Abstract
Nucleotide excision repair (NER) is a precisely coordinated process essential to avoid DNA damage-induced cellular malfunction and mutagenesis. Here, we investigate the mechanistic details and effects of the NER machinery when it is compromised by a pathologically significant mutation in a subunit of the repair/transcription factor TFIIH, namely XPD. In contrast to previous studies, we find that no single- or double-strand DNA breaks are produced at early time points after UV irradiation of cells bearing a specific XPD mutation, despite the presence of a clear histone H2AX phosphorylation (γH2AX) signal in the UV-exposed areas. We show that the observed γH2AX signal can be explained by the presence of longer single-strand gaps possibly generated by strand displacement. Our in vivo measurements also indicate a strongly reduced TFIIH-XPG binding that could promote single-strand displacement at the site of UV lesions. This finding not only highlights the crucial role of XPG’s interactions with TFIIH for proper NER, but also sheds new light on how a faulty DNA repair process can induce extreme genomic instability in human patients.
Keywords: Exo1, nucleotide excision repair, XPD helicase, XPG endonuclease, γH2AX
Introduction
The basal transcription factor TFIIH is composed of 10 proteins organized in two subcomplexes: the core and the trimeric Cdk activating kinase (CAK) complex. The TFIIH core complex is composed of seven proteins, of which XPD, p44 and TTDA are only transiently bound (Santagati et al, 2001; Giglia-Mari et al, 2006), and may therefore freely diffuse to form other complexes with distinct functions (Ito et al, 2010; Weber et al, 2010). The two DNA helicases (XPB and XPD) are responsible for its main activity, which is the opening of the DNA double helix both at promoters during transcription initiation or around helix-distorting DNA lesion within nucleotide excision repair (NER). XPD is also the bridging protein (Sandrock and Egly, 2001) between the core and the CAK complex. The CAK complex catalyses phosphorylation of the C-terminal repeat domain (CTD) of the largest subunit of the RNA polymerase II (RNA Pol II) complex (Feaver et al, 1994). In addition to the 10 canonical TFIIH components, XPG, an endonuclease belonging to the Rad2 nuclease family (Lieber, 1997), forms a complex with TFIIH and helps prevent the dissociation of the CAK from XPD (Ito et al, 2007).
Ultraviolet light induces mainly two DNA lesions: cyclopyrimidine dimers (CPDs) and 6-4 pyrimidine-pyrimidone products (6-4PPs; Douki and Cadet, 1992). The majority of these helix distorting lesions are recognized by the XPC/HRad23B/CENT2 complex in a process referred to as global genome NER (GG-NER; Sugasawa et al, 1998). Alternatively, when such lesions are located on the transcribed strand of active genes, transcription coupled NER (TC-NER) will detect them via the lesion-stalled transcription machinery (Hanawalt, 1994; Fousteri and Mullenders, 2008). In both cases, before incision of the damaged strand can take place (Oksenych and Coin, 2010), recruitment of TFIIH is indispensable for the opening of the double helix around the lesion and its subsequent verification together with XPA (Sugasawa, 2010). Interestingly, the CAK complex does not seem to be required for the GG-NER reaction since it has been found to be released from TFIIH during GG-NER (Coin et al, 2008). After unwinding of the DNA helix, the ERCC1/XPF complex and XPG (two structure-specific endonucleases) incise the damaged strand at the 5′ and 3′ sides of the lesion, respectively. The ERCC1/XPF incision creates a 3′ OH group necessary for the loading of the replication machinery that will copy, in an error-free mode, the undamaged strand of DNA to restore the genetic code. The 3′ side incision by XPG, leaving a 5′ phosphate group, is the optimal substrate for the ligation reaction and completion of repair (Moser et al, 2007; Staresincic et al, 2009).
This precisely orchestrated mechanism is essential to avoid damage-induced cellular malfunction and mutagenesis. Mutations in NER factors cause three genetic diseases: Xeroderma Pigmentosum (XP), Trichothiodystrophy (TTD) and Cockayne Syndrome (CS) (Kraemer et al, 2007). These syndromes are all characterized by a high sensitivity to UV light. Despite this common sensitivity, only XP patients are highly cancer prone, while TTD and CS patients do not develop UV-induced cancers but present premature ageing features that could be caused by the combined misfortune of accumulating unrepaired DNA damage and transcription defects (Bootsma and Hoeijmakers, 1993; Hwang et al, 1996; Coin et al, 1999; de Boer et al, 2002; Keriel et al, 2002). Strikingly, mutations in TFIIH can give rise to different NER disorders: XP, TTD and the combined forms XP/TTD (Broughton et al, 2001) and XP/CS (Dupuy et al, 1974). A recent study conducted on a mouse model mimicking one of the most common XP/CS mutations (G602D) in the XPD protein, clearly shows that these mice (hereafter XpdG602D) are among the most cancer-prone NER-deficient mice ever produced (Andressoo et al, 2006).
It is of general importance to understand the molecular mechanisms that could explain why the XpdG602D mouse model develops UV-induced cancers earlier than, for instance, a completely NER-deficient XPA mouse model (Andressoo et al, 2006). One of the explanations for this remarkable feature was offered by the observation that in XP-D/CS cells a large number of DNA breaks are induced in response to UV irradiation (Berneburg et al, 2000), suggesting that, additionally to unrepaired UV lesions, also single- or double-strand breaks will be created as a consequence of a malfunctioning TFIIH complex. Such additional genomic insults may contribute to increase genomic instability and therefore explain the extraordinary cancer susceptibility of the XpdG602D mouse model. Undoubtedly, the presence of UV-induced DNA breaks can explain the exquisite cellular UV sensitivity of XP-D/CS cells (van Hoffen et al, 1999). Remarkably, despite the strong UV sensitivity, these cells exhibit a rather high level of DNA repair synthesis after UV (unscheduled DNA synthesis (UDS)). It has been suggested that in XP-D/CS cells, the mutated TFIIH cannot be redirected from its transcriptional engagement at promoters to NER sites and that it recruits the NER machinery to promoters where it will induce illegitimate off-site incisions in a transcription-dependent fashion. Although this UV-dependent recruitment at promoters has not been formally demonstrated, the fact that DNA breaks (detected by comet assays) in XP-D/CS cells decrease after transcription inhibition reinforced this hypothesis (Theron et al, 2005). The original off-site incision model could imply the existence of a pan-nuclear transcription-dependent UV-induced DNA break formation in XP-D/CS cells. However, similarly to what has been previously shown (Theron et al, 2005), soon after localized UV irradiation, H2AX phosphorylation (γH2AX, a marker for DNA breaks) is massively present but restricted to the UV-exposed areas (Supplementary Figures S1A and B for H2A ubiquitination). Although this would argue against a pan-nuclear DNA break formation as previously noted (Theron et al, 2005), a localized γH2AX signal in XP-D/CS cells is not a sufficient proof to refute or validate the off-site incision model at promoter sites.
In order to disclose the molecular mechanism behind the cellular phenotype of XP-D/CS cells (extreme UV sensitivity, mild UDS defect, strong γH2AX signalling and formation of UV-induced DNA breaks), we studied the recruitment of several DNA repair factors to the site of UV damage and analysed the dynamics of some key NER factors in XP-D/CS cells during repair and determined the effect of transcription inhibition. In the present study, we show that only a lengthy transcription inhibition abolishes both the recruitment of RPA and the γH2AX signal in all NER-proficient and deficient cell lines tested, including XP-D/CS cells. Moreover, we show that in XP-D/CS cells, more single-strand DNA (ssDNA) is formed and more RPA molecules are loaded at local UV-exposed areas than in NER-proficient cells. Importantly, we found that Exo1 can also be recruited to the same V-exposed areas in XP-D/CS cells. Taken together, our observations suggest that, specifically in XP-D/CS cells, UV irradiation can trigger DNA strand displacements in a transcription-independent manner. We also show that DNA strand displacement could be initiated by the defective cut of the XPG endonuclease, which in the presence of the mutant XPDG602D is less efficiently bound to its protein partners (in the absence of UV) and chromatin (in presence of UV damage). We propose that such unresolved DNA strand displacements may finally lead, at later time points, to the previously documented UV-induced DNA breaks in XP-D/CS cells (Berneburg et al, 2000), thus increasing the level of genomic instability in these TFIIH mutants.
Results
Mobility of hybrid TFIIH complex (XPBYFP with XPDG602D) after DNA damage induction and transcription inhibition
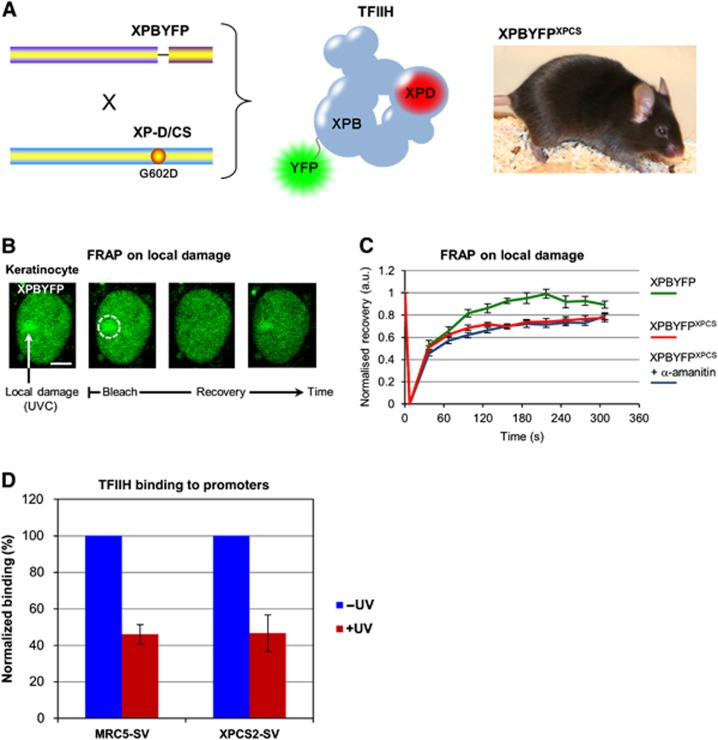
To study the consequences that the XpdG602D mutation exerts on TFIIH binding to damaged DNA, we measured the assembly and dissociation kinetics of TFIIH complexes that contain this mutation (hereafter TFIIHXP/CS). To that aim we crossed a fluorescently tagged TFIIH mouse model that expresses a yellow fluorescent protein (YFP) tagged XPB from the endogenous Xpb allele (XpbY/Y) (Giglia-Mari et al, 2009) with a mouse model expressing an XPD that bears an XP/CS causative mutation (XpdG602D), i.e. a Glycine (position 602) to Aspartic acid substitution (Figure 1A; Dupuy et al, 1974; Andressoo et al, 2006). Subsequent crossings generated mice that express (homozygously) a fluorescent tagged TFIIH with a specific mutation in another subunit: XpbY/Y and XpdG602D. This mouse model (hereafter XpbY/Y·XpdG602D) makes it possible to follow the dynamic behaviour of mutated TFIIH in living cells (different cell lines were isolated from the XpbY/Y·XpdG602D mouse model.
