Abstract
Mechanisms underlying delays in resolution programs of inflammation are of interest for many diseases. Here, we addressed delayed resolution of inflammation and identified specific microRNA (miR)-metabolipidomic signatures. Delayed resolution initiated by high-dose challenges decreased miR-219-5p expression along with increased leukotriene B4 (5-fold) and decreased (~3-fold) specialized pro-resolving mediators, e.g. protectin D1. Resolvin (Rv)E1 and RvD1 (1 nM) reduced miR-219-5p in human macrophages, not shared by RvD2 or PD1. Since mature miR-219-5p is produced from pre-miRs miR-219-1 and miR-219-2, we co-expressed in human macrophages a 5-lipoxygenase (LOX) 3′UTR-luciferase reporter vector together with either miR-219-1 or miR-219-2. Only miR-219-2 reduced luciferase activity. Apoptotic neutrophils administered into inflamed exudates in vivo increased miR-219-2-3p expression and PD1/NPD1 levels as well as decreased leukotriene B4. These results demonstrate that delayed resolution undermines endogenous resolution programs, altering miR-219-2 expression, increasing pro-inflammatory mediators and compromising SPM production that contribute to failed catabasis and homeostasis.
The acute inflammatory response is protective and evolved to repair injury and eliminate invading organisms (for further details, see ref.1). Ideally it is self-limited and leads to complete resolution of inflammatory infiltrates and clearance of cellular debris so tissues can return to homeostasis, a process historically defined as resolution1,2. Yet, when the magnitude and duration of the inflammatory insult is in excess, chronic inflammation and tissue damage can ensue that further amplifies inflammation1,3,4,5,6. The resolution phase was previously thought to be passive1,7. Evidence now indicates that resolution of inflammation is an active biosynthetic, programmed response, regulated by production and novel actions of the specialized pro-resolving mediators (SPM) as well as specific peptide mediators identified to date (recently reviewed in7,8). During self-limited inflammation, SPM are biosynthesized in resolving exudates from essential fatty acids. These include the lipoxins (LX) from arachidonic acid, as well as resolvins (Rv), protectins (PD) and maresins. These novel families of potent mediators from omega-3 eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) were originally identified in inflammatory exudates7. Each family in the SPM genus possesses distinct chemical structures, and potent bioactive members stereoselectively activate specific G-protein coupled receptors (GPCR). SPM are protective in vivo and act locally to control exudate leukocyte trafficking, pain and enhance efferocytosis (reviewed in7). Recently, intracellular signaling mechanisms of SPM were found to involve microRNAs (miRs)9,10. For example, RvD1 regulates miR-208 in human macrophages in a GPCR-dependent manner, controlling release of the anti-inflammatory cytokine IL-1010.
We introduced and defined, earlier, quantitative resolution indices that take into account temporal cellular trafficking and chemical mediators (i.e. SPM and cytokines/chemokines) within exudates11. These indices define specific actions of endogenous pro-resolving mediators as well as local actions of pharmacologic agents within resolution11,12 and are defined as ψmax, maximal neutrophil numbers that are present in the exudates; Tmax, time when ψmax occurs; and the resolution interval (Ri) from Tmax to T50 when neutrophil numbers reach half ψmax. Only a few of today's widely used pharmacopoeia have been assessed for their impact in programmed resolution11,12. Some delay resolution, e.g. NSAIDs, while others, e.g. resolvins, protectins and aspirin, stimulate resolution, shortening the resolution interval (Ri). Hence, these indices were employed recently to establish a new system to assess the actions of a wide range of drugs and endogenous mediators in resolution using differential challenges of the host with high vs. low doses (e.g. 10 vs. 1 mg) of zymosan13,14.
Mechanisms that underlie self-limited resolution versus its unwanted delay or failure are of considerable interest, because it is now well appreciated that persistent inflammation plays a central role in many diverse diseases including neuroinflammation, cardiovascular, metabolic syndrome as well as the classic inflammation-associated diseases4,7. Along these lines, miRs are involved in regulating the overall immune system15,16,17 and are thus compelling targets for investigating initiation of acute inflammation as well as resolution circuits9,10. Recently, we identified the first miR signature linked to resolution employing self-limited acute inflammation. These include miR-219-5p, miR-208, miR-146b, and miR-21, which each regulate effector molecules and were validated in both mouse exudates in vivo and human macrophages in controlling local mediators and signaling. MiRs are known to regulate several gene targets and are thus associated with networks of gene products. For example, miR-146b, miR-208, and miR-219 are associated with genes involving NF-κB, IL-10 and 5-LOX, respectively9. We identified a novel resolution circuit where the SPM resolvin D1 (7S, 8R, 17S-trihydroxy-4Z, 9E, 11E, 13Z, 15E, 19Z-docosahexaenoic acid) activates its receptors on human macrophages to regulate miR-219-5p9. Two hairpin miR-219 precursor structures are found on different chromosomes, miR-219-1 on chromosome 6 (MI0000296) and mir-219-2 on chromosome 9 (MI0000740). miR-219-1 and miR-219-2 share the mature 5p miR whereas they each generate distinct mature 3p miRs18. In the present report, we addressed whether these miRs regulated in self-limited inflammation are also involved in delayed resolution and determined differential miR expression and lipid mediator (LM)-metabolipidomics during both self-limited and delayed resolution. We also present evidence that apoptotic PMN activate resolution programs via miR-219-2 and regulate LM production in both the initiation and resolution of acute inflammatory responses.
