strongly affected, with its RNA levels being reduced by over 95% after 8 hours of treatment. These data therefore demonstrated a strong correlation between DNA damage induced by the small molecule and transcriptional repression at specific gene loci.
Pyridostatin interacts with G-quadruplexes in SRC
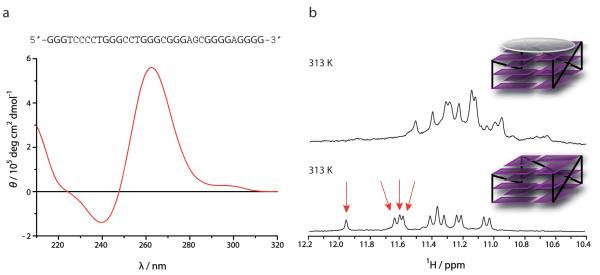
Since SRC responded particularly strongly to pyridostatin treatment, we performed circular dichroism spectroscopy (CD) and nuclear magnetic resonance (NMR) to establish whether individual PQS in this gene adopted stable G-quadruplex conformations in vitro. Out of 25 PQS identified in SRC (Fig. 6a and Supplementary Fig. 13), we observed that 23 of them adopted stable folded structures. As previously shown for other G-quadruplexes14, these sequences displayed a molar ellipticity that is characteristic of G-quadruplex structures, with maxima at 265 nm for parallel conformations, 298 nm for antiparallel conformations, or both patterns highlighting the polymorphic nature of some sequences (Supplementary Fig. 14)9. NMR spectroscopy revealed signals between 10.5 and 12.5 parts per million (ppm), demonstrating the occurrence of Hoogsteen hydrogen bond base-pairing, characteristic of stacked G-quartets that represent the core structure of G-quadruplex motifs (Fig. 6b and Supplementary Fig. 15). Similarly, some sequences displayed 12 distinct imino proton signals, showing the formation of a single species, whereas others exhibited additional signals, highlighting the presence of multiple conformers in agreement with the CD data. We next selected a PQS that formed a single folded species in vitro according to NMR spectroscopy and titrated the structured DNA with pyridostatin that was then monitored by NMR. After incubation of the DNA with 1.1 mole equivalents of the compound, we observed a global line broadening of the signals, associated with an up-field shift of the imino proton signals from 11.0-12.0 ppm to 10.6-11.5 ppm, which was particularly pronounced for four of the most shielded protons (Fig. 6b). Similar results were also observed for other PQS found in SRC. These data demonstrated that pyridostatin interacts selectively with the top G-quartet of the G-quadruplex via a stacking mode as depicted Fig. 6b35. These findings thus provided additional evidence that pyridostatin targets the common structural feature shared by G-quadruplex motifs regardless of the nature of the loop sequences, and highlighted the versatility of this small molecule. Together, the data also showed that G-quadruplex structures are prevalent in SRC, and that pyridostatin strongly interacts with such motifs, thus providing a rationale for the responsiveness of this gene to the drug.
Figure 6. Pyridostatin interacted with G-quadruplex motifs in SRC.
(a) Sequence of a PQS identified in SRC; guanines able to form G-quartet highlighted in bold. Below is a CD spectrum of this sequence pre-annealed in potassium-containing buffer; the molar ellipticity exhibits a positive signal at 265 nm and a negative signal at 240 nm characteristic of a folded parallel G-quadruplex structure (Supplementary Methods). (b) The lower panel displays the NMR spectrum of the free sequence pre-annealed in potassium containing buffer, which exhibits 12 imino proton signals between 11.0 and 12.0 ppm characteristic of a G-quadruplex structure (Supplementary Methods); red arrows indicate proton signals characteristic of the top G-quartet; schematic representation of a free parallel G-quadruplex with loops in black lines and G-quartets in purple. The upper panel displays the NMR spectrum of the G-quadruplex DNA obtained after addition of 1.1 mole equivalents of 1, which exhibits a shift up-field of the imino proton signals compared to untreated sample; schematic representation of 1 (grey) bound to the top G-quartet of the G-quadruplex motif.