Figure 1.
Transcription inhibition does not influence UV-induced TFIIH recruitment in XP-D/CS cells. (A) Scheme of the murine construction used (left), the fluorescent mutated complex produced in the mouse model (centre) and a picture of a double homozygote mouse XpbY/Y·XpdG602D (noted XPBYFPXPCS) (right). (B) Confocal images of FRAP procedure on a LD area in murine keratinocyte isolated from the XpbY/Y mouse model. Cells were damaged with UV (60 J/m2) through filters to create local XPBYFP accumulations, then XPBYFP mobility was measured by FRAP on the local damages. Scale bar, 5 μm. (C) FRAP on local damage curves of XPBYFP (green) and XPBYFPXPCS mouse keratinocytes treated (blue) or not (red) with a 16-h incubation of α-amanitin before the local UV irradiation. Vertical bars represent the standard error of the mean (s.e.m.). (D) Bar graph showing the average binding of TFIIH on four different active promoter regions, untreated (−UV in blue) and 1/2 h after 20 J/m2 global UVC exposure (+UV in red). Values are normalized to an intergenic region and the vertical bars represent the s.e.m. of the four promoters.
To determine the dynamic behaviour of TFIIHXP/CS complexes during the repair of UV lesions, we locally induced UV damage through a 5-μm pore filter (Mone et al, 2001) and measured via fluorescence recovery after photobleaching (FRAP) (Hoogstraten et al, 2002; Giglia-Mari et al, 2006) the binding properties of both wild-type TFIIH and TFIIHXP/CS complexes. In agreement with previous observations (Andressoo et al, 2006), the TFIIHXP/CS complex efficiently accumulates on locally UV-damaged areas (Figure 1B). After local damage (LD) infliction, the accumulated fluorescent proteins were photo-bleached and recovery of the fluorescent signal was measured in time. FRAP on LD data shows that wild-type TFIIH molecules are transiently bound to the damaged DNA with a complete remobilization of the entire pool of recruited TFIIH within 200 s (Figure 1C, green curve). In contrast, TFIIHXP/CS molecules do not fully recover during the measured time window (maximum recovery of 80%), thus indicating that some of the mutated complexes are immobilized in the damaged area for a longer period of time (Figure 1C, red curve). Because transcription initiation has been proposed to be the cause of the TFIIHXP/CS-specific UV-induced DNA breaks (Theron et al, 2005), we investigated whether the immobile fraction of TFIIHXP/CS molecules measured in a locally damaged area could be affected by transcription inhibition. FRAP measurements on TFIIHXP/CS complexes accumulated on locally damaged areas were hence performed in the presence of the specific RNA Pol II inhibitor α-amanitin (Figure 1C, blue curve). No difference of mobility was detected when transcription was inhibited, suggesting that the prolonged binding of TFIIHXP/CS in regions containing high concentrations of DNA lesions is independent of the transcription mechanism.
To test whether TFIIHXP/CS complexes still remain on promoters after UV irradiation, we conducted chromatin immunoprecipitation (ChIP) experiments on the promoters of four expressed genes in both MRC5 (WT) and XPCS2 (XP-D/CS) cells, prior to and 1/2 h after exposure to UV (see Supplementary Materials and Methods and Supplementary Figure S2). Our results indicate that TFIIH binding to active promoters decreases considerably and equally in both cell types after exposure to UV (Figure 1D), suggesting that the XpdG602D mutation clearly does not lead to abnormally strong or prolonged binding of TFIIHXP/CS complexes to active promoters.
Effect of transcription inhibition on UV-induced DNA damage response
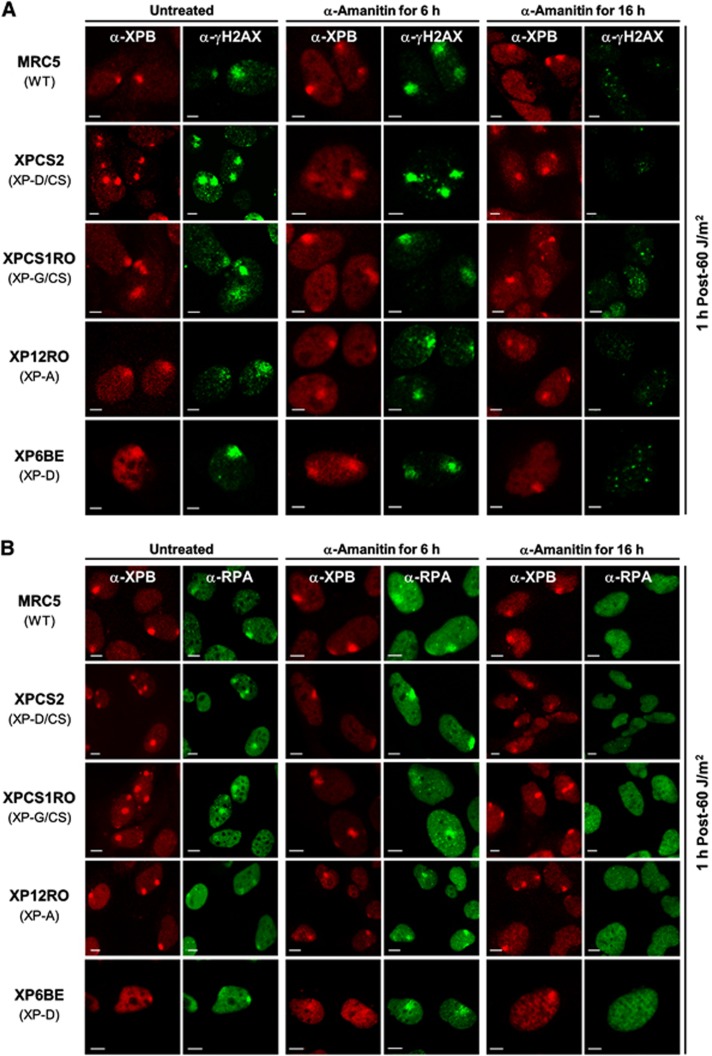
To further investigate the activation of the DNA damage signalling cascade and the recruitment of other NER partners in response to UV irradiation and transcription inhibition, we performed immunofluorescent analysis in presence of the specific RNA Pol II inhibitor, α-amanitin. For this purpose, we tested, on the different cell types used in this study, different α-amanitin concentrations and treatment durations to determine an optimum short-time low-concentration treatment still capable of providing a homogeneous transcription inhibition for all cell lines used (see Supplementary Figure S3 and data not shown). In addition to this optimum treatment condition (20 μg/ml for 6 h), we also used a more classical longer treatment (25 μg/ml for 16 h) in our experiments to conform to what was used in previous studies (up to 50 μg/ml for 24 h; Theron et al, 2005).
We first analysed the phosphorylation of histone-variant H2AX (γH2AX) after UV exposure with or without transcription inhibition by α-amanitin in different cell types, including the XP-D/CS cells. As previously shown (Theron et al, 2005), a strong γH2AX signal is observed in UV-exposed areas in XP-D/CS cells. The same strong γH2AX signal was observed in MRC5, XPCS1RO (XP-G/CS), XP12RO (XP-A) and XP6BE (XP-D) cells (Figure 2A). XP6BE cells bear the same XPD mutation (R683W) as the cells used in a previous study by Theron and collaborators (XP1BR). In XP1BR cells, they saw no γH2AX signal 2 h after global irradiation of cells at 5 J/m2. However, in XP6BE cells a strong γH2Ax signal was observed 1 h after local irradiation of cells at 60 J/m2 (Figure 2A, bottom row).
Figure 2.
Effects of transcription inhibition on H2AX phosphorylation and RPA recruitment to DNA damage are independent of the XP-D/CS mutation. (A) Images of γH2AX (green) and XPB (red) immunostained cells 1 h after local UV irradiation (60 J/m2). Cells were untreated (columns 1 and 2), treated for 6 h with α-amanitin (columns 3 and 4) or for 16 h with α-amanitin (columns 5 and 6) prior to UV exposure. (B) Images of RPA (green) and XPB (red) immunostained cells 1 h after local UV irradiation (60 J/m2). Cells were untreated (columns 1 and 2), treated for 6 h with α-amanitin (columns 3 and 4) or treated for 16 h with α-amanitin (columns 5 and 6) prior to UV exposure. All scale bars, 5 μm.
Remarkably, while short-time transcription inhibition does not affect the XPB recruitment to DNA damage or the γH2AX signal (Figure 2A, columns 3 and 4), long-time treatment strongly reduces the γH2AX signal associated to the early NER processing (Figure 2A, column 6) without affecting XPB recruitment (Figure 2A, column 5). Unexpectedly, our experiments show that significant inhibition of UV-induced γH2AX signalling via long-time transcription inhibition is also observed in wild-type, XP-G/CS, XP-A and XP-D cells (Figure 2A, rows 1, 3, 4 and 5, column 6).