Results
Delayed resolution dysregulates microRNA and lipid mediator profiles
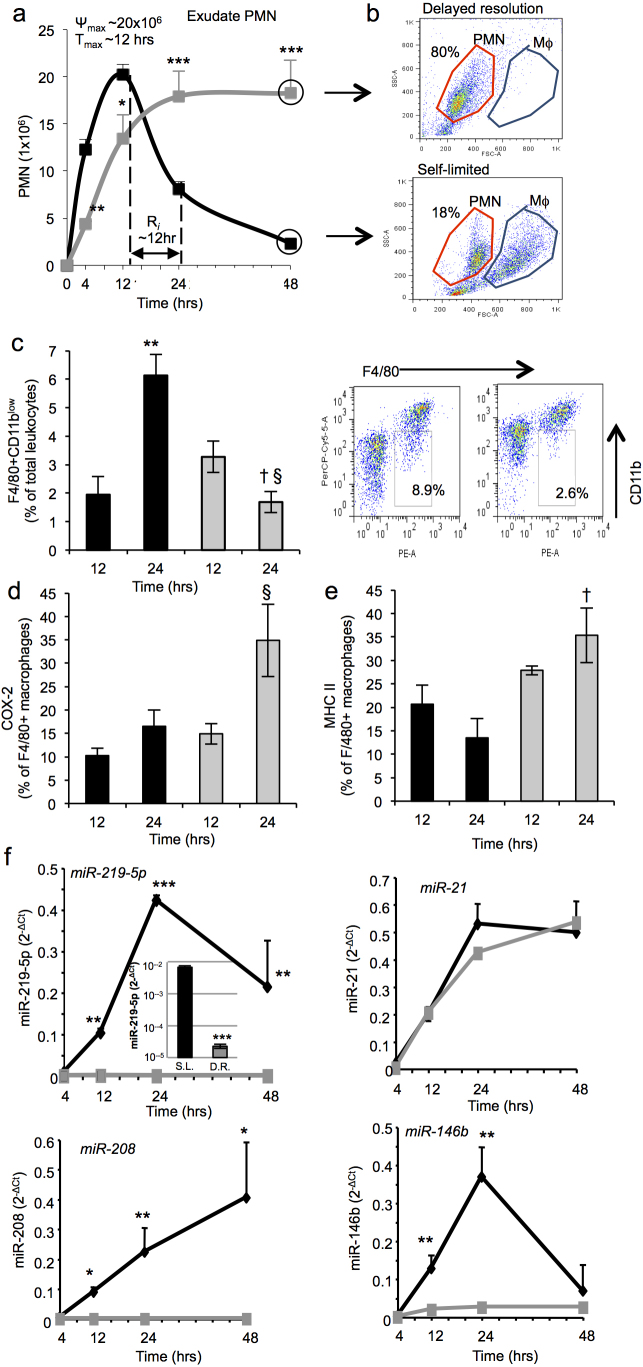
Since select miR (miR-21, miR-146b, miR-208 and miR-219-5p validated in mouse exudates and human macrophages) profiles are temporally regulated in acute self-limited inflammation9, we investigated them during delayed resolution. To this end, we used differential magnitudes of host challenge and obtained exudates from each, namely self-limited (1 mg zymosan) and delayed resolution (10 mg zymosan) with mice challenged at a time course of 4, 12, 24, and 48 hrs for direct comparison. In self-limited acute inflammation, there was a rapid and robust increase in neutrophil (PMN) numbers that peaked at 12 hrs followed by a steady decline, giving a resolution interval (Ri) = 12 hrs (Figure 1 a), results consistent with those established for acute inflammatory murine peritonitis11. The high-dose challenge group that gave delayed exudate resolution13,19,20 exhibited a less robust initial increase in PMN that remained elevated for the entire time course (4–48 hrs). Representative flow cytometry dot plots of exudate cells obtained at 48 hrs indicated that the delayed resolution challenge contained predominantly PMN, unlike the self-limited exudates that had fewer PMN (18.0 ± 3.5 × 106 vs. 2.0 ± 0.4 × 106 cells/murine exudate) and increased numbers of macrophages (Figure 1 b). We next investigated macrophage subsets in the groups of self-limited and delayed resolution. Resolving macrophages, characterized by F4/80 positive and CD11blow staining21,22,23, were found significantly increased between 12 and 24 hrs from 1.9% to 6.1% of the F4/80 cells in the self-limited exudates. In contrast, F4/80+CD11blow macrophages in the group with delayed resolution significantly decreased at these time intervals from 3.2% to 1.6% (Figure 1 c). M1 markers including COX-2 and MHC II24,25 were increased in the delayed resolution group at 24 hrs post zymosan challenge (Figure 1 d,e). Together these results indicate that the high dose challenge led to a reduction in number of resolving macrophages and an increase in M1 macrophages in the inflammatory exudates that suggests a contribution to the delayed resolution status. Moreover, these results showed that higher dose zymosan was associated with sustained PMN accumulation and reduced resolving macrophages that might contribute to the delayed or failed resolution status.
Figure 1. Specific miRs are temporally and differentially regulated in peritoneal inflammatory exudates.
Peritonitis was initiated via intra-peritoneal injection of zymosan (1 mg/mouse, black or 10 mg/mouse, gray). Exudates were collected at 4, 12, 24, and 48 hrs. (a) PMN were enumerated. (b) Representative flow cytometry dot plot of exudate cells at 48 hrs. (c,d,e) macrophages subsets were characterized using F4/80+ CD11blow (c) F4/80+COX-2+ (d) and F4/80+MHCII+(e). Black bars, self-limited; gray bars, delayed resolution. Results are mean ± SEM of n = 4, separate mice. **P<0.01 12 vs. 24 hr self-limited. § P<0.05 12 vs. 24 hr delayed resolution. † P<0.05 24 hr self-limited vs. 24 hr delayed resolution. (f) miRs were isolated from exudate cells at 4, 12, 24 and 48 hrs and analyzed by qPCR. Results are mean ± SEM, n = 4 separate mice. *P<0.05, **P<0.01, ***P<0.001, self-limited versus delayed resolution zymosan challenge. S.L., self-limited; D.R., delayed resolution.
