Abstract
The malignant transformation of human prostatic epithelium is associated with the loss of androgen receptor (AR) in the surrounding stroma. However, the function and mechanisms of AR signaling in prostate cancer (PCa) stroma remain elusive. Here we report, by using proteomics pathway array analysis (PPAA), that androgen and its receptor inhibit the proliferation of prostate stromal cells through transcriptional suppression of cyclin B1, and we confirmed our findings at mRNA and protein levels using AR-negative or -positive primary prostate stromal cells. Furthermore, AR showed a negative correlation with cyclin B1 expression in stroma of human PCa samples in vivo. Mechanistically, we identify cyclin B1 as a bona fide AR target gene in prostate stromal cells. The negative regulation of cyclin B1 by AR is mediated through switching between E2F1 and E2F4 on the promoter of cyclin B1. E2F1 binds to the cyclin B1 promoter and maintains its expression and subsequent cell cycle progression in AR-negative stromal cells or AR-positive stromal cells when androgens are depleted. Upon stimulation with androgen in AR-positive stromal cells, E2F1 is displaced from the binding site by AR and replaced with E2F4, leading to the recruitment of the silencing mediator for retinoid and thyroid hormone receptor (SMRT)/histone deacetylase 3 (HDAC3) corepressor complex and repression of cyclin B1 at the chromatin level. The switch between E2F1 and E2F4 at the E2F binding site of the cyclin B1 promoter coincides with an androgen-dependent interaction between AR and E2F1 as well as the cytoplasmic-to-nuclear translocation of E2F4. Thus, we identified a novel mechanism for E2F factors in the regulation of cell cycle gene expression and cell cycle progression under the control of AR signaling.
INTRODUCTION
E2F transcription factors control the expression of key components of the DNA replication machinery as well as the G1/S and G2/M cell cycle transitions (34, 42). Eight members of the E2F family have been characterized (28). E2F1 to E2F6 form heterodimers with either DP-1 or DP-2 protein (3) to bind DNA and activate or repress gene expression. The retinoblastoma (pRB) tumor suppressor and the related p107 and p130 proteins (termed pocket proteins) bind to particular E2Fs in vivo and recruit corepressor complexes that contain histone deacetylases (HDACs), mediating the active repression of E2F-responsive genes (1, 34).
E2F proteins are divided into two subcategories with opposing functions in transcriptional activation in vivo. E2F1, E2F2, and E2F3 (termed activator E2Fs) play a key role in promoting the activation of E2F-responsive genes, cell cycle entry, and cell growth. The conditional ablation of E2F3 in mice lacking E2F1 and E2F2 results in mouse embryonic fibroblasts (MEFs) that are unable to proliferate (38). MEFs lacking E2F3 alone display a slow-growth phenotype (11). On the other hand, E2F4 to E2F8 appear to be involved primarily in the transcriptional repression of E2F-responsive genes. The activator E2Fs contain nuclear localization signals, while E2F4 and E2F5 possess nuclear export signals. Thus, E2F4-DP and E2F5-DP complexes are primarily cytoplasmic, except in quiescent or early-G1-phase cells, wherein pocket protein binding enables the nuclear localization of E2F4 and E2F5 and recruitment of corepressor proteins to target promoters (12). E2F6 lacks sequences required for trans-activation activity and a pocket protein-binding domain (33) and represses transcription via additional factors, including the Polycomb group (25). Two recently identified family members, E2F7 and E2F8, are atypical in that they can bind to DNA without heterodimerizing with DP-1 or DP-2. These new E2F members appear to be transcriptional repressors that act in a growth-inhibitory manner (40).
Androgen and androgen receptor (AR) are essential for the development of the prostate, as well as the initiation and progression of prostate cancer (PCa), including hormone-refractory or androgen-independent cancers (18). Androgen exerts its biological effects through AR, a member of the nuclear receptor superfamily that acts as a ligand-dependent transcription factor (29). AR mediates the transcriptional activation of the general transcriptional machinery through a series of events, including ligand binding, binding to cognate androgen response elements (AREs) located in upstream regions of target genes (22), and interaction with various coactivators (4). Most of the reports on AR signaling in prostate focus on epithelial cells. AR influences the levels of expression and activity of cyclin D-CDK4, cyclin E-CDK2, and cyclin A-CDK2 complexes essential for the G1-S cell cycle transition in PCa (2). AR signaling in prostate stromal cells (composed of AR-positive and AR-negative cells) has received considerably less attention. Prostate stroma is composed of a heterogeneous population of cells, including smooth-muscle cells, fibroblasts, and myofibroblasts, which confer diverse functions on PCa growth (16). In particular, prostatic stromal cells are heterogeneous in terms of AR expression. We reported that stromal AR levels were decreased in the areas surrounding cancerous tissue, especially in androgen-independent cancer (16). Furthermore, using telomerase-immortalized human stromal cell lines which are either positive or negative for AR, we demonstrated that stromal cells lacking AR stimulated the proliferation of cocultured PCa cells in vitro and enhanced tumor growth in vivo, whereas stromal cells expressing AR suppressed PCa growth in vitro and in vivo (16).
The molecular mechanisms of AR signaling in the stroma are poorly defined. In this study, we explored the mechanism, involving E2Fs and AR, through which androgen suppresses the proliferation of stromal cells via the transcriptional inhibition of cyclin B1. The functional significance of these findings is strongly supported by a negative clinical correlation between AR and cyclin B1 expression in the stroma of human prostate cancer, leading to aggressive prostate cancer.
MATERIALS AND METHODS
Reagents.
Antibodies against AR, cyclin A2, cyclin B1, cyclin D1, cyclin E, p21, p27, p53, SKP2, p73, E2F1, E2F2, E2F3, E2F4, E2F5, E2F6, silencing mediator for retinoid and thyroid hormone receptors (SMRT), HDAC3, c-Myb, Sp1, YB-1, Rb, pRb(S780), p130, p107, acetyl-histone H3, acetyl-histone H4, polymerase II (PolII), and β-actin, as well as secondary antibodies, were purchased from Cell Signaling Technology, Inc. (Boston, MA), or Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Plasmid pCMX:cyclin B1 was a gift from T. Florin (NYUSOM). pCMV:DP:E2F4 was a gift from J. Lees (Department of Biology, MIT). Western blotting detection reagents were from Amersham Pharmacia Biotech Inc. The Lipofectamine 2000 reagent and luciferase assay kit were acquired from Invitrogen (Carlsbad, CA) and Promega, respectively. R1881 and other chemicals were purchased from Sigma Chemical Co. (St. Louis, MO).
Cell culture and cell proliferation assay.
Stromal cell lines PShTert and PShTertAR (16) were maintained in RPMI 1640 with 10% FBS, supplemented with 1% penicillin and streptomycin at 37°C under 5% CO2. For proliferation assay (16), cells were aliquoted into 6-well plates with a density of 1 × 105 in androgen-free (phenol red-free RPMI and charcoal-stripped FBS) or androgen-supplemented (10 nM R1881) medium. Cell proliferation was measured by either cell counting or WST-1 assays (Roche, Mannheim, CA) in triplicate as described previously (26). Anchorage-independent cell growth in soft agar was performed in triplicate with cells (1 × 104) suspended in 2 ml of medium containing 0.35% agar (Becton, Dickinson) spread on top of 5 ml of 0.7% solidified agar. The colony volume was calculated from the average radius of representative colonies. Three primary prostate stromal cell lines were a kind gift from D. Rowley.
Proteomic pathway array analysis (PPAA).
Total cellular proteins were extracted from PShTert or PShTertAR cells grown in androgen-free or androgen-supplemented (10 nM R1881) medium, using a lysis buffer containing 20 mmol/liter Tris-HCl (pH 7.5), 20 mmol/liter sodium pyrophosphate, 40 mmol/liter β-glycerophosphate, 30 mmol/liter sodium fluoride, 2 mmol/liter EGTA, 10 mmol/liter NaCl, and 0.5% NP-40. PPAA was performed as described previously (35, 36, 39, 41). Chemiluminescence signals were captured using the ChemiDoc XRS system. Differences in protein levels were determined by densitometric scanning and normalized using internal standards.
Construction of cyclin B1 promoter-luciferase reporter and luciferase assay.