Since γH2AX is a well-known DNA strand break marker, the observed signal might be derived from a secondary response to UV (via blocked transcription or replication forks) that induces single-strand (SSB) or double-strand DNA breaks (DSB). To check this possibility at early time points (up to 1 h after UV exposure), we stained locally UV-irradiated XP-D/CS cells with SSB- and DSB-repair markers, that is, we used the production of poly(ADP-ribose) by PARP-1 to detect SSB (Godon et al, 2008) and the recruitment of Ku80 to detect DSB (Hammel et al, 2010). Both of these markers were not detected (above threshold levels) at the local UV damage in XP-D/CS cells (Supplementary Figure S4A) at both early time points (15 min and 1 h) after UV irradiation, indicating that the strong localized γH2AX signal we have observed in these cells is not primarily due to the presence of large concentrations of SSBs or DSBs in these areas. It is however expected that DNA breaks will appear in greater numbers at later time points. We verified that the antibody staining for Ku80 and PAR functioned in our assays by staining laser-induced SSBs and DSBs in XP-D/CS cells (Supplementary Figure S4B).
To investigate whether H2AX phosphorylation was linked to replication events, we repeated the γH2AX/XPB immunofluorescence experiment in human confluent primary fibroblasts (Supplementary Figure S5A) and obtained similar results (Supplementary Figure S5B) as in SV40-transformed cells, thus demonstrating that H2AX phosphorylation after UV exposure is only NER dependent within the time frame of the experiment.
It has been proposed that the UV-induced γH2AX signal is caused by NER intermediates and likely depends on the formation of the ssDNA that allows binding of RPA and the subsequent ATRIP/ATR recruitment and activation of the latter to trigger this signalling cascade (Matsumoto et al, 2007; Vrouwe et al, 2011). To load RPA, the strand opening function of TFIIH is required. In order to verify that RPA was correctly recruited to the site of damage, (Krasikova et al, 2010), we immunostained for RPA32, one of the RPA subunits. In WT, XP-D/CS, XP-A, XP-G/CS and XP-D cells, RPA was clearly colocalized to UV-damaged areas together with XPB. However, as observed for the γH2AX signal, while short-time transcription inhibition did not affect RPA recruitment in all cell types studied (Figure 2B, columns 3 and 4), the 16-h treatment with α-amanitin hindered the recruitment of RPA into the damaged area (Figure 2B, column 6). To show that RPA accumulation on LD was not due to UV-induced replication fork arrest, we performed the same experiments in confluent primary cell lines and obtained similar results as in transformed cells (Supplementary Figure S6A). These results indicate that RPA accumulation during NER is not transcription dependent, as demonstrated by the absence of any detectable effects after 6 h of α-amanitin treatment (Figure 2B). However, the 16-h long transcription inhibition clearly affects the recruitment of RPA to the LD in all cell lines tested. A possible secondary effect of lengthy transcription inhibition could be the depletion of (undetermined) short-lived factors essential for RPA recruitment or for efficient opening of the double helix around the DNA lesions. Nevertheless, it is important to stress that these observations are seen in all cell types studied, including the XP-D cell line, hence are not XP-D/CS specific.
Incision step is partially functional in XP-D/CS cells
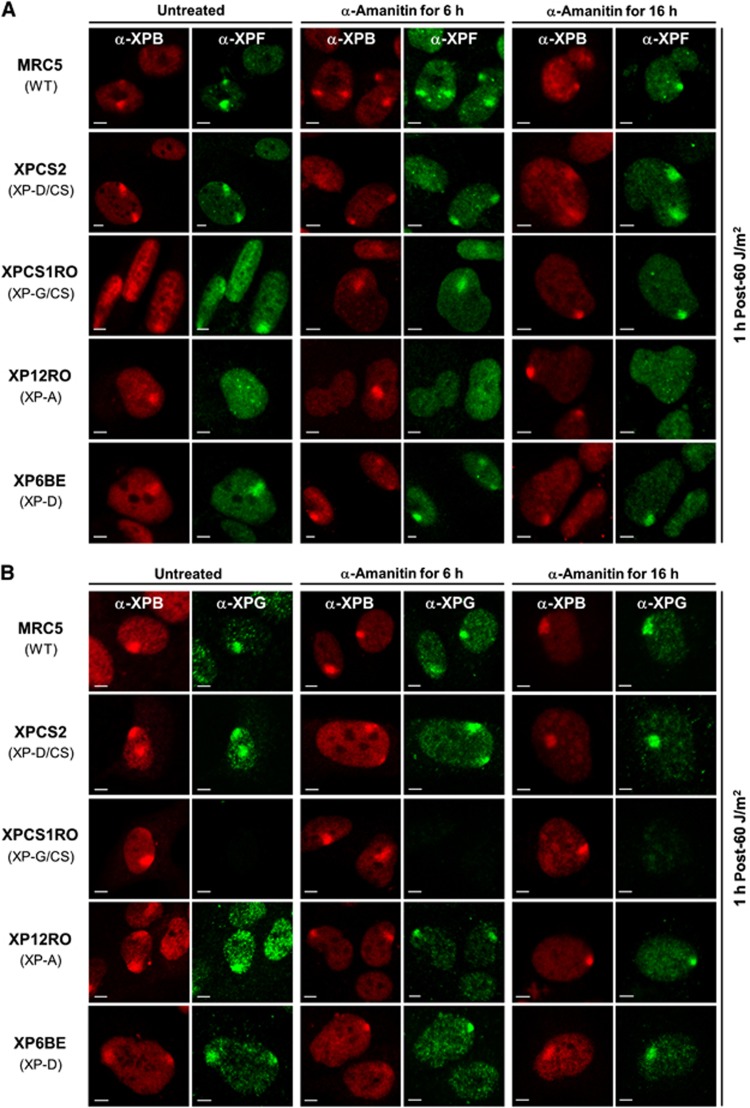
To further dissect the mechanism of aberrant repair in XP-D/CS cells after UV exposure, we studied the recruitment of other NER factors, namely the two nucleases ERCC1/XPF and XPG, which respectively cut the damaged DNA strand at 5′ and 3′ sides of the lesion.
As previously observed (Staresincic et al, 2009), we found that the ERCC1/XPF complex is recruited to locally UV-exposed areas in WT and XP-G/CS cells, but does not accumulate in the absence of XPA (Figure 3A, row 4; Volker et al, 2001). We found that in both XP-D/CS and XP-D cells, the ERCC1/XPF complex is recruited to local UV damage (Figure 3A, column 2, rows 2 and 5). Recruitment of XPF to NER complexes within XP-D/CS cells was confirmed with an adapted ChIP procedure (Supplementary Figure S6B). The XPG nuclease was also recruited to the site of damage in WT, XP-D/CS, XP-A and XP-D cells (Figure 3B, columns 1 and 2). To verify whether transcription inhibition would affect the recruitment of these NER endonucleases, we performed the same experiments in presence of α-amanitin for 6 and 16 h long treatments. As seen in Figures 3A and B (columns 3–6), for all cell lines studied, ERCC1/XPF and XPG recruitment was not affected by the transcription inhibition even after a long-time treatment.
Figure 3.
Transcription inhibition does not affect XPF and XPG recruitment to DNA damage sites. (A) Images of XPF (green) and XPB (red) immunostained cells 1 h after local UV irradiation (60 J/m2). Cells were untreated (columns 1 and 2), treated for 6 h with α-amanitin (columns 3 and 4) or treated for 16 h with α-amanitin (columns 5 and 6) prior to UV exposure. (B) Images of XPG (green) and XPB (red) immunostained cells 1 h after local UV irradiation (60 J/m2). Cells were untreated (columns 1 and 2), treated for 6 h with α-amanitin (columns 3 and 4) or treated for 16 h with α-amanitin (columns 5 and 6) prior to UV exposure. All scale bars, 5 μm.
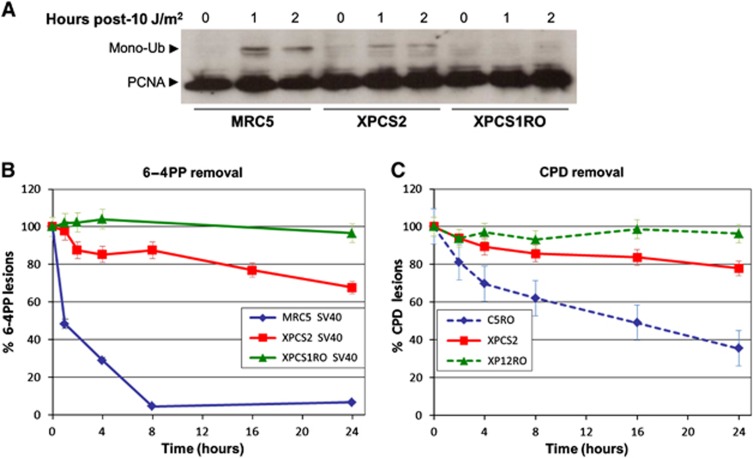
Although the ERCC1/XPF complex is recruited to the site of damage, we wanted to investigate whether its presence translates into an efficient activity, i.e., an incision which creates a 3′ OH. In order to identify such a 3′ OH in XP-D/CS cells, we analysed the presence of mono-Ubiquitinated PCNA (Ub-PCNA). PCNA is mono-ubiquitinated when loaded on the 3′ OH ends (Ogi et al, 2010) during the NER reaction or at later time points during DDR (Vrouwe et al, 2011). This post-translational modification is needed for translesion DNA polymerases to be recruited to UV damage and likely also plays a role in loading DNA polymerase-κ to NER sites (Ogi et al, 2010). As expected, very early after UV irradiation, Ub-PCNA was detected in WT cell lines but not in XP-G/CS negative cells (Figure 4A). This result is consistent with previous studies demonstrating that, in XP-G/CS deficient cells: (i) the endonuclease activity of ERCC1/XPF is abolished (Staresincic et al, 2009) and (ii) PCNA recruitment to the sites of local UV damage is severely affected (Essers et al, 2005). In contrast, for XP-D/CS cells we observed a low (intermediate) PCNA mono-Ub signal, indicating that in the presence of a TFIIHXP/CS complex, ERCC1/XPF incisions are greatly reduced in number but not completely abolished. It is important to stress that we cannot exclude that at later time points, PCNA could also be ubiquitinated in XP-G/CS cells, due to the DDR response to unrepaired damage, as it was shown previously in XPC cells (Vrouwe et al, 2011).