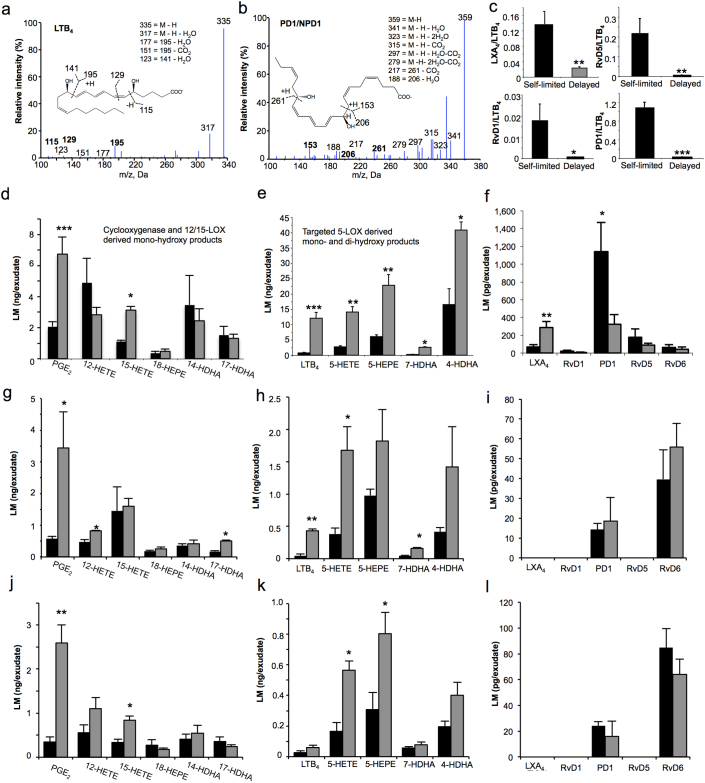
Next, the exudates from delayed resolution challenge had significantly lower expression of miR-219-5p, miR-208, but not miR-21 at the assessed time intervals (4,12,24, and 48 hrs) compared to the exudates of the self-limited group (Figure 1f). As early as 4 h after high-dose challenge, miR-219-5p expression in the exudates was ~2 log orders lower than in the self-limited resolving exudates (Figure 1f, upper left panel inset). Using lipid mediator metabolipidomics, we identified a number of cyclooxygenase (COX)- and lipoxygenase (LOX)-derived mediators including PGE2, LTB4 and PD1/NPD1 present within 3 mL aliquots of these lavage exudates (Figure 2 and Supplementary Table 1). These were identified using published physical criteria established for LM26 as shown for LTB4 and PD1/NPD1 (Figure 2 a and b) that included at least six diagnostic ions and matching chromatographic retention of authentic LM and synthetic reference materials. Using multiple reaction monitoring (MRM), we quantified the LM identified in exudates from both self-resolving and delayed resolution. We found that PGE2 (2,073±316 vs. 6,739±1,089 pg/exudate) and 15-HETE (1,097±101 vs. 3,129±232 pg/exudate) were significantly increased in delayed (i.e. high-dose challenge) compared to the self-limited response (Figure 2 d). In addition, targeted 5-LOX-derived products from each of the endogenous LM-metabolomes, namely AA, EPA and DHA metabolomes26, including LTB4 (741±168 vs. 12,070±1,904 pg/exudate), 5-HETE (2,711±365 vs. 14,068±1,773 pg/exudate) from AA, 5-HEPE (6,036±620 vs. 22,799±3,531 pg/exudate) from EPA, 7-HDHA (245±46 vs. 2,614±147 pg/exudate) and 4-HDHA (16,491±5,222 vs. 40,818±2,649 pg/exudate) from the DHA metabolome were all significantly increased in the delayed versus self-limited groups (Figure 2 e). Of interest, LXA4 levels were significantly higher in delayed (70±25 vs. 289±67 pg/exudate) compared to self-limited responses. In comparison, DHA-derived SPM including PD1/NPD1 (1,140±330 vs. 324±108 pg/exudate), RvD1 (23±10 vs. 8±1 pg/exudate) and RvD5 (177±93 vs. 86±25 pg/exudate) were all reduced in the high-dose challenge (Figure 2 f). Additionally, we determined the ratios of pro-resolving/pro-inflammatory mediators and found that these ratios were significantly lower in the peritoneal lavage obtained from mice challenged with high-dose zymosan (Figure 2 c). LMs were also quantified at 12 hrs (Fig 2 g–i) and 24 hrs (Fig 2 j–l) post zymosan initiation. In delayed resolution, PGE2, LTB4, 5-HETE and 7-HDHA were significantly higher at 12 hrs (Figure 2 g,h). Other targeted 5-LOX derived products (e.g. 7-HDHA and 4-HDHA) gave an increased trend in delayed resolution compared to self-limited challenges. PGE2, 5-HETE and 5-HEPE were significantly increased at 24 hrs in the high dose challenge (Figure 2 j,k). PD1 and RvD6 were identified but were not significantly different between the two challenges at 12 and 24 hrs (Figure 2 i,l). Notably, there was an inverse correlation between 5-LOX derived products and miR-219 expression throughout the time course. Together, these results indicate that delayed resolution from high-dose challenge was associated with both dysregulated miR and lipid mediator profiles.
Figure 2. Differential profiles of SPM and their biosynthetic pathway markers in inflammatory exudates.
Lavage exudates (3 mL) collected in 5 mL lavages from the peritoneum at 4 hrs were subjected to LC-MS-MS lipidomics. Bioactive lipid mediators and precursor/pathway marker were identified using previously established criteria26. (a,b) Representative tandem mass spectra of LTB4 and PD1 employed for identification. (c–l) Quantification was achieved by multiple reaction monitoring of Q1: M–H (parent ion), Q3: diagnostic ion in the MS-MS (daughter ion). (d–f) 4 hr, (g-i) 12 hr , (j–l) 24 hr exudates. (c) Ratios of pro-resolving versus pro-inflammatory mediators. Results are mean ± SEM, n = 4 separate murine exudates. *P<0.05, **P<0.01, ***P<0.001, self-limited versus delayed resolution challenge. Black bars, self-limited; Gray bars, delayed resolution.
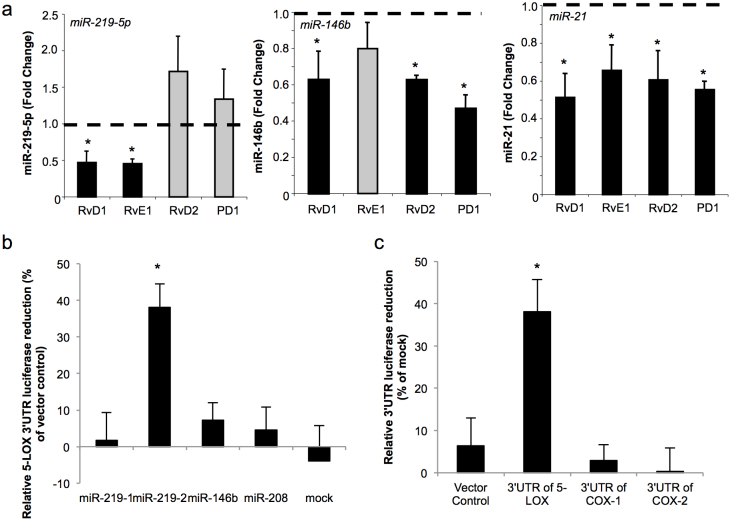
Given that RvD1 regulates expression of select miRs including miR-146b, miR-21, miR-208 and miR-219-5p9, we investigated whether this action was shared with other SPM and translates to human macrophages. RvE1 also decreased miR-219-5p and miR-21 but not miR-146b, whilst RvD2 and PD1 decreased miR-146b and miR-21 expression with no apparent regulation of miR-219-5p (Figure 3a). These results indicate that each SPM regulates a distinct panel of miRs highlighting separate mechanisms for each SPM that are produced from different precursors, EPA or DHA.