Pyridostatin reduces SRC-dependent cell motility
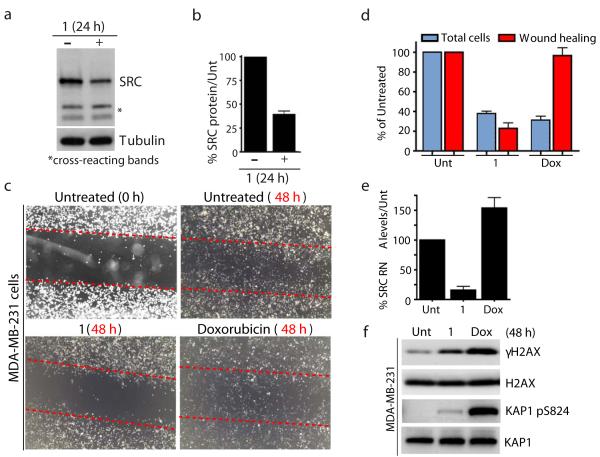
Since SRC mRNA levels were most strongly affected by the small molecule in the above analyses, we sought to confirm this result and to explore potential biological consequences. In line with pyridostatin down-regulating SRC at the mRNA level, we found that SRC protein levels were also reduced by ~60% after 24 hours of treatment in MRC5-SV40 cells (Fig. 7a; quantified in Fig. 7b). SRC is a non-receptor tyrosine kinase that plays critical roles in various cellular processes, including cell motility and invasion36-38. To evaluate whether the compound could affect cellular activities reliant on SRC, we used wound healing assays to analyze SRC-dependent cellular motility in the MDA-MB-231 breast cancer cell line39. This revealed that pyridostatin treatment greatly reduced the motility of MDA-MB-231 cells compared to untreated cells, as detected by pyridostatin strongly impairing the ability of these cells to occupy the wound space (Fig. 7c,d). Importantly, this effect did not simply reflect the ability of the small molecule to induce DNA damage and cell cycle arrest because the topoisomerase poison and DSB inducer doxorubicin (Dox) did not affect wound healing in these experiments, despite pyridostatin and doxorubicin preventing cell proliferation to similar extents (Fig. 7c,d). Furthermore, and consistent with our results with MRC5-SV40 cells, pyridostatin also reduced SRC mRNA levels in MDA-MB-231 cells, while doxorubicin did not (Fig. 7e). As shown in Fig. 7f, this occurred despite the small molecule producing lower levels of DNA damage than doxorubicin in these experiments, as revealed by analyzing the DNA damage marks γH2AX and Ser-824 phosphorylated KAP1. These findings, together with our observation that pyridostatin and doxorubicin inhibited cell growth to similar extents, suggested that pyridostatin affects wound healing at least in part through its targeting of SRC and not through DNA damage induction per se. Collectively, these data demonstrated that this small molecule targets not only SRC but also impairs SRC-dependent cellular processes. Since SRC oncogenic activities can promote tumorigenesis through effects on cellular motility38, these results suggested that pyridostatin, like bona fide SRC inhibitors37, could offer therapeutic opportunities for certain cancers.
Figure 7. Pyridostatin targeted the proto-oncogene SRC.
(a) Treatment with 1 reduced SRC protein levels; cells were analyzed as in Fig. 1d; cross-reacting bands and tubulin provide loading controls. Full gel images are displayed in Supplementary Fig. 16. (b) Quantification of SRC protein levels upon treatment with 1. SRC quantification from 3 independent experiments was performed using LI-COR Odyssey infrared imaging (LI-COR Biosciences) as described in Supplementary Methods. Error bars represent S.E.M. (c) 1 reduced migration of MDA-MB-231 cells. Migration was assessed by wound healing; dotted red lines denote the edges of the wound area; cells were analyzed 48 h after creating the wound for untreated, 1 (2 μM) and doxorubicin (Dox, 100 nM) treatments, images were captured at 20X with a light microscope. (d) Quantification of experiment in Fig. 7c; cells within equal wound areas for each treatment were trypsinized and counted in duplicate dish for each treatment; data are normalized to untreated samples and represent 3 independent experiments (error bars = S.E.M.). (e) 1 reduced SRC mRNA levels in MDA-MB-231 cells. Untreated, and cells treated with 1 (2 μM) and Dox (100 nM) were taken at 24 h and SRC RNA levels determined as in Fig. 5d. (f) 1 and Dox triggered DNA damage in MDA-MB-231 cells. Untreated, and cells treated with 1 (2 μM) and Dox (100 nM) were collected 48 h post-treatment; proteins were analyzed as in Fig. 1d with the indicated antibodies. Full gel images are displayed in Supplementary Fig. 16.