Primers P1, P2, P3, P4, P5, P6, P7, P8, and P9, with restriction sites KpnI/BglII, were designed to amplify promoter fragment of cyclin B1. The DNA fragments from P1 and P9 (−915 to +87), P2 and P9 (−715 to +87), P3 and P9 (−502 to +87), P4 and P9 (−292 to +87), P5 and P9 (−111 to +87), P5 and P8 (−111 to +14), P5 and P7 (−111 to −51), and P6 and P9 (+13 to +87) (see Table S2 in the supplemental material) were ligated into the pGL3-basic vector at the KpnI and BglII sites (Promega) and generated 1kb-LUC, 0.8 kb-LUC, 0.6 kb-LUC, 0.4 kb-LUC, 0.2 kb-LUC, 130bp-LUC, 50bp-LUC, and 70bp-LUC, respectively.
Dual-luciferase assays were performed as described previously (16, 26). The transfected stromal cells were grown in the absence or presence of 10 nM R1881 for 48 h and then harvested for the dual-luciferase reporter assay (Promega) according to the manufacturer's instructions.
Reverse transcription-PCR (RT-PCR) and RNA interference.
Total RNA was isolated with RNAqueous-4PCR kit (Ambion, Austin) by following the manufacturer's instructions. One μg total RNA was used for reverse transcription in 20-μl reaction mixtures. Five μl of the reverse transcription mixtures was used as the template in the 50-μl reaction mixtures. The PCR parameters were set as 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Fifteen μl of PCR products was separated on 2% agarose gels. Primers P10 and P11 (see Table S2 in the supplemental material), synthesized from Sigma (St. Louis, MO), were used to amplify cyclin B1 fragment (557 bp in length). A fragment of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control was amplified with primers P12 and P13 (see Table S2).
The siRNAs for AR, E2F1, E2F4, SMRT, and p21 were purchased from Ambion (Austin, TX). Transfection was carried out using Superfect (Qiagen) according to the manufacturer's instructions. Cells were harvested 24 h after transfection. Half of the samples were processed for flow cytometry analysis. The other half of the samples were used to prepare the cell lysates. Approximately 50 μg of protein was fractionated on 10% PAGE gels for Western blotting.
Western blot analysis and coimmunoprecipitation (co-IP).
Fifty μg whole-cell extract was subjected to SDS-PAGE and transferred to a nitrocellulose membrane for Western blot analysis. Immunoblots were blocked for 30 min and then incubated with primary antibodies (cyclin A2, 1:2,000; cyclin B1, 1:2,000; cyclin D1, 1:1,000; p21, 1:1,000; p27, 1:1,000; p53, 1:1,000; skp2, 1:1,000; AR, 1:1,000; E2F-1, 1:1,000; and C-myb, 1:1,000) for 2 h at room temperature and incubated for 1.5 h with the horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences) at 1:5,000 dilution in 5% nonfat dry milk. The protein bands were detected by an enhanced chemiluminescence kit (Amersham Biosciences) (19).
Stromal cells, treated with 10 nM R1881 for 24 h or mock treated with ethanol vehicle, were lysed with passive lysis buffer (Promega, WI) supplemented with protease inhibitor cocktail. Five hundred μg total proteins was precleared with protein A/G agarose, and 2 μg of antibodies was used for each immunoprecipitation. After immunoprecipitation, protein A/G agarose was washed five times with lysis buffer. Loading buffer (1×) was added to the protein A/G agarose, boiled, subjected to SDS-PAGE, and transferred to a nitrocellulose membrane for Western blot analysis.
ChIP.
Chromatin IP (ChIP) was performed as described elsewhere (42), with the following modifications. Cross-linking was initiated with 1.1% formaldehyde solution at room temperature for 10 min and then stopped by the addition of glycine to 0.125 M. The cross-linked chromatin was sonicated with a Sonifier 450 microtip (Branson Ultrasonic Corp., Danbury, CT) at power setting 2 for 12 30-s cycles on ice. This treatment produced DNA fragments with an average size of 300 bp.
For reporter ChIP, plasmids pGL34XAREE4, pGL3-basic, 1kb-LUC, 200bp-LUC, 130bp-LUC, and 70bp-LUC were transfected into PShTertAR cells with or without 10 nM R1881 for 24 h. After cross-linking, cells were lysed with buffer 1 (50 mM HEPES, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% protease inhibitor cocktail). The pellets obtained after buffer 1 treatment were lysed with buffer 2 (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitor cocktail), and the supernatant was used for reporter ChIP (42).
Primers for the amplification of E2F and ARE regions in the promoter of cyclin B1 were the following: for the E2F region, the forward primer sequence was P14 and the reverse primer sequence was P15 (see Table S2 in the supplemental material), which amplifies a 92-bp fragment from −86 to +6 upstream of the promoter region. Primers P16 and P17, from +15 to +115, were used to amplify the ARE region. A 145-bp product from −71 to −215 on the promoter of the ß-actin gene and a 215-bp product from −1 to −215 on the promoter of the p21 gene were amplified with primer sets P21 and P22 as well as P23 and P24, respectively.
Two primers, P18 and P19, located in positions +4760 to +4779 and +59 to +78 of luciferase plasmid pGL3, and primer P20, located in position +129 to +146 of pGL34XAREE4, were used for reporter ChIP.
Immunofluorescent microscopy and IHC analysis.
Stromal cells were cultured on glass coverslips treated with 10 nM R1881 or ethanol vehicle for 24 h, fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at 4°C, washed with PBS, and permeabilized in methanol-acetone (1:1) for 10 min at −20°C. The cells were incubated for 2 h at room temperature with anti-AR-specific antibody diluted 1:500 in 5% bovine serum albumin (BSA) buffer. After washing three times with Tris-buffered saline–Tween 20 (TBS-T) buffer, the cells were incubated for 45 min at room temperature with anti-rabbit IgG-Cy2-conjugated antibody (Molecular Probes, Portland, OR) diluted 1:300 in 5% BSA. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; 10 μg/ml; Molecular Probes, Carlsbad, CA) according to the manufacturer's instructions. Immunohistochemistry (IHC) for cyclin B1 and AR was performed as described previously (16).
RESULTS
AR inhibits prostate stromal cell proliferation.
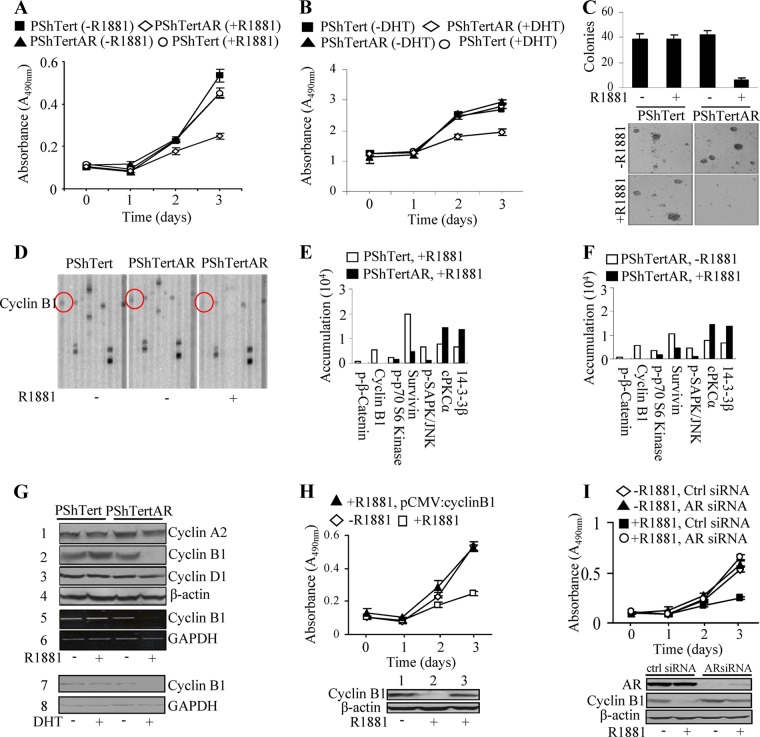
The role of stromal cells play in PCa is controversial (15). Androgen, acting through AR, promotes proliferation in PCa epithelial cells; however, its effects on prostate stromal cells are less understood. We previously established benign immortalized myofibroblast stromal cell lines from benign prostate hyperplasia that are either positive (PShTertAR) or negative (PShTert) for AR expression (16). In contrast to cancer epithelial cells, androgen suppressed the cell proliferation of AR-positive stromal cells. Using colorimetric WST-1 assays, the proliferation rate of PShTertAR cells was dramatically reduced in the presence of androgen (10 nM R1881) (P < 0.001) compared to that with androgen free medium (Fig. 1A), whereas PShTert cells showed no change in proliferation rate in androgen-free versus androgen-containing media. Growth inhibition was also observed at a lower concentration of R1881 (0.1 nM) for PShTertAR cells (see Fig. S1A in the supplemental material). The results from cell counting assays were consistent with the WST-1 assay results (see Fig. S1B). We also observed a similar degree of growth inhibition of PShTertAR cells with the natural ligand dihydrotestosterone (DHT) (Fig. 1B). Furthermore, we constructed NIH/3T3-AR cells and found that androgen inhibits the proliferation of NIH/3T3-AR cells but not NIH/3T3 cells (see Fig. S1C).