Figure 3. SPM distinctly activate select miRNAs.
(a) Human macrophages were incubated with RvD1, RvE1, RvD2, or PD1/NPD1 (each at 1 nM) or vehicle (6 hrs, 37°C) and miR levels determined by qPCR. Vehicle treatment is normalized as 1 (dashed line). Results are mean ± SEM, n = 6. *P<0.05 vehicle versus SPM. (b) Using a luciferase-based reporter system, miR-219-1, miR-219-2, miR-146b, miR-208 or mock were co-transfected with a 5-LOX 3′UTR- luciferase reporter vector into human macrophages (48 hrs, 37°C). Luminescence was monitored using SpectraMax3. Relative reduced luciferase activity of 5-LOX 3′UTR reporter was normalized with 3′UTR vector control. (c) miR-219-2 or mock were co-transfected with a 5-LOX, COX-1 or, COX-2 or control 3′UTR-luciferase reporter vector into human macrophages as in (b). Relative reduced luciferase activities of respective vectors in miR-219-2 overexpressed macrophages were normalized with mock. Results are mean ± SEM of n = 4-6 separate donors. *P<0.05 vector control versus 3′UTR of 5-LOX.
miR-219-2 directly targets the 3′UTR of 5-LOX
miR-219-5p was decreased in delayed resolution that correlated with an increase in LTB4 , a bioactive product of 5-LOX (Figure 1 and 2). Since miR-219 produces miR-219-5p and miR-219-3p, while miR-219-2 generates miR-219-5p and miR-219-2-3p (Supplementary Figure S2a online), we employed miR software (Targetscan, miRanda, miRDB, and MicroTar) to predict which miR directly targets 5-LOX. In silico analysis suggests that miR-219-2-3p targets 5-LOX with a binding context score ~71 (Supplementary Figure S2 online) whereas other select miRs (miR-21, miR-208, miR-146b, miR-219-3p, and miR-219-5p) did not have the predicted binding sites on the 3′UTR of human 5-LOX. Co-expression of miR-219-2 in human macrophages transfected with 5-LOX 3′UTR-luciferase vector resulted in a significant reduction in luminescence (~40%, p<0.05) compared to control vector-transfected macrophages (Figure 3 b). In contrast, miR-219-1, miR-146b, miR-208 or mock did not reduce luminescence, which confirmed the results of the target prediction. We also questioned whether miR-219-2 regulates COX metabolic pathway. In silico analysis predicted that there is a poorly conserved binding site on COX-2 3′UTR for miR-219-2, with a low context score ~15. Luciferase assay showed that miR-219-2 did not change the 3′UTR reporter activities of either COX-1 or COX-2 (Figure 3 c). Together, these results indicate that the 3′UTR of 5-LOX but not COX-2 is a direct target for miR-219-2.
miR-219-2 modulates expression of enzymes required for LM biosynthesis
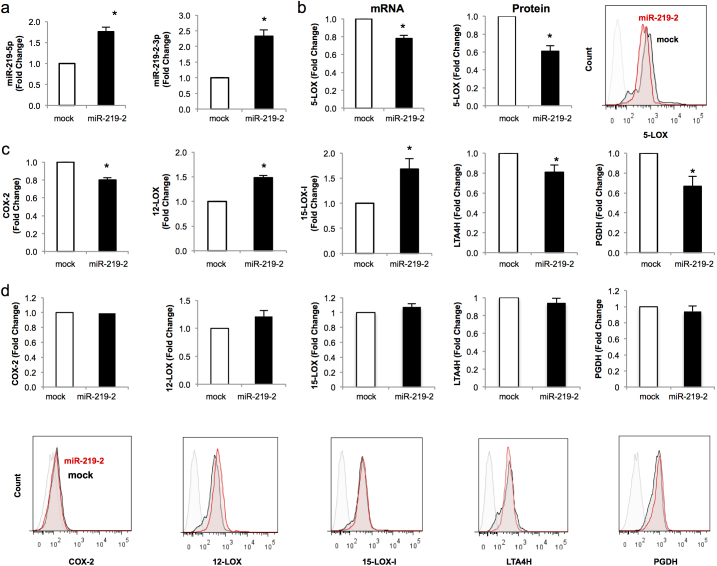
We next questioned whether miR-219-2 also regulated additional enzymes associated with LM biosynthesis and/or further metabolism (i.e. metabolic inactivation). To address this, human macrophages were transfected with either mock or miR-219-2 vectors for 72 hrs. miRs and mRNAs were then harvested from macrophages and analyzed via qPCR. miR-219-2 overexpression was validated by significant increases of miR-219-5p (~1.8 for an 80% increase above mock) and miR-219-2-3p (~2.5 or 150% increase above mock) expression (Figure 4 a). Additionally, miR-219-2 significantly decreased 5-LOX mRNA and protein (Figure 4 b). Overexpression of miR-219-2 decreased COX-2 mRNA expression by ~20%, without significant changes in protein levels (Figure 4 c,d). Overexpression of miR-219-2 led to increased mRNA 15-LOX-I (~50%), 15-LOX-II (~30%), and 12-LOX (~30%) expression compared to mock transfections (Figure 4c and S3). Decreased expression was obtained for LTA4 hydrolase (-LTA4H) by ~20%, and also the 15-hydroxyprostaglandin dehydrogenase (15-PGDH) ~40% compared to mock transfection (Figure 4c). Importantly, overexpression of miR-219-2 did not significantly modulate the protein levels of 12-LOX, 15-LOX, LTA4H and 15-PGDH 72 hrs post transfection (Figure 4 d) and suggests that regulation of these proteins may have different time courses than 5-LOX. Together, these results indicate that miR-219-2 selectively regulates 5-LOX.
Figure 4. miR-219-2 modulates expression of 5-LOX.
Human macrophages were transfected with either mock or miR-219-2 (72 hrs, 37°C). (a) miRs and mRNAs (b,c) were isolated and analyzed using qPCR. (b,d) Proteins were analyzed by flow cytometry and representative flow cytometry histograms are shown. Results are mean ± SEM of n = 6 donors. *P<0.05, mock vs. miR-219-2. Light gray IgG; black, mock; red, miR-219-2.