DISCUSSION
This study has identified novel genomic targets of the small molecule pyridostatin through the use of ChIP-Seq, thereby providing a new, unbiased approach that can be employed for identifying druggable targets of other potential therapeutic agents acting at the DNA level. We have shown that pyridostatin generates DNA damage at specific genomic loci, leading to cell cycle arrest and transcriptional down-regulation of several genes that contain PQS clusters on both of their DNA strands. Our data are consistent with the compound mediating these events by interacting with multiple G-quadruplex motifs in gene bodies during transcription and replication, thereby preventing subsequent gene expression from the affected loci. Whether the drug exerts its effects strictly through producing DNA damage and/or acts as a physical barrier to transcription and replication, however, remains to be determined. Recent physical data have shown that pyridostatin stabilizes G-quadruplexes with mechanical forces able to withstand the load forces generated by RNA and DNA polymerases40. In light of this and given the requirement for dynamic events such as transcription and replication for pyridostatin to yield DNA damage, it is possible that pyridostatin stalls polymerases during transcription and replication. These processes could in turn result in DNA breakage through physical forces imposed on the DNA substrate. In addition, DNA damage production in response to the small molecule could be mediated by the action of endonucleases, perhaps through mechanisms linked to transcription-coupled-repair poisoning as shown previously41 for the anticancer drug ecteinascidin 743.
Our use of the copper-catalyzed Huisgen reaction to fluorescently tag the drug in cells has allowed us to evaluate the cellular localization of the drug, thereby highlighting how this novel methodology could be used to trace and assess the distribution of any small molecule in cells. In our system, this approach has provided evidence for G-quadruplex structures naturally occurring in unperturbed human cells. The characteristic staining-pattern we observed for hPif1 in the absence of pyridostatin treatment, and the overlap of hPif1 staining with the labelled small molecule further demonstrated the existence of G-quadruplexes in unperturbed cells, and also implicated hPif1 in resolving these secondary structures that are known to be difficult to transcribe and replicate.
A key finding from our work is that, while pyridostatin can target telomeric loci, its most prevalent sites of genomic interaction are non-telomeric at low concentrations. Although we found that genes containing high PQS contents are more likely to be affected by the small molecule, and despite all the genes affected having higher than average PQS frequencies, not all genes with high PQS levels were demonstrably targeted by the compound in our assays. For example, we did not detect any effect on HRAS expression, even though HRAS contains one of the highest number of PQS of any human gene. Thus, in addition to there being a requirement for alternative DNA structure formation, additional mechanisms must impact on G-quadruplex folding and/or on the binding of the compound to certain G-quadruplexes. For instance, the local supercoiled nature of DNA at a particular locus is likely to modulate the dynamics of G-quadruplex folding and G-quadruplex interactions with pyridostatin, as previously reported42. Since we determined that pyridostatin acts during both transcription and replication, it is tempting to speculate that mechanisms regulating these processes might impact G-quadruplex dynamics and small molecule binding. Indeed, as double stranded DNA becomes transiently open during transcription and replication, PQS are prone to form G-quadruplexes during these processes. Consequently, the rate of transcription or replication through a particular DNA locus and/or changes in chromatin structure triggered by such events could have a marked impact on the ability of PQS in the locus to form G-quadruplex structures that can then be targeted. The propensity of PQS to form G-quadruplex structures could also depend on whether they are on the transcribed or non-transcribed strand of a gene. Similarly, whether a PQS is replicated by leading- or lagging-DNA strand synthesis could affect its propensity to form G-quadruplex structures and pyridostatin targeting43. Our studies have provided a framework upon which future work can generate more accurate predictors of whether or not particular PQS form G-quadruplex structures in vivo and what determines the druggability of these structures in cells.