Figure 4.
DNA incision and partial DNA repair occur in XP-D/CS cells in response to UV. (A) Western blot of total cell extracts from MRC5 (WT), XPCS2 (XP-D/CS) and XPCS1RO (XP-G/CS) cells stained with anti-PCNA. Cells were exposed to 10 J/m2 of UV and incubated for 0, 1 or 2 h at 37°C. Arrow indicates the mono-ubiquitinated form of PCNA. (B) 6-4PP removal kinetics via ELISA after global UV exposure (10 J/m2) in SV40-transformed cell lines MRC5 (WT) in blue, XPCS2 (XP-D/CS) in red and XPCS1RO (XP-G/CS) in green. Vertical bars represent the s.e.m. of three independent experiments. (C) CPD removal kinetics via ELISA after global UV exposure (10 J/m2) in human primary cell lines C5RO (WT) in dashed blue, XPCS2 (XP-D/CS) in red and XP12RO (XP-A) in dashed green. Vertical bars represent the s.e.m. of three independent experiments.
It has been proposed that UV lesions are not processed in XP-D/CS cells (van Hoffen et al, 1999) and that the relatively high level of UDS measured in these cells is a consequence of off-site repair reactions (Berneburg et al, 2000). However, because of the recruitment of the two NER endonucleases to the UV-induced LD and the presence of the 3′ OH substrate in XP-D/CS cells, we speculated that perhaps a certain level of lesion removal could be observed in these cells. Hence, we measured the removal of UV-induced 6-4PPs (Figure 4B) and CPDs (Figure 4C) in XP-D/CS cells and compared this to the lesion removal in WT and XP-G/CS or XP-A cells. Within the first 4 h after UV irradiation, 20% of 6-4PP and 10% of CPDs are repaired in XP-D/CS cells, while respectively 70% of 6-4PP and 30% of CPDs are removed in WT cells (MRC5 and C5RO), whereas no removal was observed in XP-G/CS (XPCS1RO) nor in XP-A (XP12RO) cells, as expected (Figures 4B and C). These results show that at least 15% of lesions (20% of 6-4PPs and 10% of CPDs) are actually removed confirming that a certain amount of NER proteins are recruited to UV-lesion sites. Although seemingly in disagreement with previous data, indicating that there is no quantifiable incision activity in XP-D/CS cell extracts on a naked DNA bearing a single cisplatin lesion, our results show that in living cells, in the proper chromatin context, a small proportion of UV lesions are in fact removed by the NER machinery. This would imply that at least some UV-induced lesions will lead to the proper positioning of the NER machinery and the excision of the damaged strand.
RPA accumulation and single-strand formation on UV-induced damages containing a TFIIHXP/CS complex
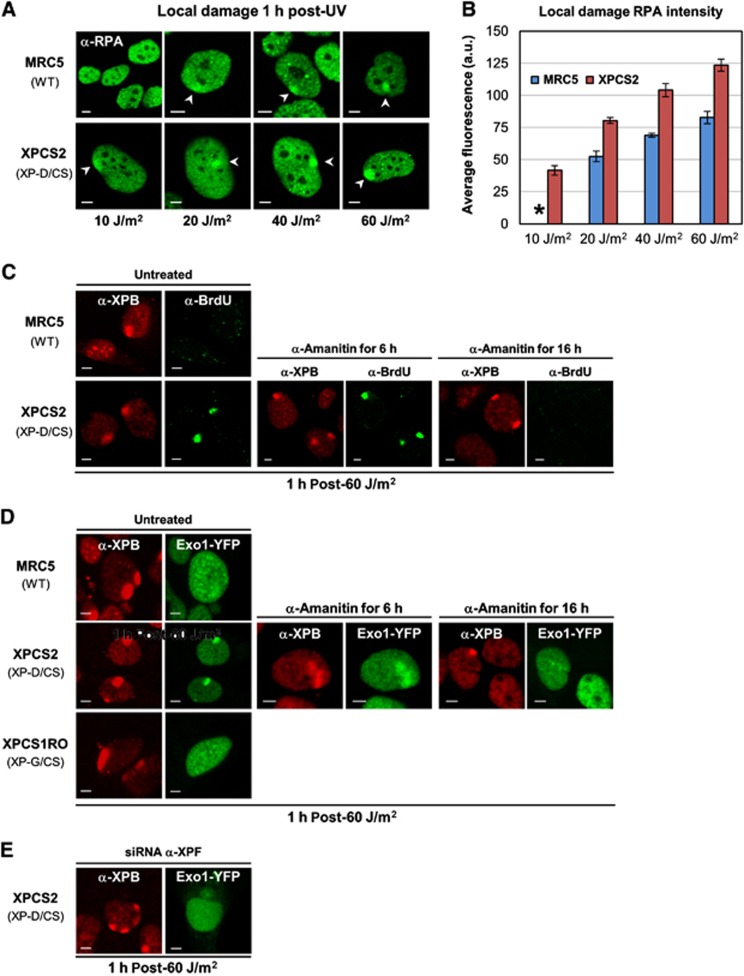
The above results suggest that in contrast with the striking UV sensitivity of XP-D/CS cells, limited UV-lesion removal still occurs. This removal could concur with the partial UDS measured in these cells. To investigate whether incision and damage removal may still take place to some extent via a normal NER reaction (with direct coupling of repair synthesis to 5′ incision (Staresincic et al, 2009) and NER intermediates similar to the WT situation), we measured the amount of RPA being recruited to local UV damage in XP-D/CS cells. To do so, we produced increasing levels of local UV damage in WT and XP-D/CS cells, and estimated (1 h after exposure) the amount of RPA recruitment as a function of the UV dose (Figure 5A) by calculating the average fluorescence intensity of the local RPA signal (Figure 5B). Our results show that at an exposure of 10 J/m2, while RPA recruitment remains essentially undetectable in WT cells, RPA is clearly seen to accumulate in XP-D/CS cells. For higher UV doses, the intensity of RPA accumulation is significantly higher than in WT (MRC5) cells at the corresponding UV dose: a 20-J/m2 (local) irradiation in XP-D/CS cells is similar to a 60 J/m2 exposure in WT cells. Since the RPA accumulation signal increases as a function of the UV dose, that is, as a function of increasing numbers of UV-induced lesions (Figure 5B), these data would indicate that more RPA molecules are recruited per UV lesion in XP-D/CS cells compared to WT cells.
Figure 5.
Single-strand DNA stretches are generated in XP-D/CS cells in response to UV. (A) Confocal images after RPA immunostaining in MRC5 and XPCS2 cells, 1 h after local UV irradiation (10–60 J/m2). (B) Bar graph showing the increasing levels of RPA accumulation in MRC5 (blue) and XPCS2 (red) cells as a function of increasing local UV exposure. The data were obtained by quantifying at least 10 localized RPA accumulations from images as shown in (A). The * at 10 J/m2 indicates an undetectable RPA accumulation signal in MRC5 cells. Vertical bars represent the s.e.m. (C) Confocal images of XPB (red) and BrdU (green) immunofluorescence in MRC5 (WT) and XPCS2 (XP-D/CS) cells, 1 h after local UV exposure (60 J/m2). Cells were either untreated, treated for 6 h with α-amanitin or for 16 h prior to UV exposure (left, middle and right panels, respectively). (D) Confocal images of XPB immunofluorescence (red) and Exo1-YFP (green) in MRC5 (WT), XPCS2 (XP-D/CS), and XPCS1RO (XP-G/CS) cells after local UV exposure (60 J/m2). Cells were either untreated, treated for 6 h with α-amanitin or for 16 h prior to UV exposure (left, middle, and right panels, respectively). (E) Confocal images of XPB immunofluorescence (red) and Exo1-YFP (green) in XPCS2 (XP-D/CS) after XPF depletion by siRNA silencing. All scale bars, 5 μm.
Because more RPA molecules bind to the UV-damaged sites, we hypothesized that also more RPA substrate is created during the faulty NER reaction in XP-D/CS cells as compared to NER-proficient cells. We hence verified whether more ssDNA is available for RPA binding after UV damage in these mutant cells. In order to measure ssDNA, we incubated WT and XP-D/CS cells with BrdU for 48 h and we immunodetected the incorporated BrdU under non-denaturing conditions to quantify the amount of UV-induced open single-strand produced in XP-D/CS cells. Our results (Figure 5C) show that at a local UV dose of 60 J/m2, the NER-dependent single-strand formed is easily detectable in XP-D/CS cells but not in WT cells, indicating that more ssDNA substrate per lesion is formed in these TFIIH mutant cells. In WT cells, NER-dependent ssDNA can be visualized only at higher UV doses (Supplementary Figure S7). Interestingly, while short-time transcription inhibition does not affect ssDNA production (Figure 5C, columns 3 and 4), a 16 h treatment with α-amanitin inhibits the formation of ssDNA in XP-D/CS cells (Figure 5C, columns 5 and 6). These results are in accordance with the inhibition of RPA recruitment under the same transcription inhibition conditions observed in all cell types studied (Figure 2B, columns 5 and 6).