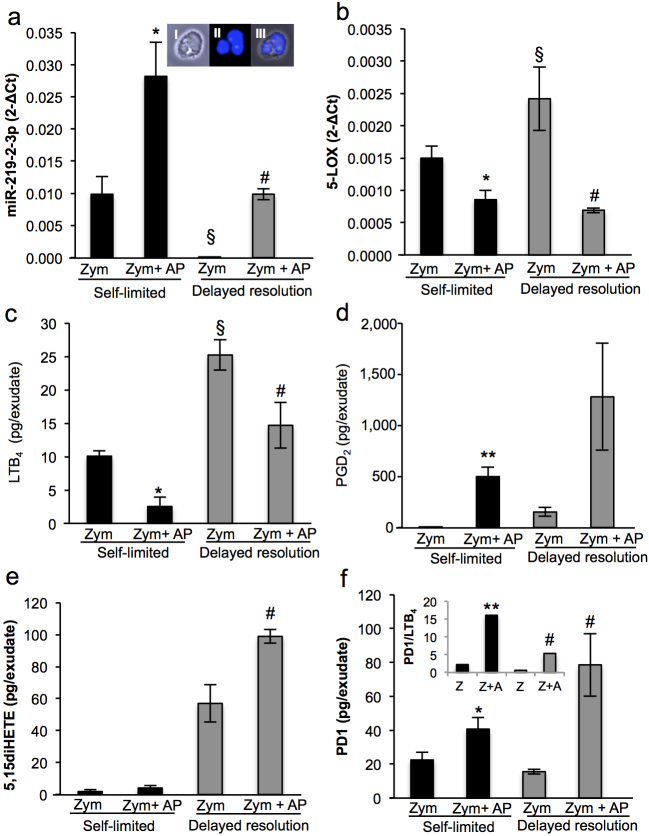
Apoptotic PMN enhance miR-219-2, decrease 5-LOX expression and restore pro-resolving lipid mediators in exudates
Apoptotic PMN are protective in vivo27,28; therefore, we addressed whether apoptotic PMN activate resolution programs via regulating miR-219-2 and 5-LOX. Administration of apoptotic PMN (5×106 cells/per mouse) at the height of inflammation (12 hrs post initiation) significantly increased miR-219-2-3p expression (Figure 5 a) and decreased 5-LOX expression (Figure 5 b) in both self-limited and delayed resolution challenges. LM metabolipidomics demonstrated that the decreased 5-LOX expression also led to a reduction of LTB4 in both groups (Figure 5 c). Conversely, when compared in both groups, PGD2 (5±1 vs. 498±95 pg/exudate; 151±43 vs. 1,279±558 pg/exudate), 5,15-diHETE (1.8±1.0 vs. 4.0±1.4; 56.9±11.0 vs. 98.2±4.3 pg/exudate) and PD1 (22.4±4.4 vs. 40.6±6.9; 15.5±1.3 vs. 78.5±18.0 pg/exudate) biosynthesis was significantly increased after apoptotic PMN administration. (Figure 5 d-f and Supplementary Table S2 online). The inset in Figure 5f displays increased PD1/NPD1:LTB4 ratio produced by the addition of apoptotic PMN. Together, these results indicate that apoptotic PMN orchestrated the onset of resolution programs in part via upregulation of miR-219-2 that led to the downregulation of pro-inflammatory LM and concomitant upregulation of pro-resolving LM production.
Figure 5. Apoptotic PMN stimulate resolution programs during acute inflammation.
At 12 hrs, apoptotic human PMN (5×106 PMN/mouse) were injected into mouse peritonitis exudates initiated by zymosan (1 mg or 10 mg/mouse; see Fig. 1) injection. Exudates were harvested at 24 hours. (a) miR-219-2-3p and (b) 5-LOX expression were quantified via qPCR. (A, inset) Representative macrophage and PMN from exudate (I) bright field, (II) DAPI, (III) Merged image. Magnification 40X. Representative of n = 4 separate murine exudates. (c–f) Bioactive lipid mediators and precursor/pathway markers were identified and quantified (see Materials and Methods). Results are mean ± SEM of n = 4 separate murine exudates. *P<0.05, **P<0.01. Self-limited zymosan vs. zymosan plus apoptotic PMN. #p<0.05 delayed resolution zymosan vs. zymosan plus apoptotic PMN. §P<0.05 self-limited zymosan versus delayed resolution zymosan challenge. Black bars, self-limited; Gray bars, delayed resolution. Z: zymosan, AP: apoptotic PMN.
Discussion
In the present report, we investigated the relationship between LM and resolution phase miR in resolving inflammatory exudates vs. a delayed resolution system in vivo in murine peritonitis and their translation to human macrophages. The results document that 1) miR-219-2 expression is decreased and directly regulates 5-LOX, a key enzyme in the biosynthesis of LMs, 2) exudate pro-inflammatory mediators are increased while pro-resolving lipid mediators decrease in delayed resolution compared to self-limited responses, and 3) apoptotic PMN rescue miR-219-2 expression, reducing LT and enhancing local SPM production in exudates.
To date, mechanisms associated with the temporal regulation of unwanted delays in resolution remain of wide interest. Results presented herein are the first integration of miR expression profiling with metabolipidomics in inflammatory exudates to define a specific miR-LM signature associated with delayed resolution of the acute inflammatory response. Additionally, these results provide a molecular basis to the burgeoning concept that the onset of resolution is programmed during initial events of inflammation29. Results in Figure 1 indicated that, as early as 4 hrs, miR-219-5p expression is decreased by 2 log orders compared to self-limited challenges. This decrease in miR-219-5p expression was associated with significant increases in targeted 5-LOX-derived products including LTB4 and decreases in SPM including PD1/NPD1 (Figure 2). Of note, SPM exert their actions in picogram to nanogram range (see reviews 5 and 7 and those references within) and hence are biologically relevant in low amounts. In this regard, picogram levels of SPM accumulated in mouse exudates, e.g. 70 pg of LXA4, 1,140 pg of PD1 and 177 pg of RvD5 in self-limited inflammation, which are functionally relevant. Importantly, miR-219-5p can be produced by either miR-219-1 or miR-219-2 and this miR-metabolipidomics approach at initial events in acute inflammation and its resolution revealed that miR-219-1 or miR-219-2 may play a pivotal role in determining whether inflammation resolves or is delayed and hence has potential for chronicity.