Fig 1.
AR inhibits proliferation of prostate stromal cells. (A) Reduced cell proliferation of PShTertAR cells in the presence of androgen (10 nM R1881) by WST-1 assays. (B) DHT (10 nM) also reduced cell proliferation of PShTertAR cells by WST-1 assays. (C) Reduction in number and size of colonies in anchorage-independent assays of PShTertAR cells in the presence of androgen. Both PShTert and PShTertAR cells were incubated in androgen-free medium for 48 h before proliferation assay and anchorage-independent assay. (D) Representative immunoblot indicates the inhibition of cyclin B1 expression in PShTertAR cells upon androgen stimulation detected by PPAA. (E and F) Quantification of immunoblot shows the difference in the levels of signal pathway proteins and phosphoproteins of PShTert and PShTertAR cells in the presence and absence of androgen. Cyclin B1 expression was inhibited in the presence of androgen for PShTertAR cells. (G) Androgen repressed the expression of cyclin B1 at both the protein level by Western blotting (lane 2) and the mRNA level by RT-PCR (lane 5) analysis. DHT repressed cyclin B1 expression (lane 7). (H) The introduction of cyclin B1 (lower) reversed the growth inhibition mediated by androgen in PShTertAR cells (upper). (I) Cyclin B1 expression was inhibited in the presence of androgen. Androgen receptor knockdown by AR siRNA leads to reversed cyclin B1 expression in the presence of androgen (lower) and the subsequent reversal of growth inhibition (upper). ctrl, control.
We next measured anchorage-independent growth in soft agar for PShTert and PShTertAR cells in the presence and absence of androgen. In the absence of androgen, PShTertAR cells have the same capacity to form colonies as PShTert cells. However, in the presence of androgen, there is a significant decrease in colony number (Fig. 1C, upper). In addition, the size of colonies for PShTertAR cells in the presence of androgen was considerably reduced (Fig. 1C, lower).
Identification of signaling network proteins regulated by AR in prostate stromal cells.
To determine the underlying signaling pathways of stromal cell growth inhibition by AR, we performed PPAA (35, 36, 41) on PShTertAR and PShTert cells in the presence and absence of androgen. PPAA makes use of one-dimensional gel electrophoresis followed by multiplex immunoblot arrays for the majority of signaling pathway proteins and phosphoproteins to allow the global evaluation of their changes, and thereby it establishes an interactive network of the pathways. Among 159 antibodies tested (see Table S1 in the supplemental material), 16 proteins showed more than a 2-fold change between androgen-stimulated and androgen-free conditions. Most of these pathways relate to cell survival and proliferation, as reflected in alterations of cell cycle-related proteins (Fig. 1E and F). Of the 16 proteins, 14 showed reduced expression after androgen treatment. Among the most dramatic changes observed (p-β-catenin, p-p70S6K, p-SAPK, and survivin), strikingly, cyclin B1 completely disappeared in the presence of androgen in PShTertAR cells, as shown by immunoblotting in Fig. 1D and quantification in Fig. 1E and F. In addition, other proteins (14-3-3β and cPKCα) showed significant increases in expression after androgen treatment. These differentially expressed proteins formed an interrelated network (see Fig. S2A in the supplemental material) using IPA software (Ingenuity). Consistently, the changes in the levels of proteins and phosphoproteins in AR-positive stromal cells in the presence and absence of androgen is almost identical to that between AR-positive and -negative stromal cells in the presence of androgen (Fig. 1E and F), indicating that these changes in protein levels are AR dependent. This result was further confirmed by comparing AR-positive and -negative stromal cells in the absence of androgen (see Fig. S2B). The experiment was performed in triplicate with consistent results.
Transcriptional repression of cyclin B1 expression by AR.
Since we observed a dramatic change in the expression of cyclin B1 coincidently with the AR-dependent regulation of PShTertAR cell growth by PPAA, we performed Western blot analysis for cyclin B1 expression along with that of several other cell cycle genes (Fig. 1G) in the presence or absence of androgen. Similarly to our PPAA results, cyclin B1 was completely repressed in PShTertAR cells in the presence of androgen (Fig. 1G). RT-PCR and quantitative PCR (qPCR) revealed that the inhibition of cyclinB1 expression by androgen in PShTertAR cells was at the transcriptional level (Fig. 1G; also see Fig. S3A in the supplemental material). We examined cyclin B1 repression by androgen during the time course of 0, 6, 12, 24, 36, 48, 72, and 96 h and found that cyclin B1 was completely repressed by 24 h (see Fig. S3B). The natural ligand DHT also inhibits the transcriptional activation of cyclin B1 as well as cell proliferation in PShTertAR cells, similarly to our results with R1881 (Fig. 1G). Interestingly, further studies showed that androgen inhibits cyclin B1 as well as proliferation in NIH/3T3-AR cells (see Fig. S3C).
To further confirm that the blockage of cyclin B1 expression is responsible for androgen-mediated growth inhibition, pCMV::cyclinB1 was transfected into PShTertAR cells in androgen medium. The growth inhibition of PShTertAR cells was reversed in androgen-containing medium with the forced expression of cyclin B1 to an extent observed in androgen-free medium (Fig. 1H), indicating that growth suppression by androgen in PShTertAR cells is mediated via the blockage of cyclinB1 expression. Fluorescent-activated cell sorting (FACS) analysis also showed that cell cycle conditions are regulated by androgen and cyclin B1 repression (see Fig. S4A and B in the supplemental material).
To ensure that androgen mediates the inhibition of cyclin B1 expression through AR rather than the genomic effects of the random integration of pBabe-AR in the cells, we knocked down AR expression with an AR-specific short interfering RNA (siRNA) in PShTertAR cells and performed Western blot analysis for cyclin B1 expression. AR knockdown reversed the suppression of cyclin B1 expression by androgen (Fig. 1I, lower) and promoted the increased proliferation of PShTertAR cells (Fig. 1I, upper) in the presence of androgen. Further, we did not observe differential expression of other cyclins, including cyclin A2 and cyclin D1, by Western blot analysis in the presence or absence of androgen for PShTert and PShTertAR cells (Fig. 1G), indicating specificity through which AR functions to suppress gene expression associated with cell proliferation.
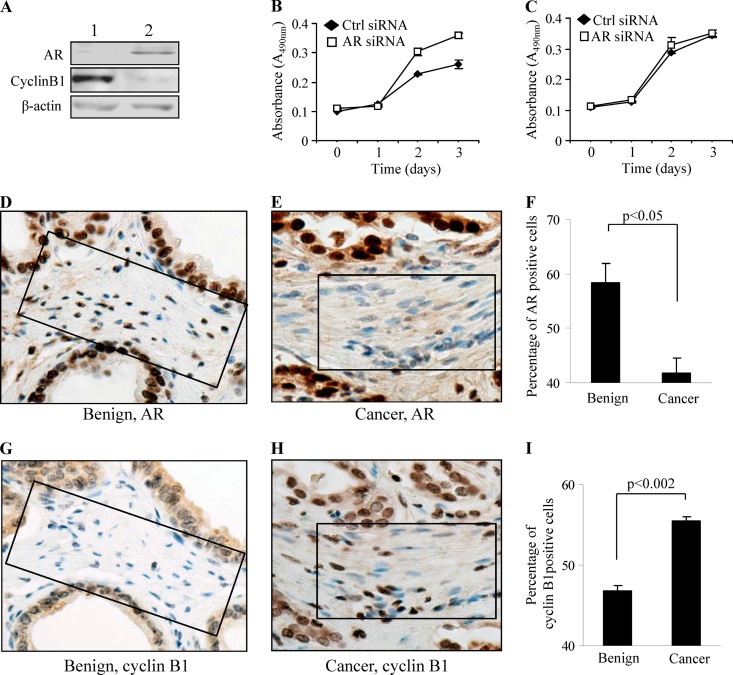
Our in vitro data clearly showed that AR negatively regulates the expression of cyclin B1. To further determine whether the inhibitory activity of AR is a general phenomenon in prostate stromal cells, we investigated the growth of three primary prostate stromal cell lines, one negative and two positive for AR expression (Fig. 2A; also see Fig. S3D in the supplemental material), by silencing AR expression with siRNAs. AR knockdown in the AR-positive cells promoted their growth in androgen-containing medium (Fig. 2B), while no effect was observed in AR-negative cells treated with an AR siRNA (Fig. 2C), indicating that the growth-inhibitory property of AR is a general effect in prostate stromal cells.