It will clearly be of interest to explore whether pyridostatin affects the activities of proteins that operate on G-quadruplex structures and, conversely, whether the actions of such proteins influence the targeting of the small molecule to certain PQS regions. In this regard, we note that DNA helicases have been proposed to regulate G-quadruplex formation and processing because these enzymes are known to catalyze the unwinding of duplex DNA. For example, DNA helicases including hPif1, BLM, WRN and FANCJ can unwind G-quadruplex motifs in vitro28,44-46, while the ATR-X helicase interacts with PQS clusters and has been linked to transcriptional regulation of genes containing these sequences47. By establishing a genome-wide map of pyridostatin target sites, our work provides a basis for further defining the molecular mechanisms and consequences of G-quadruplex binding by these and other cellular proteins. Our findings will also facilitate future studies assessing how these enzymes might influence G-quadruplex formation and thereby affect these structures during transcription, replication and potentially DNA damage signalling and repair. Finally, our results highlight the potential druggability of G-quadruplex structures and suggest how pyridostatin, as well as other compounds with similar modes of action, could be exploited as tools for genomic studies and for therapeutic benefit. In particular, the observation that this small molecule can selectively down-regulate the proto-oncogene SRC and induce DNA damage suggests that pyridostatin and its derivatives could exhibit potential as anticancer agents.
METHODS
Chemical synthesis of 1 and 2
Pyridostatin was synthesized as described17. Pyridostatin-α was synthesized as described in Supplementary Methods.
Cell culture, reagents and treatments
Detailed information is provided in Supplementary Methods.
Cell growth assays
Cells were plated at equal confluence and either untreated or treated with 2 μM 1 continually for 72 h. Cells from individual plates were trypsinized and counted in a Coultercounter (Beckman Coulter, Inc). Graphs represent total cell numbers at each time interval and error bars represent S.E.M. Data represent three independent experiments.
Protein extracts and western blotting
Whole cell extracts were prepared and analyzed as described in Supplementary Methods.
Immunofluorescence analyses
Cells were grown on poly-L-lysine-treated coverslips. Coverslips were washed twice with PBS at room temperature (rt). Cells were pre-extracted by incubating coverslips in cold CSK buffer (10 mM PIPES pH 6.8, 100 NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% (v/v) Triton X-100) for 5 min at rt. Cells were washed twice with PBS and fixed with 2% (v/v) formaldehyde (PFA) for 12 min at rt followed by two washes with PBS. For non-extracted samples, cells were fixed with PFA for 12 min and then treated with 0.2% Triton-X100 for 10 min at rt. Primary antibodies were incubated for 1 h at rt in PBS with 5% fetal bovine serum (FBS). Cells were then washed twice with PBS before incubation with Alexa Fluor-conjugated secondary antibodies (Molecular Probes) in PBS + 5% FBS for 30 min at rt. Cells were again washed twice with PBS. Coverslips were then mounted on slides in Vectashield containing DAPI (Vector Laboratories). Cells were imaged with an inverted FV1000 confocal microscope (Olympus). Primary antibodies used were γH2AX (Upstate), TRF1 (Abcam) and Cyclin A (Santa Cruz Biotechnology, Inc). EdU staining was performed using Invitrogen Click-iT Alexa Fluor 594 or 647 imaging kit according to the manufacturer’s instructions (Invitrogen). EdU was pulsed at 10 μM for 10 min before incubation with 1 and for another 10 min before harvesting the cells. In cellulo labelling of 2 was performed with a novel protocol adapted for the Invitrogen Click-iT reagents to yield 3. In a typical experiment, cells were first CSK treated (5 mins), fixed with 2% PFA (12 mins), incubated with 2 in PBS containing 5% FBS (1 h), and Alexa Fluor 594 was clicked using Invitrogen Click-iT reagents. For IF analysis of mitotic spreads, MRC5-SV40 cells were untreated or treated with 2 μM 1 for 24 h in 6 cm dishes. Cells were treated with 0.1 μg/ml colcemid (Gibco) for 1 h followed by incubation with Chk1/Chk2i for 2 h to abrogate the G2/M checkpoint. Cells were then processed and analyzed as described in Supplementary Methods.