Of interest, the actions of LMs identified originally with human cells in vitro30 and their roles in resolution were demonstrated in humans31. Cantharidin-induced blisters were raised to emulate a sterile self-limited inflammatory event. Two groups emerged; those individuals that were early resolvers and those that were delayed resolvers31. Temporal formation of pro-resolving lipoxin A4 and the aspirin-triggered 15-epi-LXA4 produced with low-dose aspirin in these subjects determined whether an individual was an early or delayed resolver31. These findings in humans demonstrated that low-dose aspirin is pro-resolving and anti-inflammatory, limiting PMN entry to the inflammatory loci. Together the present results underscore that timely controlled production of SPM is an essential component of successful complete tissue resolution and termination of the acute inflammatory response.
Evidence in humans with non-resolving diseases such as asthma32, cystic fibrosis33,34, or localized aggressive periodontitis (LAP) show impaired production of SPM such as LXA4, RvE1 and PD1. Specifically, breath condensates from patients with severe asthma show significantly less PD1/NPD1 than healthy control individuals32. Also, LAP patients have a significant reduction of LXA4 as well as the DHA metabolome markers 14-HDHA and 17-HDHA in activated whole blood compared to those of matched healthy subjects35. Recent evidence from a pre-clinical model of obesity and insulin resistance indicates that lacking PD1/NPD1 in muscle and adipose may exacerbate obesity-linked inflammation36. Also, reduced NPD1 and 15-LOX in human Alzheimer's disease37 and estrogen downregulation of the LXA4 circuit38 provide evidence that failed resolution mechanisms are demonstrable in many organ systems throughout the body. These findings are consistent with the present results, which demonstrate that delayed resolution (in the high-dose challenge model) is associated with reduced production of local SPM (Figures 1 and 2). Hence, diminished local levels of individual SPM may, in part, explain the persistent uncontrolled inflammation associated with these diseases.
miRs generally act as posttranscriptional or translational repressors of gene transcripts39. Notably, the miR-219-2 but not miR-219-1′s binding with the 3′ UTR of 5-LOX (Figure 3 b) provides direct evidence for decreased miR-219-2 expression in the delayed resolution challenges (Figures 1 b, 2 d). miR-219-2 also increased both 15-LOX-I and 15-LOX-II mRNA expression (Figure 4d and Supplementary Figure 3 online), critical enzymes in the biosynthesis of protectins as well as lipoxins and resolvins in human tissues7. This was corroborated in vivo, where increased miR-219-2 expression correlated with increased PD1/NPD1 (Figures 1b, 2 c, and f). However, unlike 5-LOX, in silico analysis (Targetscan, miRanda, miR database, and MicroTar) suggested that miR-219-2 does not have predicted pairing sequences with the 3′UTR of 15-LOX-I or 15-LOX-II, implicating that miR-219-2′s regulatory actions of 15-LOX likely result from indirect mechanisms. In this regard, it is plausible that miR-219-2 tempers the system by decreasing pro-inflammatory LMs via direct regulation of 5-LOX and LTB4 and increasing pro-resolving LMs including PD1/NPD1, leading to a resolving milieu.
It is widely appreciated that apoptotic PMN are an integral component of the inflammatory response for complete resolution of acute inflammation40 and are protective in vivo27,28. In the present experiments, apoptotic PMN activated resolution in vivo via the miR-219-2/5-LOX axis. Along these lines, in both self-limited and delayed resolution challenges, the addition of apoptotic PMN gave an increase in miR-219-2 expression and a concomitant reduction in 5-LOX expression (Figure 5 a, b). In the self-limited inflammation, there was an increased trend in COX-2 expression at 24 hrs when apoptotic cells were given (2-ΔCt = 0.05 ± 0.02; mean ± SEM), compared to zymosan alone (2-ΔCt = 0.03 ± 0.01). In the delayed resolution COX-2 mRNA levels were modestly increased in the presence of apoptotic PMN (2-ΔCt = 0.08 ± 0.02) compared to zymosan alone (2-ΔCt = 0.04 ± 0.02). However, the changes in mRNA levels were not statistically significant. These results indicate that COX-2 expression was not significantly regulated at 24 hrs by apoptotic PMN in both the self-limited inflammation and delayed resolution. LTB4 levels were also decreased in these exudates, whereas both PGD2 and PD1/NPD1 (pro-resolving mediators) were increased (Figure 5 c,d,f). Of note, PGD2 can participate in the control of the onset of resolution in vivo41. Also, PD1/NPD1 enhances resolution11,12 and has pro-resolving actions in peritonitis, renal ischemic injury42, and liver steatosis43 as well as action in many other animal disease models reviewed in7,26. Hence, the ability of apoptotic PMN to regulate miR and LM regulation provides a clear example demonstrating that miR-219-2 regulates the local inflammatory milieu by decreasing LTB4 and increasing SPM, illustrating a new checkpoint control mechanism44 in resolution.
Taken together, high-dose challenges of the innate immune system that give delayed resolution undermine the endogenous resolution programs and lead to increased pro-inflammatory mediators and compromised SPM generation in this experimental in vivo system. Moreover, the present results provide evidence that in inflammatory exudates miR-219-2 is a key regulator of LM and a component of the active resolution programs that may be relevant in the pathogenesis of inflammatory diseases and potentially failed resolution checkpoint mechanisms that can contribute to disease.
Methods
Materials
Recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) was purchased from R&D Systems (Minneapolis, MN, USA). JetPei macrophage transfection reagent was purchased from PolyPlus Transfection (Illkirch, France). Primers were either designed or bought from Invitrogen (Carlsbad, CA) or Qiagen (Germantown, MD). miRNA expression vectors were purchased from Origene (Rockville, MD). See Supplementary Tables S3 and S4 online for primer and plasmid sequences, antibodies, and catalog numbers. Resolvin D1 (7S,8R,17S-trihydroxy-4Z,9E,11E,13Z,15E,19Z-docosahexaenoic acid) and deuterated labeled PGE2-d4, LTB4-d4 and 5S-HETE-d8 were purchased from Cayman Chemical (Ann Arbor, MI, USA). miScript Reverse Transcription and Omniscript RT Kits were from Qiagen.