Strand displacement substrate is formed in XP-D/CS cells after UV-damage induction
Thus far, our data seem to present an apparent contradiction, with only 15% of UV lesions being removed in XP-D/CS cells, accompanied with a rather high level of residual UDS (40% compared to WT; Broughton et al, 1995). In view of the fact that more RPA molecules are recruited to locally damaged areas in these cells and more ssDNA is formed, we postulated that faulty UV lesion processing in XP-D/CS cells could induce 5′ incisions (by ERCC1/XPF) that are competent to initiate replication but that the subsequent second incision at the 3′ of the lesion (by XPG) would be hindered. The observed higher amount of repair replication per lesion would predict larger repair patch sizes and DNA repair synthesis measured by UDS in XP-D/CS cells could be derived from strand displacement synthesis beyond the usual XPG incision site. If indeed repair synthesis were initiated in these mutants without a proper second incision, then 5′ flap structures would be formed and be targeted by structure-specific endonucleases (e.g., FEN1) or exonucleases (e.g., Exo1). Recently, it was shown that Exo1 functions in problematic repair reactions of UV lesions, such as clashed NER intermediates when, for example, lesions are located in close proximity (Giannattasio et al, 2010; Sertic et al, 2011). To test whether indeed Exo1 could be involved in the aberrant processing of UV-induced lesions in XP-D/CS cells, we produced stably expressing Exo1-YFP WT, XP-D/CS and XP-G/CS cells. After local UV-damage induction, we observed Exo1 localization specifically in XP-D/CS cells, but not in WT or in XP-G/CS cells (Figure 5D). These data suggest that in the presence of a TFIIHXP/CS complex, after UV irradiation an Exo1 compatible substrate is formed. As expected, Exo1-YFP recruitment was affected when transcription was inhibited for 16 h (Figure 5D, columns 5 and 6) while it was unchanged after the 6-h transcription inhibition (Figure 5D, columns 3 and 4).
To demonstrate that recruitment of Exo1 is dependent on the endonuclease activity of the ERCC1/XPF complex in XP-D/CS cells, we knocked down XPF by RNA interference in these cells (Supplementary Figure S8). Because under these conditions Exo1 accumulation on locally damaged DNA was also abolished (Figure 5E), we confirmed that the ERCC1/XPF endonuclease activity is required to produce the Exo1 substrate in XP-D/CS cells. The Exo1 substrate was also recognized by the flap endonuclease 1 (Fen1, see Supplementary Figure S9).
Together, these results indicate that while transcription inhibition per se does not affect ssDNA production, RPA recruitment and Exo1-substrate formation in XP-D/CS cells, a longer treatment with α-amanitin somehow affects not only RPA recruitment and γH2Ax signal in all cell types but also formation of long ssDNA and Exo1-substrate production which is observed specifically in XP-D/CS cells.
XPDG602D mutation hinders the proper binding of XPG to the TFIIH complex
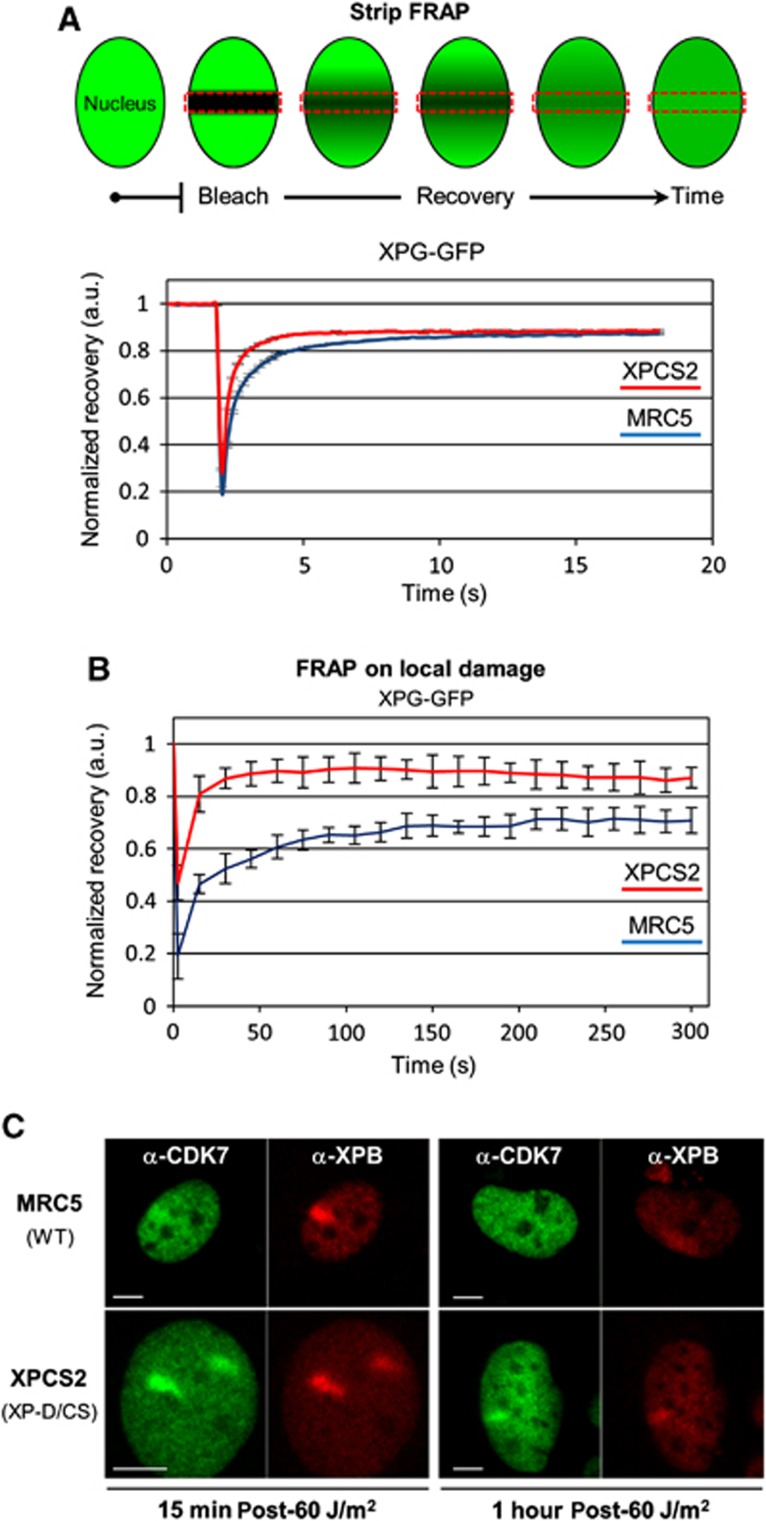
We have shown that after UV, more ssDNA is formed in XP-D/CS cells in comparison to WT cells. This observation would argue for an improper functioning of the XPG endonuclease in these mutant cells. Hence, we hypothesized that the XpdG602D mutation negatively influences the XPG endonuclease activity either by partially inhibiting XPG binding to TFIIH (Ito et al, 2007) or by improper positioning of the endonuclease due to mutation-induced structural changes in TFIIH. In order to verify whether XPDG602D influences the stability of the XPG binding to TFIIH, we produced WT and XP-D/CS cells that stably express XPG fused to the GFP marker (XPG-GFP; Zotter et al, 2006) and measured by FRAP experiments the mobility of XPG-GFP in the absence or presence of DNA damage. Our results show that without any induction of damage, XPG mobility in cells expressing the XpdG602D mutation is increased compared to XPG mobility in WT cells, suggesting that without any damage, the binding of XPG to TFIIH is reduced when XPD is mutated in G602D and that, in comparison with WT cells, more XPG molecules are freely diffusing in XP-D/CS cells (Figure 6A). Interestingly, although XPG recruitment to the locally damaged DNA is not hindered by the presence of the mutated XPDG602D in TFIIH (Figure 3B), the stability of XPG in the chromatin-bound complex is greatly reduced in XP-D/CS cells compared to WT cells (Figure 6B). The estimated XPG-GFP turnover time (TT) on UV lesions in WT cells is around 4 min (TT: 240 s), which corresponds to the average TT (or residence time) of many NER proteins on UV lesions, including XPG (Zotter et al, 2006; Hoogstraten et al, 2008). However, in XP-D/CS cells, XPG-GFP TT in the locally UV-exposed area is greatly reduced (TT: 40 s) demonstrating that the XpdG602D mutation significantly reduces the capacity of XPG to bind properly (or stay bound) to TFIIHXP/CS.
Figure 6.
XpdG602D mutation decreases affinity between XPG and TFIIH. (A) Top panel: Strip-FRAP scheme (see Materials and methods). Bottom panel: Strip-FRAP curves of XPG-GFP protein stably expressed in WT cells (MRC5, blue curve) and XP-D/CS cells (XPCS2, red curve). S.e.m. bars (obtained from >10 cells) are hidden by the plot line thickness. (B) FRAP on local damage curves of XPG-GFP WT cells (MRC5, blue curve) and XP-D/CS cells (XPCS2, red curve). Local damaged areas were obtained after a local UV irradiation of 60 J/m2 through a porous filter. Vertical bars represent the s.e.m. of at least 10 measured cells. (C) Confocal images of XPB (red) and CDK7 (green) immunofluorescence in MRC5 cells (top row) and XPCS2 cells (XP-D/CS) (bottom row), 15 min (left panels) and 1 h (right panels) after local UV exposure (60 J/m2). All scale bars, 5 μm.
Because the binding of XPG to TFIIHXP/CS is hindered, we questioned whether other XPD interacting proteins could be affected by the mutation G602D. One of the interacting partners of XPD within TFIIH is CDK7 (Sandrock and Egly, 2001) which during GG-NER has been shown to be released from TFIIH (Coin et al, 2008). To test whether the G602D mutation could influence the stability of the CAK-TFIIH complex during NER, we performed a series of immunofluorescence assays on WT and XP-D/CS cells, 15 min and 1 h after local UV irradiation (Figure 6C). We found that in WT cells, CDK7 accumulation was barely detectable at 15 min post-UV and not at all 1 h later (Figure 6C, top row), in agreement with previously published in-vitro data (Coin et al, 2008). In contrast, localized CDK7 accumulation was very clear in XP-D/CS cells at both early time points, yet strongest at the 15-min time point (Figure 6C, bottom row). These results suggest that the XpdG602D mutation abnormally retains the CAK subcomplex to the core TFIIH complex during GG-NER.