Neutral comet assays
MRC5-SV40 cells were analyzed essentially as described48 with detailed information provided in Supplementary Methods.
FRET-melting assay, CD and NMR spectroscopy
Detailed information is provided in Supplementary Methods.
Chromatin immunoprecipitation (ChIP)
ChIP analyses were performed as previously described48.
ChIP-Sequencing and bioinformatics
ChIPed DNA was amplified using an Illumina ChIP-Seq DNA sample prep kit (IP-102-1001) according to the manufacturer’s protocol. Sequences were generated using the Illumina HiSeq 2000 genome analyzer. Resulting 36 bp reads were mapped against the Human genome (GRCh37) with bwa49. Reads with bwa quality scores > 13 were extended to the average library length (300bp). Only one representative read per genomic location (unique reads) was considered. The resulting data was binned to 50 bp regions for display on the UCSC Genome Browser50. PQS were mapped using QuadParser6 with default settings against the GRCh37 release. A bed file was created from this data (source data in Supplementary Dataset 1) for display on the UCSC Genome Browser50. ChIP-Seq libraries of γH2AX were analyzed as described in Supplementary Methods.
RNA analysis
MRC5-SV40 or MDA-MB-231 cells were untreated or treated with 2 μM 1 or 100 nM doxorubicin (Dox) for the indicated times. Total RNA was purified and analyzed by quantitative Real-Time PCR (qRT-PCR) as described in Supplementary Methods. For qRT-PCR analysis, all individual samples were analyzed in duplicate and the mean CT (cycle threshold) values were first normalized to the value of two housekeeping genes, ALAS1 and B2M, and then to the untreated sample. Values are expressed as the geometric mean of both ALAS1 and B2M normalized to the untreated sample. Data from three independent experiments for each time point are graphed and error bars are S.E.M.
Cell migration assays
Cell migration was measured in MDA-MB-231 cells by wound healing assays. Cells were grown to 80% confluency before being streaked with a sterile pipette tip followed by addition of new media containing 2 μM 1 or 100 nM doxorubicin. Representative images were captured 48 hours post-treatment. Cells were counted from an equal area within the wound for each treatment. Data is normalized to untreated cell samples and the graph represents 3 independent experiments with error bars representing S.E.M. Identical dishes for each treatment were trypsinized and counted to determine cell numbers. Cell number and wound healing data are normalized to untreated cell samples and the graph represents data collected from 3 independent experiments with error bars representing S.E.M.
Supplementary Material
Acknowledgements
We thank AstraZeneca for providing the Chk1/Chk2 inhibitor (AZD7762), Dr. S. Müller for technical assistance with FRET-melting experiments and the NCI for screening 1 on a panel of 60 cancer cell lines. R.R. is a Herchel Smith Research Fellow, K.M.M. is funded by a Wellcome Trust project grant 086861/Z/08/Z, and J.V.F. is funded by CRUK program grant C6/A11224. We thank CRUK for programme funding and the BBSRC for project funding (M.N.). S.B. is funded by an EMBO Long-term fellowship (ALTF 93-2010). Research in the S.P.J. laboratory is funded by CRUK program grant C6/A11224, the European Research Council and the European Community Seventh Framework Programme (GENICA and DDResponse). Core infrastructure funding is provided by CRUK and the Wellcome Trust. S.P.J. receives his salary from the University of Cambridge, supplemented by CRUK.
Footnotes
Author Contributions R.R., K.M.M., S.B. and S.P.J. conceptualized the study. R.R., K.M.M. and S.P.J. designed the experiments. R.R. and K.M.M. performed all the experiments unless started otherwise. J.V.F. provided assistance with western blotting and FACS analyses. R.R. invented 1 and 2 and developed the cellular labelling strategy. M.N. performed the chemical synthesis of 2. T.O. and S.B. performed hPif1 experiments. R.R. and M.N. performed CD and NMR spectroscopy. C.R.B. performed computational data analysis. B.X. provided assistance with ChIP-Seq, qRT-PCR, and wound healing assays. All the authors analyzed the data. K.M.M., R.R. and S.P.J. wrote the manuscript.