Fig 2.
AR expression negatively correlates with cyclin B1 expression in vivo. (A) Inverse relationship of AR and cyclin B1 expression exists in AR-negative (lane 1) and AR-positive (lane 2) primary prostate stromal cells. (B and C) The AR knockdown by siRNA leads to increased growth of AR-positive primary cells (B) but has no effect on the growth of AR-negative primary cells (C). (D and E) Decreased AR expression in cancer stromal cells (E) compared to that of stromal cells in benign matrix (D) by IHC. (F) Decrease (16.55%) of AR-positive cells was observed in the areas surrounding cancerous tissue compared to a benign area (n = 20) (P < 0.05). (G and H) Increased cyclin B1 expression was detected in cancer stromal cells (H) compared to stromal cells in benign matrix (G) by IHC. (I) Increase (9.2%) of cyclin B1-positive cells was obtained in the areas surrounding cancerous tissue compared to a benign area (n = 20) (P < 0.05). Panels D, G, E, and H are serial sections. Square boxes outline stromal cells.
To further investigate if there is a negative correlation between AR and cyclin B1 in vivo, we examined their expression within areas surrounding cancerous and adjacent benign tissue from 14 formalin-fixed, paraffin-embedded human PCa cases. The PCa patients ranged in age from 63 to 84 years old (mean, 70), and the Gleason score ranged from 6 to 9. Nine of the 14 cases were stage II cancer (64%), and 5 were stage III (36%). AR and cyclin B1 expression were scored in three stromal areas surrounding cancerous tissue and three areas of benign tissue in each case. In every scored area, 100 cells were counted to determine the relative percentages of AR or cyclin B1-positive cells. The levels of AR or cyclin B1 were expressed as an average percentage of cyclin B1-positive stromal cells. A 16.55% decrease in stromal AR expression was observed in the areas surrounding cancerous tissue compared to that in benign areas (P < 0.05) (Fig. 2D, E, and F), which is consistent with our previous data (16). On the contrary, a 9.2% increase in cyclin B1 was observed in the areas surrounding cancerous tissue compared to that of the benign area (P < 0.002) (Fig. 2G, H, and I), confirming the negative regulation of cyclin B1 by AR in vivo. Relevant to its clinical importance, the decreased stromal AR is associated with aggressive cancer, including high-grade and androgen-independent PCa (16). Here, we also showed that there is a greater decrease in stromal AR expression in association with PCa in African American (AA) patients than in Caucasian (CA) patients. There is a 1.52-fold greater decrease in AA PCa (n = 69) than in CA PCa (n = 46) for stromal AR (P = 0.0066).
Identification of a novel ARE negatively regulating the transcription of cyclin B1.
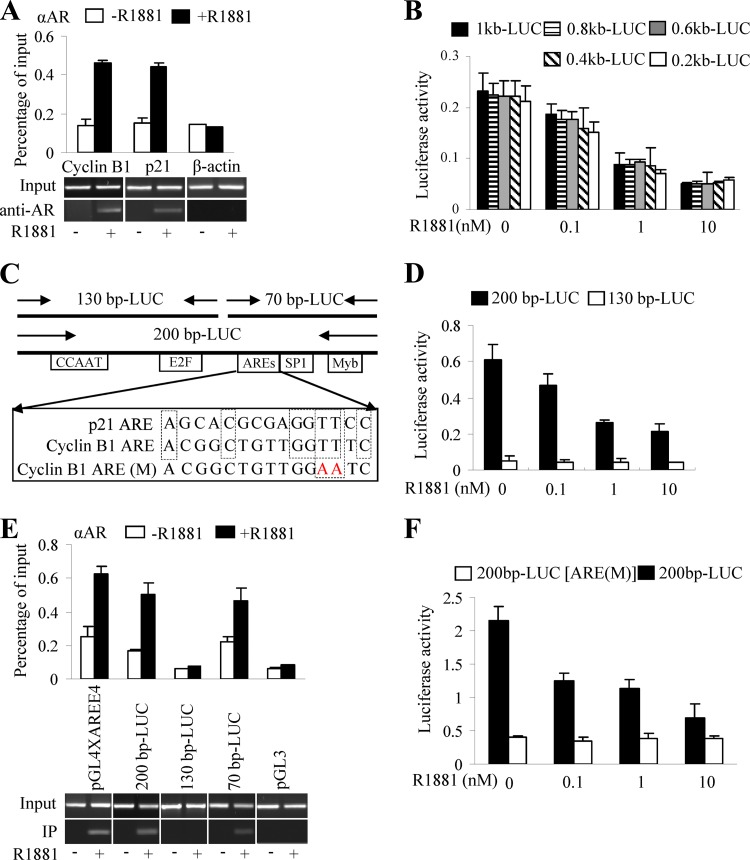
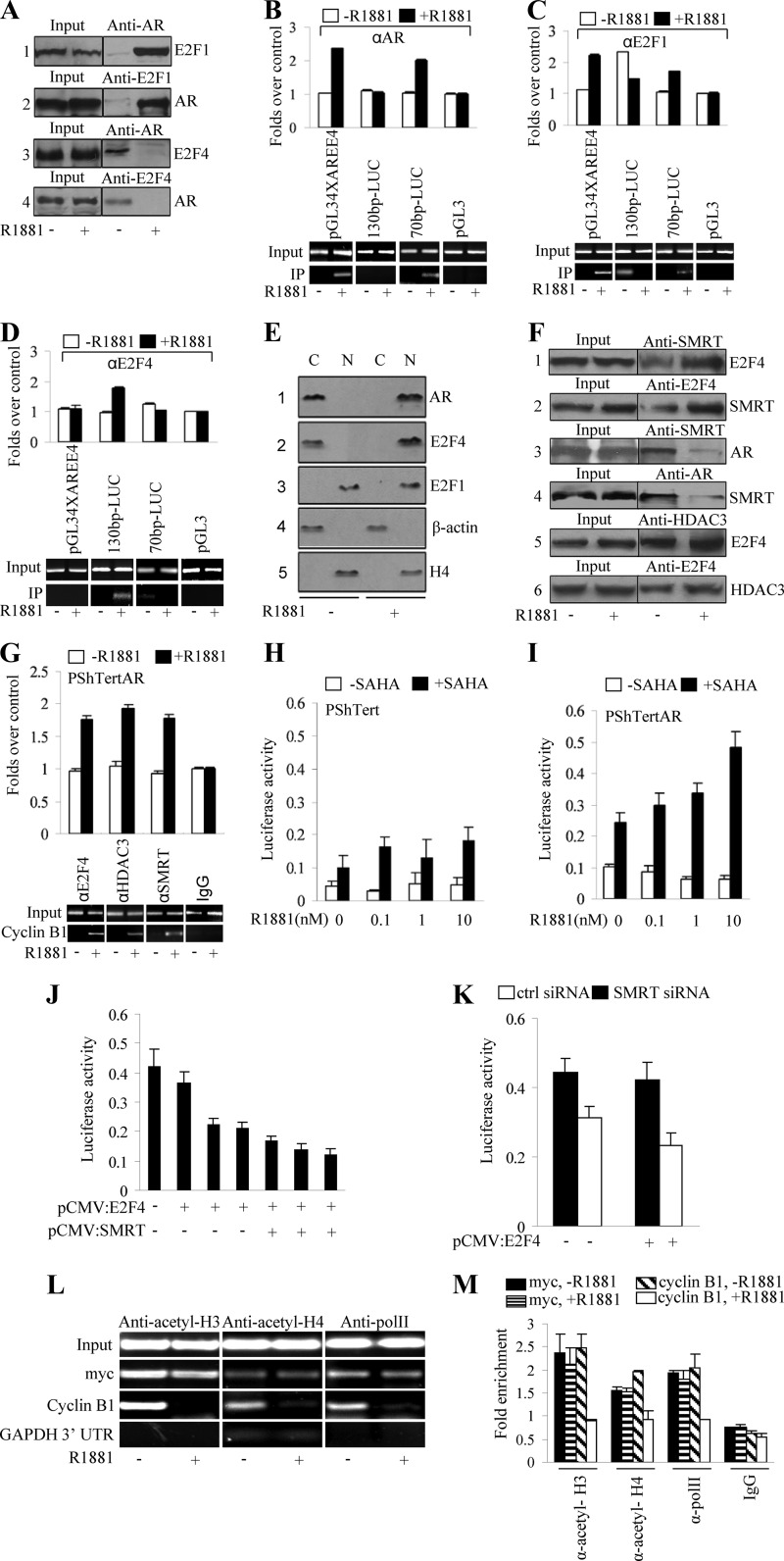
To examine whether the androgen receptor-mediated transcriptional repression of cyclin B1 in vivo is directed through AREs (32) in the cyclin B1 promoter, ChIP assays were performed with anti-AR antibody. The promoter regions of known AR target genes, p21 and c-flip (20), were selected as positive controls. The promoter region of β-actin, which lacks an ARE, served as a negative control. Our data showed the increased recruitment of AR to the promoter regions of p21 (Fig. 3A) and c-flip (see Fig. S5A and B in the supplemental material) upon androgen stimulation in PShTertAR cells. In addition, AR recruitment to the promoter region of cyclin B1 was enhanced in the presence of androgen (Fig. 3A), suggesting that cyclin B1 is regulated directly by AR.