Murine peritonitis
Male FVB mice (6-8 week old, Charles River Laboratories, Wilmington, MA, USA) were administered zymosan A at either 1 mg/mouse or 10 mg/mouse, suspended in 1 ml of sterile saline to initiate peritonitis11,13,14. At selected time intervals, mice were euthanized, peritoneal exudates were collected by lavaging the peritoneum with Ca2+/Mg2+ free phosphate buffered saline (PBS−/−, 5 ml), and leukocytes were enumerated using a hemocytometer and light microscopy. Differential cell counts were assessed with Wright-Giemsa staining for microscopy and flow cytometry. Fluorochrome-conjugated antibodies for flow cytometry were CD16/32-blocking Ab (clone 2.462) and rat anti-mouse CD11b (Mac-1 α chain, clone M1/70) from BD Bioscience (San Jose, CA, USA); Gr-1 (Ly-6G; clone RB6–8C5), and isotype-matched IgG controls were purchased from eBioscience (San Diego, CA, USA). All procedures were conducted in accordance with protocols approved by the Harvard Medical School Standing Committee on Animals guidelines for animal care (Protocol 02570).
Peripheral blood PMN and preparation of apoptotic PMN
Fresh human neutrophils (PMN) were isolated by dextran–Histopaque double gradient from whole blood from healthy volunteers (deidentified) with no medication intake for 2 weeks before donation (Partners Human Research Committee Protocol no. 88–02642), and heparin (1 U/mL) was added as anti-coagulant. Informed consents were obtained from healthy volunteers and PMN were freshly isolated and subjected to incubation conditions to produce apoptosis (16-20 hrs, 37°C, 5% CO2).
LC-MS/MS-based lipid mediator lipidomics
LC-MS/MS was performed with a Shimadzu LC-20AD HPLC (Shimadzu Scientific Instruments, Columbia, MD) equipped with an Agilent Eclipse Plus C18 column (4.6 mm × 50mm × 1.8µm) paired with an ABI Sciex Instruments 3200 Qtrap linear ion trap triple quadrupole mass spectrometer (Applied Biosystems, Foster City, CA). Instrument control and data acquisition were carried out using AnalystTM 1.5 software (Applied Biosystems). The mobile phase consisted of methanol/water/acetic acid (60/40/0.01; v/v/v) and was ramped to 80/20/0.01 (v/v/v) after 10 min, 100/0/0.01 (v/v/v) after 12 min, and 90/10/0.01 (v/v/v) after 1.5 minutes to wash and equilibrate the column. Ion pairs from reported multiple reaction monitoring (MRM) methods carried out earlier26 were used for profiling and quantification of individual lipid mediators. See Supplementary Table S1 online for individual MRM transitions. Criteria used for identification of each LM and pathway markers were carried out as in26. Briefly, each was matched using retention time and at least 6 diagnostic ions compared to synthetic standards where available and authentic standards26. Quantification was performed using calibration curves for each product and LM, and recoveries determined using deuterated internal standards (PGE2-d4, LTB4-d4 and 5(S)-HETE-d8) for each chromatographic region of interest (for further details, see ref.26).
PBMC isolation and cell culture
Peripheral blood mononuclear cells (PBMC) were isolated from human whole venous blood by density gradient centrifugation using Histopaque®-1077 (Sigma-Aldrich, Inc., St. Louis, MO, USA) and cultured in RPMI with 10 ng/mL human recombinant GM-CSF (37°C, 5% CO2, 7 days) as in9.
Transfection of miRs, RNA isolation and real-time PCR
Human macrophages (2 × 106 cells/10 ml) were transfected with pCMV-miR (mock) or miR-219 plasmid (5 μg) using a macrophage JetPei macrophage transfection reagent (10 μL) and incubated (72 h, 37°C, pH 7.45)9. miRNA fractions and large RNAs from human macrophages were isolated using High Pure miRNA Isolation (Roche Applied Science, Indianapolis, IN, USA)9. Omniscript and miScript (Qiagen, Germantown, MD, USA) were used for reverse transcriptions for mRNAs and miRs. Real-time PCR reactions were performed with a 7900HT Real-Time PCR thermal cycler (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR Master Mix (Qiagen). Data analysis was carried out using Real-Time PCR software 7900HT version 2.4 (Applied Biosystems). Relative concentrations of genes of interest were determined using the comparative Ct method after normalizing to the endogenous control. For analysis of human miRNAs, small nucleolar RNA U1 (RNAU1A; NCBI accession no. NR_004421) was selected as reference miRNA; for mRNAs, GAPDH (NCBI accession no. NM_002046.3) was used as a control and direct comparison.
Cloning of miR vectors and 3′-UTR luciferase reporter
Human macrophages were co-transfected with miR expression plasmids and luciferase reporter plasmid DNAs (2:1 ratio of microRNA vs. reporter) according to manufacturer's instructions. After incubation (37°C, 5% CO2, 48 hrs), luciferase activity was assayed using SuperLightTM Luciferase Reporter Gene Assay (BioAssay Systems, Hayward, CA) and measured using a Spectra Max M3 microplate reader (Molecular Devices, Inc., Sunnyvale, CA, USA).
Statistics
Statistical significance was assessed using Student's t-test. P-values <0.05 were deemed statistically significant.
Author Contributions
GF, YL, JD and CNS designed research, performed experiments, analyzed data, interpreted results, contributed to manuscript preparation. JD performed LC-MS-MS-based metabololipidomics. NC analyzed data, interpreted results and wrote the manuscript. CNS conceived overall research plan.
Supplementary Material
Supplemental Figures S1-S3 and Tables S1-S4
Acknowledgments
We thank Mary H. Small for expert help in manuscript preparation. This work was supported by National Institutes of Health grant no. R01-GM038765. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or NIH.
Footnotes
CNS is an inventor on patents [resolvins] assigned to BWH and licensed to Resolvyx Pharmaceuticals. CNS was scientific founder of Resolvyx Pharmaceuticals and owns founder stock in the company. CNS' interests were reviewed and are managed by the Brigham and Women's Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- Majno G. & Joris I. Cells, Tissues, and Disease: Principles of General Pathology. 2nd edn, (Oxford University Press, 2004). [Google Scholar]
- Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell 140, 771–776 (2010). [DOI] [PubMed] [Google Scholar]
- Lawrence T. & Gilroy D. W. Chronic inflammation: a failure of resolution? Int. J. Exp. Pathol. 88, 85–94 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. & Ding A. Nonresolving inflammation. Cell 140, 871–882 (2010). [DOI] [PubMed] [Google Scholar]
- Serhan C. N. A search for endogenous mechanisms of anti-inflammation uncovers novel chemical mediators: missing links to resolution. Histochem. Cell Biol. 122, 305–321 (2004). [DOI] [PubMed] [Google Scholar]
- Serhan C. N. P. I. Molecular Mechanisms in Leukocyte-Mediated Tissue Injury, P01DE013499 (2000–2005), NIDCR/National Institutes of Health.