Discussion
XP-D/CS cells present a peculiar cellular defect in DNA repair which gives rise to a very high UV sensitivity while maintaining a surprisingly high level of repair replication (30–40%) (Broughton et al, 1995). It has been shown that in XP-D/CS cells, UV irradiation triggers the formation of NER-dependent DNA breaks (Berneburg et al, 2000) and a consequent signalling cascade revealed by γH2AX (Theron et al, 2005). In this study, we investigated the upstream molecular mechanism which ultimately leads to the formation of DNA breaks giving rise to the high genomic instability observed in these XP-D/CS cells (Berneburg et al, 2000). To achieve this, we explored in detail the UV-dependent early events in the faulty NER process that is driven by the mutant TFIIHXP/CS complex.
One of the puzzling issues that needed to be initially solved was to know whether the massive early γH2AX signal observed in XP-D/CS cells was due to the presence of UV-induced DNA breaks. Recently, it has been shown that γH2AX signal is not just triggered by DNA breaks but can also be caused by the NER machinery processing itself (Matsumoto et al, 2007; Vrouwe et al, 2011). It has been shown that this signal is associated to TFIIH-operated DNA unwinding around the lesion and to the presence of RPA on the ssDNA formed (Vrouwe et al, 2011). In this study, we showed that also in XP-D/CS cells, the strong γH2AX signal seen at early time points after local UV irradiation is not predominantly due to the presence of concentrated DNA breaks but possibly set off by faulty NER-processing intermediates. However, transcription inhibition-dependent DNA breaks are formed in XP-D/CS cells after exposure to UV and have been observed by comet assays and PAR staining in previous studies (Berneburg et al, 2000; Theron et al, 2005), therefore the question that remains to be answered is: how are DNA breaks formed at ‘later’ time points and what is the relation between their formation and transcription?
Transcription inhibition has been shown to affect DNA break formation in XP-D/CS cells (Theron et al, 2005). In this study, we used two different durations (6 and 16 h) of treatment with the specific RNA Pol II inhibitor α-amanitin to investigate the transcription dependence of UV-induced events in XP-D/CS cells. Having observed that certain cell types are more resistant to α-amanitin inhibition of transcription, we verified that both treatments were sufficient to induce a block of transcription in all the cell types used. Surprisingly, we found that the short-time treatment did not affect the local UV-induced recruitment of key NER factors (XPB, RPA, XPF and XPG) and subsequent γH2AX signalling in all cell types, including XP-D/CS cells. In contrast, regardless of the cell type we studied, the 16-h long transcription inhibition abolished both RPA accumulation and γH2AX, without affecting the recruitment of XPB, XPF and XPG. Specifically in XP-D/CS cells, the abundant UV-induced formation of ssDNA and recruitment of Exo1 to compatible substrates were not affected by the short-time α-amanitin treatment but again abolished under the 16-h treatment.
The striking difference observed between the short and long treatment effects suggests that these events are not transcription dependent per se but transcription inhibition dependent. The exact mechanism that would explain the effect of lengthy transcription inhibition on RPA recruitment and H2AX phosphorylation in all tested cell types, and on ssDNA formation with Exo1 recruitment specifically in XP-D/CS cells, remains unclear. We propose that during extended transcription inhibition mRNA levels will drop significantly which may lead to unwanted secondary effects such as depletion of short-lived factors that facilitate RPA recruitment or DNA unwinding with the formation of suitable RPA substrate.
Although several studies have proposed a general mechanism of action for TFIIHXP/CS complexes (Berneburg et al, 2000; Theron et al, 2005), a clear molecular mechanism has yet to be revealed. We have shown that more RPA accumulates in locally UV-irradiated areas in XP-D/CS compared to WT cells which could imply that more ssDNA is formed after UV irradiation in these cells. Indeed, we detected larger amounts of ssDNA in XP-D/CS cells compared to WT cells. To explain this, we looked into the recruitment and activities of NER factors in XP-D/CS cells. We found that the presence of a TFIIHXP/CS complex does not impede the recruitment of both NER endonucleases (ERCC1/XPF and XPG) and does not abolish the endonuclease activity of ERCC1-XPF. This observation is compatible with our results showing a reduced level of lesion removal (20% for the 6-4PPs and 10% for the CPDs, 16 h after UV irradiation) and can partly explain the 40% UDS measured in XP-D/CS versus WT cells. Moreover, our live cell protein mobility measurements showed that both before and after UV, XPG binding to TFIIHXP/CS is reduced, which may diminish XPG incision efficiency. Concomitantly with this observation, we found that CDK7 is inefficiently released from TFIIHXP/CS after UV exposure. These results are in agreement with XPD structural data (Fan et al, 2008; Liu et al, 2008; Wolski et al, 2008) which predict that the XP-D/CS mutation confers a certain rigidity to XPD, influencing mainly the dynamic protein–protein interactions within TFIIH, such as CDK7, and possibly with closely interacting proteins such as XPG (Ito et al, 2007). It is possible to imagine that XPDG602D would change the conformation of TFIIH in such a way that CDK7 (and probably the CAK) would be more strongly retained to the core, while XPG would on the contrary be less bound to TFIIH.
A reduced XPG activity in XP-D/CS cells could be the trigger for the lengthening of the single-strand substrate and formation of extended single-strand flaps, although repair replication may be the actual driving force in the extension the ssDNA by strand displacement. We detected such 5′ flaps via recruitment of an exogenously expressed Exo1 protein. While Exo1 can recognize and process single-strand 5′ overhang ends, their resolution would most likely not be achieved because of low endogenous Exo1 concentrations (Tishkoff et al, 1998). A scheme recapitulating this proposed molecular mechanism is presented in Figure 7.
Figure 7.
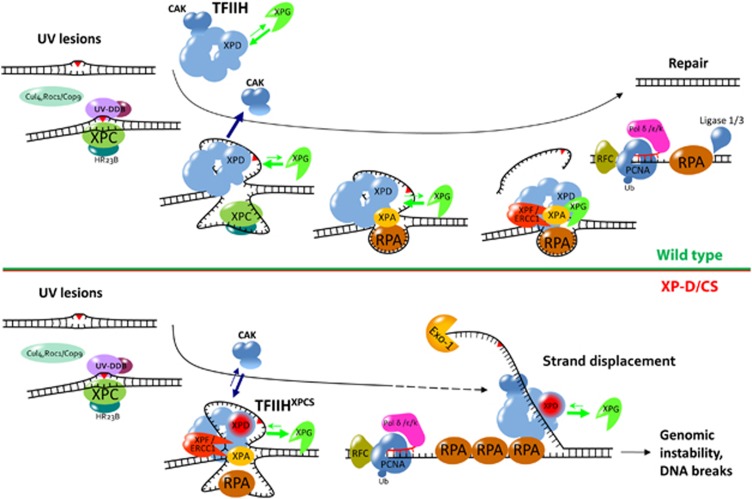
Global genome NER model in wild-type versus XP-D/CS cells. While in WT cells the interaction between TFIIH and XPG allows a normal incision in 3′ of the lesion carrying DNA strand and the release of the CAK complex, in XP-D/CS cells, the weaker affinity between XPG and the TFIIH complex could significantly reduce the rate of successful incisions by the XPG endonuclease. This could then result in a DNA strand displacement mechanism, ultimately responsible for the advent of DNA breaks at later time points and the observed increased genomic instability. Moreover, in the presence of XPDG602D, the CAK subcomplex does not seem to be properly released from TFIIH during GG-NER.
Our mechanistic model is not intrinsically incompatible with the so-called ‘off-site incision’ model (Berneburg et al, 2000; Theron et al, 2005): under similar conditions of transcription inhibition, both models predict the disappearance of the XP-D/CS-specific, UV-induced DNA breaks. However, while the ‘off-site incision’ model is completely transcription dependent, our model does not require transcription. Moreover, this study has provided strong evidence indicating that transcription dependence must be inferred with caution and that transcription inhibition dependence should be considered as an alternative. Our final argument in favour of faulty NER processing of DNA UV lesions in XP-D/CS cells (as opposed to off-site incision at promoters or at transcription sites) comes in the form of a numerical estimate. Indeed, when performing a localized UVC exposure at 60 J/m2, the total number of UV lesions dealt by GG-NER (in the order of 100 000) far exceeds the number of promoters (in the order of 1000 within the locally UV-exposed area) or lesions located on active genes and processed by TC-NER. Consequently, the overwhelming majority of events visualized via NER-related factors, within locally UV-irradiated areas must statistically be associated to (transcription independent) GG-NER processing.
Although, XP-D/CS patients are rare and it is epidemiologically difficult to assess their level of cancer predisposition, the XpdG602D mouse model mimicking the most common patient mutation (Andressoo et al, 2006) is the most skin cancer-prone NER model ever generated. Because of its high sensitivity to damage, this mouse model can be used to score weak carcinogenic potential of chemicals. Disclosing the molecular mechanism underlying this damage sensitivity represents a new step forward into the understanding of NER processing and may help validate the use of the XpdG602D mouse model in drug testing assays.
Materials and methods
Mice models
The XpbY/Y mouse model was created as described previously (Giglia-Mari et al, 2009). A double homozygote XpbY/Y·XpdG602D mouse model was obtained by crossing the XpbY/Y mouse model with an XpdG602D mouse model (Andressoo et al, 2006), mimicking the XP-D/CS patient mutation.
Cell culture
Cells used in this study were SV40 transformed human fibroblasts: MRC5 (WT); XPCS2 (XP-D/CS), XPCS1RO (XP-G/CS), XP12RO (XP-A), XP6BE (XP-D); human primary fibroblast: C5RO (WT), XPCS2 (XP-D/CS), XPCS1RO (XP-G/CS), XP12RO (XP-A); murine keratinocytes: XPBYFP (WT), XPBYFPXPCS (XP-D/CS).