Fig 3.
Novel androgen-responsive element identified on cyclin B1 promoter. (A) Promoter occupancy of AR on cyclin B1 promoter by ChIP. Immunoprecipitation was performed with 2 μg of anti-AR antibody. The p21 promoter region was selected as a positive control for AR recruitment, and the ß-actin promoter region served as a negative control. AR was recruited to the promoter of cyclin B1 and p21 in the presence of androgen, while no recruitment to the promoter region of ß-actin upon androgen treatment was seen. (B) Dual-luciferase assays with 1kb-LUC, 0.8 kb-LUC, 0.6 kb-LUC, 0.4 kb-LUC, and 0.2 kb-LUC reporter constructs showed transcriptional repression by AR in the presence of androgen. (C) Schematic representation of a putative binding site for transcription factor, ARE sequences on the cyclin B1 promoter, and construction of luciferase reporter (200bp-LUC, 130bp-LUC, 70bp-LUC) with fragments of the cyclin B1 promoter. (D) Luciferase assay for reporter constructions 200bp-LUC and 130bp-LUC. One hundred ng 200bp-LUC or 130bp-LUC was transfected into PShTertAR cells in the absence or presence of ligand (R1881; 0.1, 1, and 10 nM). 200bp-LUC showed induction, while the 130bp-LUC luciferase reporter lost induction upon treatment with R1881. (E) Reporter ChIP was performed with the 200bp-LUC, 130bp-LUC, and 70bp-LUC luciferase reporter plasmids. The luciferase reporter pGL4AREE4 (containing four AREs upstream of the E4 promoter) served as a positive control and the empty luciferase pGL3 basic as a negative control. The results show that AR binds to the 200-bp (left) and the proximal 70-bp (left) but not the distal 130-bp cyclin B1 promoter in the presence of androgen. (F) Luciferase assay was performed with wild-type 200bp-LUC and mutated 200bp-LUC AREs. Two mutations (TT to AA) in the putative ARE abolish the AR-mediated transcriptional repression of the cyclin B1 promoter.
To delineate potential AREs, we generated cyclin B1 promoter-luciferase reporter plasmids with 200-bp serial deletions of the 1-kb proximal promoter region. All constructs resulted in equal levels of transcriptional repression, showing that one or more potential AREs are located in a 200-bp (−111 to +87) proximal promoter region (Fig. 3B). This 200-bp region was further dissected into a 70-bp (+10 to +87) proximal (70bp-LUC) and a 130-bp (−111 to +15) distal fragment (130bp-LUC) (Fig. 3C). Luciferase assays indicated that the 130bp-LUC luciferase reporter lost inducibility upon treatment with R1881 (Fig. 3D), suggesting that the ARE is located in the 70-bp region.
To confirm that the potential ARE is located in the 70-bp region, we performed ChIP with the 200bp-LUC, 130bp-LUC, and 70bp-LUC reporter plasmids. The luciferase reporter pGL4AREE4 (containing 4 ARE sequences upstream of the E4 promoter) served as a positive control, and the empty luciferase pGL3-basic was a negative control. Using anti-AR antibody and primers specific for the reporter and cyclin B1 promoter, we showed that regions from both 200bp-LUC and 70bp-LUC could be amplified, while no band was detected for the 130bp-LUC reporter (Fig. 3E) These results indicate that a potential ARE is located within the 70-bp region. The comparison of the 70-bp DNA sequence to known AREs suggested that a 16-bp region (from +17 to +33) resembles the ARE of the p21 gene (20) (Fig. 3C). To further verify that this consensus sequence represents an authentic ARE, site-directed mutations (Fig. 3C) were introduced in this region of the 200bp-LUC reporter. The mutant 200bp-LUC reporter was no longer repressed by androgen (Fig. 3F), indicating that the 16-bp region (from +17 to +33) in the cyclin B1 promoter is a bona fide ARE.
Switch between E2F1 and E2F4 binding to E2F binding site regulates the transcriptional repression of cyclin B1.
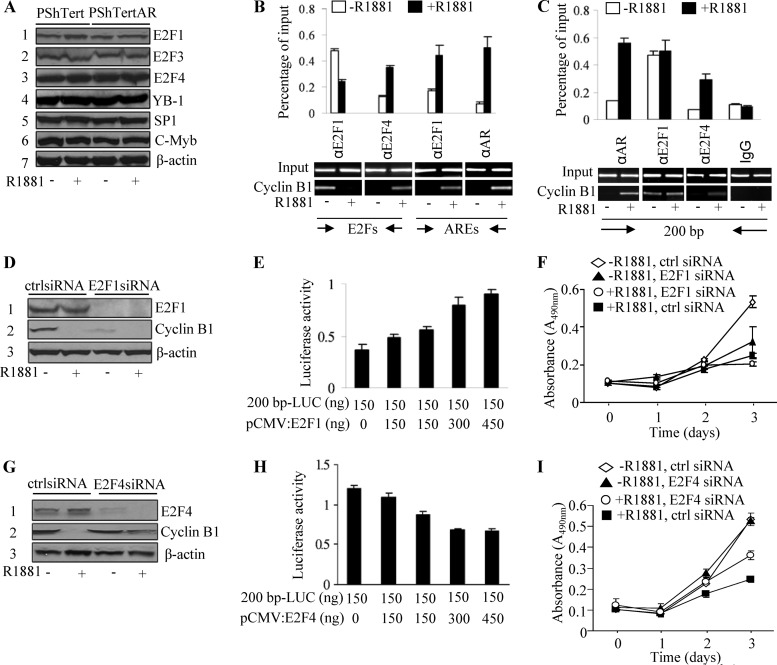
Sequence inspection revealed the presence of an E2F regulatory element, CCAAT elements, a CHR element, a Myb binding site, and a Sp1 binding site in the 200-bp promoter region of cyclin B1 (Fig. 3C) that may be critical for the transcription of cyclin B1 (42). To investigate the possibility that AR regulates the transcription of cyclin B1 cooperatively with these transcriptional factors, we examined the expression of E2F1, E2F2, E2F3, E2F4, E2F5, E2F6, YB-1, SP1, and c-Myb by Western blotting in the presence and absence of androgen. With the exception of E2F2, E2F5, E2F6, and E2F8, which were undetectable, the expression of the remaining factors was similar in PShTert and PShTertAR cells in the presence or absence of androgen, indicating that cyclin B1 repression mediated by androgen was not caused by the differential expression of these factors (Fig. 4A).
Fig 4.
Switch between E2F1 and E2F4 leads to repression of cyclin B1 transcription. (A) Expression of E2F1, E2F3, E2F4, YB-1, Sp1, and c-Myb in PShTert and PShTertAR cells in the presence and absence of androgen. (B) E2F1 decreases recruitment while E2F4 increases recruitment at the E2F binding site in the presence of androgen with primers P14 and P15. Both AR and E2F1 increase recruitment at the ARE region of the cyclin B1 promoter in the presence of androgen with primers P16 and P17. P14 and P15 were primers designed for the E2F site, and primers P16 and P17 were used to amplify the ARE region. (C) ChIP showed that E2Fs bind the E2F binding site on the promoter region of cyclin B1. Both AR and E2F4 increased recruitment in the presence of androgen, but E2F1 does not show different recruitment between the absence and presence of androgen with primers P14 and P17 (fragment from −86 to +115, including the E2F regulatory element, CCAAT elements, the CHR element, an ARE, the Myb binding site, and the Sp1 binding site). (D) Knockdown of E2F1 by siRNA decreased cyclin B1 expression in the absence of androgen. (E) E2F1 increases transactional activation of cyclin B1 in dual-luciferase assays with 200bp-LUC cyclin B1. (F) WST-1 assay indicated a decreased rate of cell proliferation with siRNA knockdown of E2F1 even in androgen-free media for PShTertAR cells. (G) Knockdown of E2F4 by siRNA increased cyclin B1 expression even in the presence of androgen. (H) E2F4 decreases transactional activation of cyclin B1 in dual-luciferase assays with 200bp-LUC cyclin B1. (F) WST-1 assay showed that the increased rate of cell proliferation with the siRNA knockdown of E2F4 in the presence of androgen for PShTertAR cells.