- Serhan C. N. Novel resolution mechanisms in acute inflammation: to resolve or not? Am. J. Pathol. 177, 1576–1591 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perretti M. & D'Acquisto F. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat. Rev. Immunol. 9, 62–70 (2009). [DOI] [PubMed] [Google Scholar]
- Recchiuti A., Krishnamoorthy S., Fredman G., Chiang N. & Serhan C. N. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J. 25, 544–560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy S., Recchiuti A., Chiang N., Fredman G. & Serhan C. N. Resolvin D1 receptor stereoselectivity and regulation of inflammation and pro-resolving microRNAs. Am. J. Pathol. 180, 2018–2027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannenberg G. L. et al. Molecular circuits of resolution: Formation and actions of resolvins and protectins. J. Immunol. 174, 4345–4355 (2005). [DOI] [PubMed] [Google Scholar]
- Schwab J. M., Chiang N., Arita M. & Serhan C. N. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature 447, 869–874 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Xavier R. A. et al. A new strategy for the identification of novel molecules with targeted proresolution of inflammation properties. J. Immunol. 184, 1516–1525 (2010). [DOI] [PubMed] [Google Scholar]
- Bystrom J. et al. Resolution-phase macrophages possess a unique inflammatory phenotype that is controlled by cAMP. Blood 112, 4117–4127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy F. J. & O'Neill L. A. J. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann. Rheum. Dis. 67 Suppl 3, iii50–iii55 (2008). [DOI] [PubMed] [Google Scholar]
- O'Neill L. A., Sheedy F. J. & McCoy C. E. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175 (2011). [DOI] [PubMed] [Google Scholar]
- Roy S. & Sen C. K. MiRNA in innate immune responses: novel players in wound inflammation. Physiol. Genomics 43, 557–565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. P., Glasner M. E., Yekta S., Burge C. B. & Bartel D. P. Vertebrate microRNA genes. Science 299, 1540 (2003). [DOI] [PubMed] [Google Scholar]
- Winyard P. G. & Willoughby D. A. in Methods in Molecular Biology (ed J. M. Walker) (Humana, Totowa, NJ, 2003). [Google Scholar]
- Sampaio A. L. F., Dufton N. & Perretti M. in Fundamentals of Inflammation (eds C.N. Serhan, P.A. Ward & D.W. Gilroy) 329–337 (Cambridge University Press, 2010). [Google Scholar]
- Bannenberg G. L. et al. Molecular circuits of resolution: formation and actions of resolvins and protectins. J Immunol 174, 4345–4355 (2005). [DOI] [PubMed] [Google Scholar]
- Schif-Zuck S. et al. Saturated-efferocytosis generates pro-resolving CD11b low macrophages: modulation by resolvins and glucocorticoids. Eur J Immunol 41, 366–379 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stables M. J. et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood 118, e192–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F. O., Gordon S., Locati M. & Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177, 7303–7311 (2006). [DOI] [PubMed] [Google Scholar]
- Lawrence T. & Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 11, 750–761 (2011). [DOI] [PubMed] [Google Scholar]
- Yang R., Chiang N., Oh S. F. & Serhan C. N. Metabolomics-lipidomics of eicosanoids and docosanoids generated by phagocytes. Curr. Protoc. Immunol. 95, 14.26.11–14.26.26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel A. et al. Apoptotic neutrophils and T cells sequester chemokines during immune response resolution via modulation of CCR5 expression. Nat. Immunol. 7, 1209–1216 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y. et al. Apoptotic cells protect mice against lipopolysaccharide-induced shock. J. Immunol. 180, 4978–4985 (2008). [DOI] [PubMed] [Google Scholar]
- Serhan C. N. & Savill J. Resolution of inflammation: the beginning programs the end. Nat. Immunol. 6, 1191–1197 (2005). [DOI] [PubMed] [Google Scholar]
- Clària J. & Serhan C. N. Aspirin triggers previously undescribed bioactive eicosanoids by human endothelial cell-leukocyte interactions. Proc. Natl. Acad. Sci. U.S.A. 92, 9475–9479 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris T. et al. Dichotomy in duration and severity of acute inflammatory responses in humans arising from differentially expressed proresolution pathways. Proc. Natl. Acad. Sci. USA 107, 8842–8847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B. D. et al. Protectin D1 is generated in asthma and dampens airway inflammation and hyper-responsiveness. J. Immunol. 178, 496–502 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp C. L. et al. Defective lipoxin-mediated anti-inflammatory activity in the cystic fibrosis airway. Nat. Immunol. 5, 388–392 (2004). [DOI] [PubMed] [Google Scholar]
- Romano M. Lipoxin and aspirin-triggered lipoxins. ScientificWorldJournal 10, 1048–1064 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredman G. et al. Impaired phagocytosis in localized aggressive periodontitis: rescue by resolvin E1. PLoS One 6, e24422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. J., Arita M., Taguchi R., Kang J. X. & Marette A. Transgenic restoration of long-chain n-3 fatty acids in insulin target tissues improves resolution capacity and alleviates obesity-linked inflammation and insulin resistance in high-fat-fed mice. Diabetes 59, 3066–3073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw W. J. et al. A role for docosahexaenoic acid-derived neuroprotectin D1 in neural cell survival and Alzheimer disease. J. Clin. Invest. 115, 2774–2783 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. B. et al. Estrogen negatively regulates epithelial wound healing and protective lipid mediator circuits in the cornea. FASEB J. 26, 1506–1516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslett C. Resolution of acute inflammation and the role of apoptosis in the tissue fate of granulocytes. Clin. Sci. (Lond.) 83, 639–648 (1992). [DOI] [PubMed] [Google Scholar]
- Rajakariar R. et al. Hematopoietic prostaglandin D2 synthase controls the onset and resolution of acute inflammation through PGD2 and 15-deoxyDelta12 14 PGJ2. Proc. Natl. Acad. Sci. U.S.A. 104, 20979–20984 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield J. S. et al. Resolvin D series and protectin D1 mitigate acute kidney injury. J. Immunol. 177, 5902–5911 (2006). [DOI] [PubMed] [Google Scholar]
- González-Périz A. et al. Obesity-induced insulin resistance and hepatic steatosis are alleviated by omega-3 fatty acids: a role for resolvins and protectins. FASEB J. 23, 1946–1957 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C. N. & Chiang N. Novel endogenous small molecules as the checkpoint controllers in inflammation and resolution: entrée for resoleomics. Rheum. Dis. Clin. N. Am. 30, 69–95 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures S1-S3 and Tables S1-S4