Primary human cell lines were maintained in Ham’s F-10 (Lonza) supplemented with 10% fetal bovine serum (FBS, Fisher Bioblock Scientific) and antibiotics (Lonza), cultured in 3% O2, 5% CO2 at 37°C. The SV40-transformed cell lines were cultivated in 45% DMEM (Lonza), 45% Ham’s F-10, 10% FBS and antibiotics at 37°C and 5% CO2. Mouse keratinocytes were cultivated in CnT-57 (Cell’N Tec), 10% FBS with antibiotics at 37°C and 5% CO2.
Transfection of RNA interference sequences
XPF siRNA oligos were a SmartPool mixture (Dharmacon). Cells were transfected using Lipofectamine 2000 (Invitrogen) transfection reagent according to manufacturer’s instruction. Immunostaining was performed 48 h after transfection. Knockdown efficiencies were confirmed by western blot and/or immunofluorescence.
Plasmids transfection
Cells were grown on 24 mm coverslips. Twenty-four hours after seeding, cells were transfected with JetPEI (PolyPlus) transfection reagent according to manufacturer’s instructions. Cells were then incubated for 48 h. For the co-transfection Exo1-YFP/siRNA α-XPF, the cells were plated on 24 mm coverslips, transfected 8 h later with Exo1-YFP expressing vector and 24 h later with the XPF siRNA SmartPool.
Local UV irradiation and immunofluorescence
SV40-transformed cells were grown on 24 mm coverslips for 24 h before the experiment. Human primary fibroblasts were seeded >2 weeks before the experiment. Coverslips were washed twice with PBS then exposed to UVC light (254 nm, 6 W UVC lamp) through a 5-μm pore polycarbonate membrane filter (Millipore). One hour after irradiation, cells were fixed with 2% paraformaldehyde for 20 min at RT, followed by five washes with PBS 0.1% Triton X-100 (PBST). Fixed cells were treated with a PBS-blocking solution (PBS+: PBS containing 0.15% glycine and 0.5% bovine serum albumin) for 1 h, subsequently incubated with the indicated primary antibodies diluted in PBS+ for 2 h, followed by extensive PBST washes. Cells were then incubated for 1 h with secondary antibodies conjugated with Alexa Fluor 488 and/or 633 fluorescent dyes (Molecular Probes, 1:400 dilution in PBS+) After extensive washing, the coverslips were mounted with Vectashield liquid (Vector) (containing DAPI for Ki67). Images of the cells were acquired with a Zeiss LSM 710 inverted confocal microscope, using a × 40/1.3 objective. Antibodies used for the IF analyses were (i) mouse monoclonal anti-XPG (Upstate, clone 8H7), (ii) mouse monoclonal anti-RPA32/RPA2 (Abcam, clone 9H8), (iii) rabbit polyclonal anti-XPB (p89) (Santa Cruz, clone S19), (iv) rabbit polyclonal anti-ERCC1 (van Vuuren et al, 1993), (v) mouse monoclonal anti-XPF (NeoMarkers, clone 219), (vi) mouse monoclonal anti-γH2AX (Ser139) (Upstate, clone JBW301), (vii) rabbit polyclonal anti-Ki67 (Abcam, ab833), (viii) mouse monoclonal anti-BrdU (Roche, clone BMC9318) and (ix) mouse monoclonal anti CDK7 (Euromedex, clone 2F8). To induce transcription inhibition, cells were incubated in 20 μg/ml α-amanitin for 6 h or 25 μg/ml for 16 h prior to UVC irradiation. To detect ssDNA formation, cells were grown in presence of 20 μm BrdU during 48 h prior the UV local irradiation (60 J/m2).
LD quantification
For the quantification of the RPA LD accumulation signal in cells exposed to UV through a porous filter, we employed a direct approach using the ImageJ software (NIH, Bethesda, MD, USA). Confocal images (LSM 710 confocal laser scanning microscope, Zeiss) of the UV-exposed cells were acquired (all settings identical for all the different conditions and cell lines) with the focal plane set as close as possible to the RPA LD maximum within each cell nucleus. The images were imported into ImageJ and the average RPA fluorescence of the nucleus (F_nucleus) was measured using the polygonal selection tool. In parallel, the average RPA fluorescence signal in the LD area (F_LD) was also measured with the same selection tool. Finally, F_nucleus was subtracted from F_LD to obtain the net fluorescence signal (in arbitrary units) due to RPA accumulation. At least 10 cells were analysed per condition and cell line.
Fluorescence recovery after photobleaching
FRAP experiments were performed on a Zeiss LSM 710 confocal laser scanning microscope (Zeiss), using a × 40/1.3 objective, under a controlled environment (37°C, 5% CO2). A small area of the nucleus was photobleached for 60 ms (100% of 25 mW Argon laser intensity). Recovery of fluorescence was monitored (1% of Argon laser intensity) every 200 ms for 20 s. FRAP data were exported using the ZEN software (Zeiss) and normalized to the average fluorescence measured before the bleach. Every plotted FRAP curve is an average of at least 10 cells.
FRAP on LD
One hour before FRAP experiment, cells were locally damaged with 60 J/m2 UVC through a 5-μm pore filter. All recordings were made at 512 × 512 pixels with the zoom set to 6 and an Argon laser line adapted to the fluorescent protein of interest (488 nm for GFP, 514 nm for YFP). After one prebleach image, a region of interest (ROI) encircling the LD region was photobleached (three iterations at 100% intensity, 25 mW, Argon laser). Recovery of the fluorescence in the ROI was measured by imaging the cell every 10 s for 120–300 s at 1% laser intensity. Analysis was made using the ZEN software (Zeiss). All FRAP data were normalized to the average prebleached ROI fluorescence after removal of the background signal. Every FRAP curve is an average of at least 10 measured cells.
ELISA
Human SV40-transformed cell lines (6-4PP removal) or human primary fibroblasts (CPD removal) were grown in 10 cm dishes; UVC irradiated (10 J/m2) and incubated from 0 to 24 h before being harvested. The DNA of each sample was extracted with the DNA blood mini kit (Qiagen). The 96-well plates were coated two nights consecutively at 37°C with, 0.003% Protamine Sulphate in PBS and, then with denaturated DNA samples (10 min at 95°C) with 15 and 300 ng of DNA, respectively, for the CPDs and 6-4PPs. After five washes with PBS 0.05% Tween-20 (PBST) plates were treated during 30 min at 37°C with PBS 2% FBS then washed five times with PBST and incubated 1 h at 37°C with primary antibodies solutions: anti-6-4PP or anti-CPD (Bioconnect/MBL), followed by extensive PBST washes and a secondary antibody coupled to horseradish peroxidase (SouthernBiotech) incubation for 30 min at 37°C. The signal was revealed with 100 μl of o-Phenylenediamine (Sigma-Aldrich) in Citrate-Phosphate buffer during 30 min at 37°C. The reaction was stopped with 50 μl of 2 M HCl and absorbance was measured at 492 nm with a MicroQuant plate reader.
Western blot
Cells were grown in 10 cm dishes to confluent density. After UVC irradiation (10 J/m2) and incubation from 0 to 2 h, whole cell proteins were extracted with the ProteoJET Mammalian Cell Lysis reagent (Fermentas) according to manufacturer’s instruction in presence of a protease inhibitors cocktail (Roche). Protein concentrations were measured by the Bradford method using the Bio-Rad protein assay. 30 μg of total cell lysates was resolved by 12% SDS–PAGE followed by transfer onto a nitrocellulose membrane. After 1 h blockage at room temperature with PBS and 3% milk, the membrane was incubated with primary antibodies overnight at 4°C and with the secondary antibodies coupled to HRP (Bio-Rad) for 1 h at room temperature. Membranes were probed with an enhanced chemiluminescence reagent (Pierce). PCNA and Ubiquitinated PCNA were detected by mouse monoclonal anti-PCNA antibody (Abcam, PC-10).
ChIP for qPCR analysis
MRC5 and XPCS2 cells were grown in 14.5 cm culture dishes. From both cell lines, one dish was UVC irradiated with 20 J/m2 and incubated under normal conditions for 30 min before fixation. Cells were fixed with 1% formaldehyde (Sigma) in PBS for 10 min at room temperature. Fixed cells were harvested and cell lysis was performed with lysis buffer (50 mM Hepes-KOH pH 8, 1 mM EDTA, 500 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100 and protease inhibitor). Nuclei were resuspended in immunoprecipitation (IP) buffer (50 mM Hepes-KOH pH 7.5, 1 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS and protease inhibitor) and sonicated in a Bioruptor UDC-200 (Diagenode, set-up high for 30 min, with cycles of 30 s on/1 min off) to yield DNA fragments with an average size of 300 bp. In all, 50 μg extract was incubated with 0.5 μg anti-XPB (S-19, Santa Cruz) (ChIP) or no antibody (mock), overnight at 4°C. IP was performed for 1 h at 4°C rotating with Protein G beads (Ademtech). After IP, the beads were washed and proteins were eluted with elution buffer (50 mM Tris–HCl pH 8.0, 10 mM EDTA and 1% SDS) by incubation on a thermomixer (Eppendorf) at 37°C, 1400, r.p.m. DNA from ChIP, mock and input preparations was decrosslinked and purified by phenol-chloroform extraction. Samples were quantified by real-time PCR (qPCR) using the Power SYBR Green PVR master mix (Applied Biosystems) on a 7300 real-time PCR system (Applied Biosystems). ChIP data were depicted as fold enrichment of four different promoters (see Supplementary data) normalized to an intergenic region (Supplementary Figure S2). Final ChIP data were calculated as the average of the four different promoter regions with –UV of respectively MRC5 or XPCS2 set at 100%.