We next performed ChIP experiments to detect promoter occupancies at their predicted binding sites. First, as expected, AR occupied the ARE region (+15 to +115) in the presence of androgen (Fig. 4B). Interestingly, the recruitment of E2F1 to the ARE region increased in an androgen-dependent manner (Fig. 4B). We also examined the recruitment of E2F1, E2F3, and E2F4 to E2F sites (−86 to +6) and found that E2F4 was exclusively recruited to this region in the presence of androgen (Fig. 4C). In contrast, E2F1 exhibited decreased recruitment to the E2F binding site in the presence of androgen (Fig. 4B), suggesting that the occupancy on the E2F site by E2F1 or E2F4 is critical for cyclin B1 transcription. Finally, using primers spanning the region from −86 to +115, we showed that the recruitment of E2F3, YB-1, SP1, and c-Myb did not change irrespective of androgen treatment (see Fig. S5C in the supplemental material).
To verify the functional role of E2F1 in cyclin B1 transcription and subsequent cell proliferation, we first performed dual-luciferase assays with the 200bp-LUC cyclin B1 reporter and increasing amounts of E2F1 expression vector. As expected, E2F1 increased the expression of the reporter gene in a dose-dependent manner (Fig. 4E). Cell proliferation then was examined after E2F1 knockdown with siRNA in PShTertAR cells. The levels of cyclin B1 were reduced (Fig. 4D), and the growth rate of PShTertAR cells was diminished even in androgen-free medium (Fig. 4F), indicating that E2F1 binding to the E2F regulatory element is essential for cyclin B1 transcription and cell cycle progression.
In contrast to the role of E2F1 in transcriptional activation, E2F4 represses transcription (31). We examined the possibility that E2F4 is involved in cyclin B1 repression in the presence of androgen using luciferase assays with cyclin B1 200bp-LUC reporter and vector expressing DP-1 fused to E2F4 protein. As indicated in Fig. 4H, E2F4 expression led to the repression of the reporter gene in a dose-dependent manner. In contrast, E2F4 knockdown reversed the repression of cyclin B1 (Fig. 4G) and partially restored the decreased proliferation of PShTertAR cells in the presence of androgen (Fig. 4I; P = 0.014), indicating that E2F4 is involved in cyclin B1 expression and the subsequent inhibition of cell proliferation.
To further confirm that the E2F site is important for the androgen-mediated transcriptional repression of cyclin B1, we mutated the E2F site in the 200bp-LUC reporter [5-TAGGCTGGCTCTTCTCG(GC→TT)GTGCTGCGGCGGAA-3]. Luciferase assays showed that the 200bp-LUC bearing an E2F mutation lost the ability to be transactivated by androgen (see Fig. S6A in the supplemental material) or the ectopic expression of E2F1, as well as the ability to be repressed by E2F4 (see Fig. S6B and C).
Physical interaction between E2F1 and AR exposes E2F binding site to nuclear E2F4, resulting in androgen-mediated repression.
Our data indicated that E2F4 occupied the E2F binding site, while E2F1, as well as AR, were recruited to the ARE of the cyclin B1 promoter in an androgen-dependent manner. The interaction between AR and E2F1 then was examined with coimmunoprecipitation. Interestingly, AR interacted with E2F1 in PShTertAR cells only in the presence of androgen (Fig. 5A). Reciprocal coimmunoprecipitation experiments confirmed the androgen-dependent interaction between AR and E2F1 (Fig. 5A). In contrast to the interaction between AR and E2F1, AR interacted with E2F4 only in the androgen-free condition (Fig. 5A).
Fig 5.
Interaction between AR and E2F1 facilitates nuclear E2F4 binding to E2F binding site. (A) AR interacts with E2F1 only in the presence of androgen by co-IP. Using AR as a pulldown antibody and E2F1 as detection antibody (lane 1) or using E2F1 as the pulldown antibody and AR as the detection antibody (lane 2), AR interacts with E2F1 only in the presence of androgen (10 nM). (B) AR interacts with E2F4 only in the absence of androgen by co-IP. Interaction between AR and E2F4 was not detected (lanes 3 and 4) in the presence of androgen and was present only in the absence of androgen. (B to D) Recruitment of E2F1 and E2F4 on the E2F site and the ARE region was detected by reporter ChIP with 130bp-LUC (containing an E2F site but without AREs) and 70bp-LUC (including AREs but excluding an E2F site). pGL34XAREE4 (containing four AREs upstream of the E4 promoter) and pGL3-basic served as positive and negative controls, respectively. (B) With AR immunoprecipitation, the 70bp-LUC gave positive signal only in the presence of androgen, while the 130bp-LUC showed no signal regardless of androgen. With E2F1 immunoprecipitation, the 70bp-LUC gave signal in the presence of androgen as that from AR. (C) Meanwhile, E2F1 showed dramatically decreased recruitment on the 130bp-LUC in the presence of androgen. (D) With E2F4 immunoprecipitation, the 130bp-LUC gave a positive signal, while the 70bp-LUC showed no signal in the presence of androgen. (E) Cell fraction showed nuclear (N) and cytoplasmic (C) localization of AR (nuclear localization in the presence of androgen and cytoplasmic localization in the absence of androgen; lane 1), E2F1 (nuclear localization in the presence and absence of androgen; lane 2), and E2F4 (nuclear localization in the presence of androgen and cytoplasmic localization in the absence of androgen; lane 3) by cell fractionation. H4 serves as a control for nuclear protein, and β-actin serves as a control for cytoplasmic protein. (F) E2F4 shows interaction with SMRT by co-IP regardless of androgen, using SMRT as the pulldown antibody and E2F4 as detection antibody (lane 1) or using E2F4 as pulldown antibody and SMRT as the detection antibody (lane 2). The interaction between E2F4 and SMRT appears to be stronger in the presence of androgen. SMRT shows interaction with AR in the absence of androgen, either using SMRT as the pulldown antibody and AR as detection antibody (lane 3) or using AR as the pulldown antibody and SMRT as the detection antibody (lane 4). E2F4 shows interaction with HDAC3 by co-IP regardless of androgen status, using HDAC3 as the pulldown antibody and E2F4 as the detection antibody (lane 5) or using E2F4 as the pulldown antibody and HDAC3 as the detection antibody (lane 6). (G) SMRT and HDAC3 were recruited to the E2F binding site in the presence of androgen by ChIP with primers for E2F site amplification. (H and I) HDAC inhibitor (SAHA) reverses the androgen-mediated transcription repression of cyclin B1 promoter in PShTertAR cells (I) that is seen in PShTert cells (H). (J) SMRT serves as a corepressor for E2F4 repression in the luciferase assay with increased SMRT. The 130-bp cyclin B1 promoter reporter (only containing the E2F binding site and lacking ARE) was used in the assay. (K) SMRT knockdown by siRNA released the transcriptional repression by E2F4. (L and M) ChIP assay showed that the recruitment of acetyl-histone H3, acetyl-histone H4, and PolII to the cyclin B1 promoter region was dramatically reduced in the presence of androgen. UTR, untranslated region.
To further clarify the mechanism of recruitment of E2F1 and E2F4 on the E2F site and the ARE region upon androgen treatment, we performed ChIP with 130bp-LUC (containing the E2F site but without ARE) and 70bp-LUC (including ARE but excluding the E2F site). Luciferase reporters pGL34XAREE4 (containing 4 ARE sequences upstream of the E4 promoter) and pGL3-basic served as positive and negative controls, respectively. Using anti-AR antibodies, 70bp-LUC was enriched only in the presence of androgen, whereas 130bp-LUC showed no signal regardless of androgen addition (Fig. 5B). In E2F1 ChIP assays, the enrichment of 70bp-LUC was comparable to AR enrichment in the presence of androgen. At the same time, E2F1 showed dramatically little recruitment on 130bp-LUC in the presence of androgen (Fig. 5C). On the other hand, E2F4 was enriched on the 130bp-LUC but not the 70bp-LUC reporter in the presence of androgen (Fig. 5D). Together, these data further indicated that E2F1 associated with AR is recruited to the ARE region while E2F4 binds to the E2F site on the cyclin B1 promoter in the presence of androgen.