Competing financial interests The authors declare no competing financial interests.
Additional Information Supplementary information is available online at http://www.nature.com/naturechemicalbiology/. Reprints and permissions information are available online at http://www.nature.com/reprints/index.html.
References
- 1.Dervan PB. Design of sequence-specific DNA-binding molecules. Science. 1986;232:464–71. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- 2.Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nat. Rev. Genet. 2000;1:116–25. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 4.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Regulation of gene expression by small molecules. Nature. 1997;387:202–5. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 5.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–7. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–16. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun D, et al. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–6. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 8.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11593–8. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–55. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller KM, Rodriguez R. G-quadruplexes: selective DNA targeting for cancer therapeutics? Expert Review of Clinical Pharmacology. 2011;4:139–42. doi: 10.1586/ecp.11.4. [DOI] [PubMed] [Google Scholar]
- 11.Gomez D, et al. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J. Biol. Chem. 2006;281:38721–9. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- 12.Salvati E, et al. PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene. 2010;29:6280–93. doi: 10.1038/onc.2010.344. [DOI] [PubMed] [Google Scholar]
- 13.Drygin D, et al. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–61. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 14.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–7. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–54. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 16.Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Mol. Cell. 2010;40:703–13. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez R, et al. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008;130:15758–9. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Müller S, Kumari S, Rodriguez R, Balasubramanian S. Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. 2010;2:1095–8. doi: 10.1038/nchem.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zabludoff SD, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol. Cancer Ther. 2008;7:2955–66. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 22.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 23.Leahy JJ, et al. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004;14:6083–7. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 24.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–9. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Mergny JL, Maurizot JC. Fluorescence resonance energy transfer as a probe for G-quartet formation by a telomeric repeat. Chembiochem. 2001;2:124–32. doi: 10.1002/1439-7633(20010202)2:2<124::AID-CBIC124>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 26.Ribeyre C, et al. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paeschke K, Capra JA, Zakian VA. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell. 2011;145:678–91. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders CM. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 2010;430:119–28. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- 29.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–33. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 32.Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:721–6. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massip L, Caron P, Iacovoni JS, Trouche D, Legube G. Deciphering the chromatin landscape induced around DNA double strand breaks. Cell Cycle. 2010;9:2963–72. doi: 10.4161/cc.9.15.12412. [DOI] [PubMed] [Google Scholar]
- 34.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–81. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavathiotis E, Heald RA, Stevens MF, Searle MS. Recognition and Stabilization of Quadruplex DNA by a Potent New Telomerase Inhibitor: NMR Studies of the 2:1 Complex of a Pentacyclic Methylacridinium Cation with d(TTAGGGT)4. Angew. Chem. Int. Ed. 2001;40:4749–51. doi: 10.1002/1521-3773(20011217)40:24<4749::aid-anie4749>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 36.Martin GS. The hunting of the Src. Nat. Rev. Mol. Cell Biol. 2001;2:467–75. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- 37.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat. Rev. Clin. Oncol. 2009;6:587–95. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 38.Yeatman TJ. A renaissance for SRC. Nat. Rev. Cancer. 2004;4:470–80. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 39.Pichot CS, et al. Dasatinib synergizes with doxorubicin to block growth, migration, and invasion of breast cancer cells. Br. J. Cancer. 2009;101:38–47. doi: 10.1038/sj.bjc.6605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koirala D, et al. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat. Chem. 2011;3:782–787. doi: 10.1038/nchem.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takebayashi Y, et al. Antiproliferative activity of ecteinascidin 743 is dependent upon transcription-coupled nucleotide-excision repair. Nat. Med. 2001;7:961–6. doi: 10.1038/91008. [DOI] [PubMed] [Google Scholar]
- 42.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. Cancer. 2009;9:849–61. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 43.Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002;31:405–9. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 44.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–92. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 45.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 46.London TB, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008;283:36132–9. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Law MJ, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–78. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 48.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010;17:1144–51. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.