Supplementary Material
Acknowledgments
This work was supported by La Ligue Nationale Contre le Cancer (CG), the Association for International Cancer Research (AICR: 07-0129) (CG, POM), ATIP InCa/CNRS contract no 039438 (POM, GGM), ANR blanc FRETNET (SM) and the foundation RITC (AM). This work also benefited from the TRI Optical Imaging Platform at IPBS (Genotoul, Toulouse, France) supported by grants from the Région Midi-Pyrénées (CPER), the Grand Toulouse community, the ARC (ARC Equipement No 8505), the CNRS and the EU through the FEDER program. We are grateful to Dr J-S Hoffmann and Dr M-J Pillaire for helpful discussions. We would like to thank Drs A Gourdin and LG Rasmussen, respectively, for kindly providing us with the RPA-GFP and Exo1-YFP plasmids.
Author contributions: GGM designed the experiments and supervised the work. CG, SM, JN, AM and FC performed the experiments. GGM, POM and FC analysed the data. WV provided valuable reagents and cell lines. POM and GGM wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andressoo JO, Mitchell JR, de Wit J, Hoogstraten D, Volker M, Toussaint W, Speksnijder E, Beems RB, van Steeg H, Jans J, de Zeeuw CI, Jaspers NG, Raams A, Lehmann AR, Vermeulen W, Hoeijmakers JH, van der Horst GT (2006) An Xpd mouse model for the combined xeroderma pigmentosum/Cockayne syndrome exhibiting both cancer predisposition and segmental progeria. Cancer Cell 10: 121–132 [DOI] [PubMed] [Google Scholar]
- Berneburg M, Lowe JE, Nardo T, Araujo S, Fousteri MI, Green MH, Krutmann J, Wood RD, Stefanini M, Lehmann AR (2000) UV damage causes uncontrolled DNA breakage in cells from patients with combined features of XP-D and Cockayne syndrome. EMBO J 19: 1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootsma D, Hoeijmakers JH (1993) DNA repair. Engagement with transcription. Nature 363: 114–115 [DOI] [PubMed] [Google Scholar]
- Broughton BC, Berneburg M, Fawcett H, Taylor EM, Arlett CF, Nardo T, Stefanini M, Menefee E, Price VH, Queille S, Sarasin A, Bohnert E, Krutmann J, Davidson R, Kraemer KH, Lehmann AR (2001) Two individuals with features of both xeroderma pigmentosum and trichothiodystrophy highlight the complexity of the clinical outcomes of mutations in the XPD gene. Hum Mol Genet 10: 2539–2547 [DOI] [PubMed] [Google Scholar]
- Broughton BC, Thompson AF, Harcourt SA, Vermeulen W, Hoeijmakers JH, Botta E, Stefanini M, King MD, Weber CA, Cole J et al. (1995) Molecular and cellular analysis of the DNA repair defect in a patient in xeroderma pigmentosum complementation group D who has the clinical features of xeroderma pigmentosum and Cockayne syndrome. Am J Hum Genet 56: 167–174 [PMC free article] [PubMed] [Google Scholar]
- Coin F, Bergmann E, Tremeau-Bravard A, Egly JM (1999) Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J 18: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly JM (2008) Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol Cell 31: 9–20 [DOI] [PubMed] [Google Scholar]
- de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, Meradji M, Hoeijmakers JH (2002) Premature aging in mice deficient in DNA repair and transcription. Science 296: 1276–1279 [DOI] [PubMed] [Google Scholar]
- Douki T, Cadet J (1992) Far-UV photochemistry and photosensitization of 2′-deoxycytidylyl-(3′-5′)-thymidine: isolation and characterization of the main photoproducts. J Photochem Photobiol B 15: 199–213 [DOI] [PubMed] [Google Scholar]
- Dupuy JM, Lafforet D, Rachman F (1974) Xeroderma pigmentosum with liver involvement. Helv Paediatr Acta 29: 213–219 [PubMed] [Google Scholar]
- Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W (2005) Nuclear dynamics of PCNA in DNA replication and repair. Mol Cell Biol 25: 9350–9359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA (2008) XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell 133: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD (1994) Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH (2008) Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res 18: 73–84 [DOI] [PubMed] [Google Scholar]
- Giannattasio M, Follonier C, Tourriere H, Puddu F, Lazzaro F, Pasero P, Lopes M, Plevani P, Muzi-Falconi M (2010) Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell 40: 50–62 [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G, Miquel C, Theil AF, Mari PO, Hoogstraten D, Ng JM, Dinant C, Hoeijmakers JH, Vermeulen W (2006) Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biol 4: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Theil AF, Mari PO, Mourgues S, Nonnekens J, Andrieux LO, de Wit J, Miquel C, Wijgers N, Maas A, Fousteri M, Hoeijmakers JH, Vermeulen W (2009) Differentiation driven changes in the dynamic organization of Basal transcription initiation. PLoS Biol 7: e1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, Favaudon V (2008) PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res 36: 4454–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, Tainer JA (2010) Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem 285: 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC (1994) Transcription-coupled repair and human disease. Science 266: 1957–1958 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D, Bergink S, Ng JM, Verbiest VH, Luijsterburg MS, Geverts B, Raams A, Dinant C, Hoeijmakers JH, Vermeulen W, Houtsmuller AB (2008) Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J Cell Sci 121: 2850–2859 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D, Nigg AL, Heath H, Mullenders LH, van Driel R, Hoeijmakers JH, Vermeulen W, Houtsmuller AB (2002) Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol Cell 10: 1163–1174 [DOI] [PubMed] [Google Scholar]
- Hwang JR, Moncollin V, Vermeulen W, Seroz T, van Vuuren H, Hoeijmakers JH, Egly JM (1996) A 3′→5′ XPB helicase defect in repair/transcription factor TFIIH of xeroderma pigmentosum group B affects both DNA repair and transcription. J Biol Chem 271: 15898–15904 [DOI] [PubMed] [Google Scholar]
- Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K (2007) XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell 26: 231–243 [DOI] [PubMed] [Google Scholar]
- Ito S, Tan LJ, Andoh D, Narita T, Seki M, Hirano Y, Narita K, Kuraoka I, Hiraoka Y, Tanaka K (2010) MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell 39: 632–640 [DOI] [PubMed] [Google Scholar]
- Keriel A, Stary A, Sarasin A, Rochette-Egly C, Egly JM (2002) XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell 109: 125–135 [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, Digiovanna JJ (2007) Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship.. Neuroscience 145: 1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasikova YS, Rechkunova NI, Maltseva EA, Petruseva IO, Lavrik OI (2010) Localization of xeroderma pigmentosum group A protein and replication protein A on damaged DNA in nucleotide excision repair. Nucleic Acids Res 38: 8083–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19: 233–240 [DOI] [PubMed] [Google Scholar]
- Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF (2008) Structure of the DNA repair helicase XPD. Cell 133: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, Inobe M, Matsunaga T (2007) Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J Cell Sci 120: 1104–1112 [DOI] [PubMed] [Google Scholar]
- Mone MJ, Volker M, Nikaido O, Mullenders LH, van Zeeland AA, Verschure PJ, Manders EM, van Driel R (2001) Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep 2: 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, Fousteri MI (2007) Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell 27: 311–323 [DOI] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LH, Yamashita S, Fousteri MI, Lehmann AR (2010) Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell 37: 714–727 [DOI] [PubMed] [Google Scholar]
- Oksenych V, Coin F (2010) The long unwinding road: XPB and XPD helicases in damaged DNA opening. Cell Cycle 9: 90–96 [DOI] [PubMed] [Google Scholar]
- Sandrock B, Egly JM (2001) A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J Biol Chem 276: 35328–35333 [DOI] [PubMed] [Google Scholar]
- Santagati F, Botta E, Stefanini M, Pedrini AM (2001) Different dynamics in nuclear entry of subunits of the repair/transcription factor TFIIH. Nucleic Acids Res 29: 1574–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M (2011) Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc Natl Acad Sci USA 108: 13647–13652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Scharer OD (2009) Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J 28: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K (2010) Regulation of damage recognition in mammalian global genomic nucleotide excision repair. Mutat Res 685: 29–37 [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell 2: 223–232 [DOI] [PubMed] [Google Scholar]
- Theron T, Fousteri MI, Volker M, Harries LW, Botta E, Stefanini M, Fujimoto M, Andressoo JO, Mitchell J, Jaspers NG, McDaniel LD, Mullenders LH, Lehmann AR (2005) Transcription-associated breaks in xeroderma pigmentosum group D cells from patients with combined features of xeroderma pigmentosum and Cockayne syndrome. Mol Cell Biol 25: 8368–8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD (1998) Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res 58: 5027–5031 [PubMed] [Google Scholar]
- van Hoffen A, Kalle WH, de Jong-Versteeg A, Lehmann AR, van Zeeland AA, Mullenders LH (1999) Cells from XP-D and XP-D-CS patients exhibit equally inefficient repair of UV-induced damage in transcribed genes but different capacity to recover UV-inhibited transcription. Nucleic Acids Res 27: 2898–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vuuren AJ, Appeldoorn E, Odijk H, Yasui A, Jaspers NG, Bootsma D, Hoeijmakers JH (1993) Evidence for a repair enzyme complex involving ERCC1 and complementing activities of ERCC4, ERCC11 and xeroderma pigmentosum group F. EMBO J 12: 3693–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 8: 213–224 [DOI] [PubMed] [Google Scholar]
- Vrouwe MG, Pines A, Overmeer RM, Hanada K, Mullenders LH (2011) UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J Cell Sci 124: 435–446 [DOI] [PubMed] [Google Scholar]
- Weber A, Chung HJ, Springer E, Heitzmann D, Warth R (2010) The TFIIH subunit p89 (XPB) localizes to the centrosome during mitosis. Cell Oncol 32: 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C (2008) Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol 6: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotter A, Luijsterburg MS, Warmerdam DO, Ibrahim S, Nigg A, van Cappellen WA, Hoeijmakers JH, van Driel R, Vermeulen W, Houtsmuller AB (2006) Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced dna damage depends on functional TFIIH. Mol Cell Biol 26: 8868–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.