E2F4 is known to translocate from the cytoplasm to the nucleus in nonproliferative cells (12, 31). Thus, we examined the localization of E2F1 and E2F4 in the presence and absence of androgen using subcellular fractionation. E2F4 partitioned to the cytoplasm in the absence of androgen. In the presence of androgen, E2F4 translocated from cytoplasm to nucleus (Fig. 5E). E2F4 localized to the cytoplasm in PShTert cells either in the presence or absence of androgen (see Fig. S6D in the supplemental material). E2F4 subcellular localization mirrored that of AR, since both proteins exhibited primarily cytoplasmic localization in the absence of androgen and nuclear localization in the presence of androgen. As expected, E2F1 localized to the nucleus irrespective of androgen treatment (Fig. 5E).
E2F4 recruits corepressor complex repressing cyclin B1 transcription.
E2F4 recruits corepressors to repress transcription (21, 27). The corepressor complexes include Sin3A/Sin3B/HDAC1/HDAC2, HDAC3/GPS2/TBL1/NCoR/SMRT and HDAC4/HDAC5/SMRT/NCoR complexes (6). SMRT and NCoR have been shown to be corepressors for AR (9). Here, we performed coimmunoprecipitation experiments to determine whether E2F4 interacts with Sin3B, SMRT, NCoR, HDAC1, and HDAC3. Reciprocal co-IP experiments revealed an interaction between E2F4 and SMRT (Fig. 5F) in the presence of androgen. Similarly, we were able to detect interactions between E2F4 and HDAC3 (Fig. 5F) but not HDAC1. Thus, it is highly likely that E2F4 represses cyclin B1 transcription through the recruitment of an SMRT/HDAC3 corepressor complex (Fig. 5F). We also performed coimmunoprecipitations to examine the interaction between AR and E2F4 or SMRT. The interaction between AR and E2F4 was only detected in the absence of androgen. AR also showed a strong interaction with SMRT in the absence of androgen, and the interaction was diminished in the presence of androgen (Fig. 5F).
To verify the possibility that E2F4, in association with SMRT/HDAC3, represses cyclin B1 transcription, we measured factor occupancy on 130bp-LUC with E2F4, SMRT, and HDAC3 antibodies in PShTertAR cells. The results indicated that SMRT and HDAC3 were recruited to E2F binding sites in the presence of androgen, confirming that E2F4 is present in a complex with SMRT/HDAC3 corepressors (Fig. 5G).
To link the functional relevance with physical interactions between E2F4 and the SMRT/HDAC3 corepressor complex, we determined whether the HDAC inhibitor SAHA could reverse the androgen-mediated transcriptional repression of cyclin B1 in luciferase assays using the 200-bp cyclin B1 promoter reporter. The inhibition of cyclin B1 transcription by androgen was reversed by the addition of SAHA (Fig. 5H and I; also see Fig. S7A in the supplemental material) in PShTertAR cells. To determine whether SMRT acts as a corepressor for E2F4 repression, we also performed a luciferase assay with the 130-bp cyclin B1 promoter reporter (containing only the E2F binding site but without the ARE). As expected, the luciferase activity decreased with increasing amounts of SMRT, indicating that SMRT serves as a corepressor for E2F4 repression (Fig. 5J). Furthermore, we showed that siRNAs targeting SMRT relieved the transcriptional repression by E2F4 (Fig. 5K) and partially reversed the proliferation block of PShTertAR cells in the presence of androgen (see Fig. S7B).
To evaluate histone modification patterns in the cyclin B1 promoter region, we performed ChIP assays with anti-acetylated-histone H3, anti-acetylated-histone H4, and anti-PolII in PShTertAR cells in the presence and absence of androgen. Upon androgen treatment, the acetylation of histones H3 and H4 dramatically diminished concomitantly with PolII recruitment to the cyclin B1 promoter region, confirming gene repression (Fig. 5L and M).
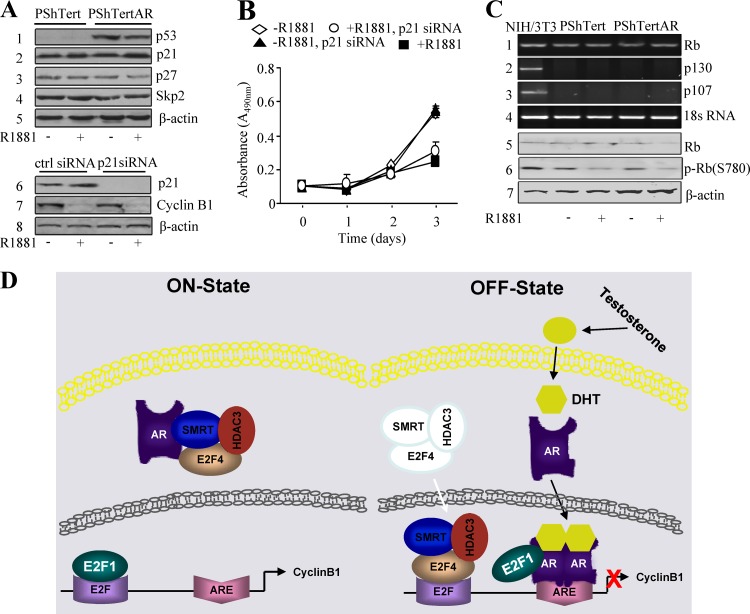
pRB family proteins and the cell cycle inhibitors p21 and p53 do not mediate the regulation of AR-mediated cyclin B1 repression.
Several studies demonstrated that p21WAF1/Cip1, as well as p53, are involved in AR signaling (23, 24, 30) and the regulation of cyclin B1 expression (14). In PShTertAR cells, endogenous p21 increases by 22% in the presence of androgen according to Western blotting (Fig. 6A, upper). Luciferase assays with a p21 promoter-containing reporter also showed increased transactivation by androgen, which is consistent with Western blotting results (see Fig. S7C in the supplemental material). However, p21 knockdown in these cells did not alter the expression of cyclin B1 (Fig. 6A, lower) and did not affect cell proliferation regardless of androgen addition (Fig. 6B), indicating that p21 is not involved in androgen-mediated repression in prostate stromal cells. In addition to activating transcription, p53 is able to repress the transcription of certain genes, including cyclin B1, through interaction with Sp1 bound to the cyclin B1 promoter (13). In PShTert cells, p53 showed minimal expression (Fig. 6A). In PShTertAR cells, the overexpression of AR led to the upregulation of p53 in both androgen-containing and androgen-free medium (Fig. 6A). Since the expression of cyclin B1 is only inhibited in androgen medium, p53-mediated repression of cyclin B1 expression in PShTertAR cells upon androgen treatment does not appear to be a primary mechanism for growth suppression.
Fig 6.
Model for cooperative regulation of cyclinB1 expression by AR, E2F1, and E2F4 in cell growth suppression. (A) Increased p53 expression was detected in PShTertAR cells for both androgen and androgen-free medium. A slight increase (2-fold) of p21 expression (lane 2) was obtained in the presence of androgen. There is no change in the levels of p27 (lane 3) and Skp2 (lane 4) expression. The p21 knockdown by siRNA did not alter the expression of cyclin B1 in PShTertAR cells regardless of androgen status (lanes 5 and 6). (B) The p21 knockdown did not increase the rate of PShTertAR cell proliferation, especially in the presence of androgen. (C) The p107 and p130 proteins were not detectable by RT-PCR in PShTert or PShTertAR cells in the presence or absence of androgen (lanes 2 and 3). Rb was expressed in PShTert and PShTertAR cells as determined by RT-PCR and Western blotting (lanes 1 and 5). However, the levels of phosphorylated Rb also decreased in stromal cells negative for AR at similar levels (lane 6). (D) Model for cooperative regulation of cyclin B1 expression by AR, E2F1, and E2F4 in cell growth suppression. In the absence of androgen, E2F1 binds to the E2F binding site, while AR, E2F4, SMRT, and HDAC3 may form a complex in the cytoplasm. In the presence of androgen, AR, E2F4, SMRT, and HDAC3 translocate into the nucleus. AR binds to the ARE on the cyclin B1 promoter and interacts with E2F1, exposing the E2F binding site for E2F4 binding. E2F4 recruits the corepressors SMRT and HDAC3 and exerts the repression of cyclin B1 transcription, leading to the inhibition of cell proliferation.
E2F activity is regulated through the pRB family of proteins, which includes p107 and p130 (8, 34). The p107 and p130 proteins were not detectable (Fig. 6C) by RT-PCR analysis in PShTert or PShTertAR cells in the presence or absence of androgen. pRB was expressed in PShTert and PShTertAR cells as determined by RT-PCR and Western blotting (Fig. 6C). However, the levels of phosphorylated pRB also decreased in stromal cells negative for AR at similar levels (Fig. 6C). In addition, coimmunoprecipitations failed to reveal an interaction between pRB and E2F1 (data not shown), and ChIP assays indicated that pRB was not recruited to the promoter region of cyclin B1 (see Fig. S7D in the supplemental material). These results suggest that the regulation of E2F function is variable in different cell types.
DISCUSSION
In this study, we describe a novel type of cooperative negative regulation of cyclin B1 expression, involving hormone receptor AR, transcription factors E2F1 and E2F4, and corepressor proteins SMRT and HDAC3, in prostate stromal cells.
Androgen and AR inhibit prostate stromal cell growth, and growth suppression is mediated by the repression of cyclin B1 at the transcriptional level. Using a combination of approaches, we demonstrate that AR directly inhibits cyclin B1 expression through a novel ARE on the cyclin B1 promoter in AR-positive prostate stromal cells. To address the underlying mechanism, the recruitment of E2F1 and E2F4 to the E2F binding site of the cyclin B1 promoter was investigated. We showed that E2F1 is recruited to the E2F binding site of the cyclin B1 promoter in the absence of androgen in PShTertAR cells, and it was displaced from the E2F binding site upon androgen treatment (Fig. 6D). Interestingly, ChIP experiments also indicated that the transcriptional repressor E2F4 was specifically recruited to the E2F binding site of the cyclin B1 promoter only in the presence of androgen. This result indicates that E2F1 and E2F4 competitively bind to the E2F site to activate or repress the cyclin B1 gene under different conditions. Reciprocal coimmunoprecipitation experiments showed the interaction between AR and E2F1 only in the presence of androgen (Fig. 6D). In the presence of androgen, both E2F1 and E2F4 are localized in the nucleus. The preferential binding of AR to E2F1 rather than E2F4 may be determined at least in part by the affinities between AR and E2F1 or E2F4. Under these conditions, E2F1 binds to the cyclin B1 promoter through interactions with AR at the ARE and does not occupy the E2F binding site of the cyclin B1 promoter. Concomitantly, E2F4 translocates from the cytoplasm to the nucleus, binds to the E2F site, and subsequently represses cyclin B1 transcription and cell cycle progression. Upon androgen treatment, both AR and E2F4 showed nuclear localization in PShTertAR cells. AR could facilitate (directly or indirectly) the nuclear localization of E2F4 upon R1881 treatment. It will be of great interest to determine the precise mechanism for the nuclear localization of E2F4 by AR in the future. SMRT (5, 10) has been reported to function as an AR corepressor. The overexpression of SMRT inhibits dihydrotestosterone-dependent transactivation by AR and further suppresses the anti-androgen (flutamide)-mediated inhibition of AR activity (17). In our study, coimmunoprecipitation experiments revealed that SMRT complexes with E2F4 instead of AR. Functional analysis also indicated that SMRT serves as a corepressor for the E2F4 transcriptional repression of cyclin B1 in PShTertAR cells. Thus, SMRT interacts with E2F4 rather than AR to suppress gene expression in the presence of androgen in AR-positive prostate stromal cells, suggesting a novel mechanism for cell type-specific E2F family member recruitment to the cyclin B1 promoter.
We propose that the E2F4 replacement of E2F1 on the cyclin B1 promoter is facilitated by a two-step, androgen-dependent mechanism (Fig. 6D). First, AR interacts with E2F1 and removes it from the E2F binding site at the cyclin B1 promoter. Second, E2F4 translocates into the nucleus in the presence of androgen, allowing it to occupy the vacated E2F binding site. Since we detected interactions between E2F4 and SMRT and between E2F4 and HDAC3, we speculate that E2F4 recruits an SMRT/HDAC3 complex to the cyclin B1 promoter in the presence of androgen.
We observed interactions between AR and E2F4 only in the absence of androgen. Similarly, SMRT showed strong interaction with AR in androgen-free conditions but minimal interaction in the presence of androgen. Co-IPs also suggested interaction between E2F4 and SMRT or HDAC3 both in the presence and absence of androgen (Fig. 5H). Given that AR, SMRT, HDAC3, and E2F4 localized in the cytoplasm in the absence of androgen, we speculate that unliganded AR, E2F4, SMRT, and HDAC3 exist in the same cytoplasmic complexes.
One other interesting finding is that the E2F regulation of cyclin B1 expression is not likely to be mediated through pRB family proteins. Although pRB is expressed, p107 and p130 proteins were not detected. Rb and its phosphorylated forms are detected in PShTertAR cells in the presence or absence of androgen. The changes are similar to that in PShTert cells. Although p21 and p53 are well-established cell cycle regulators, our studies failed to implicate them in the regulation of prostate stromal cell proliferation (Fig. 6B), and they may not play a primary role in androgen-dependent growth suppression in prostate stromal cells.
Epithelial AR is essential for the initiation and progression of PCa. Stromal AR also plays a critical role in hormonal carcinogenesis of the prostate (7). The dysregulation of stromal-epithelial interactions mediated by stromal AR signaling contributes not only to the initiation and progression of PCa but also to late stages of carcinogenesis (37). PShTert and PShTertAR have been characterized as myofibroblastic in nature with the endogenous expression of vimentin (see Fig. S7E in the supplemental material). In this study, we describe a novel type of cooperative negative regulation of cyclin B1 expression involving hormone receptor AR, transcription factors E2F1 and E2F4, and corepressor proteins, including SMRT and HDAC3, in PshTertAR cells. It will be interesting to stably suppress cyclin B1 to test a long-term effect on PCa growth. In addition, it will be worthwhile to investigate the molecular signals that mediate the cross-talk between epithelial and stromal cells and the consequent control of cyclin B1 expression in stromal cells. We examined the regulation of cyclin B1 by androgens in LNCaP cells (PCa epithelial cells) and did not observe the negative regulation of cyclin B1 by androgens. Interestingly, androgen increased the transactivation of the cyclin B1 promoter in epithelial cells (see Fig. S7F) but showed repression with the same reporter in stromal cells, suggesting that the regulation of cyclin B1 by AR is cell type specific. Stromal and epithelial AR signaling therefore exerts different effects on the regulation of cyclin B1.
In summary, our data suggest a model for the androgen-mediated repression of cyclin B1 transcription and cell proliferation (Fig. 6D). In the absence of androgen, E2F4, SMRT, and HDAC3 exist in complexes with unliganded AR located in the cytoplasm. E2F1 binds to the E2F site on the promoter of cyclin B1 and activates the transcription of cyclin B1, promoting cell cycle progression. In the presence of androgen, activated AR forms a complex with E2F1 and removes it from the E2F binding site. At the same time, E2F4, SMRT, and HDAC3 translocate from the cytoplasm to the nucleus and bind to the available E2F binding site in the cyclin B1 promoter, subsequently blocking the transcription of cyclin B1 and repressing cell proliferation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael J. Garabedian and Zhengxin Wang for helpful discussions of the experiments. The Department of Defense PCa Biorepository Network (PCBN) is thanked for tissue requisition.
This study is supported by NIH 1U01CA149556-01, DOD PCRP (PC080010 and PC111624), NYUSOM CTSI (1UL1RR029893), and NYUSOM Center of Excellence on Urologic Disease Fund to P.L.; DOD (PC051346) and NYU Molecular Oncology and Immunology Training grant (T32 CA009161) for predoctoral fellowships to G.D.; DOD postdoctoral fellowship (PC081578) to Y.R.L.; and NIH R01 grant to B.D. (5R01 CA077245-10).
Footnotes
Published ahead of print 16 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Balciunaite E, et al. 2005. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 25: 8166–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balk SP, Knudsen KE. 2008. AR, the cell cycle, and prostate cancer. Nucl. Recept. Signal. 6: e001 doi:10.1621/nrs.06001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bandara LR, Buck VM, Zamanian M, Johnston LH, La Thangue NB. 1993. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 12: 4317–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brinkmann AO, et al. 1999. Mechanisms of androgen receptor activation and function. J. Steroid Biochem. Mol. Biol. 69: 307–313 [DOI] [PubMed] [Google Scholar]
- 5. Chen JD, Evans RM. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377: 454–457 [DOI] [PubMed] [Google Scholar]
- 6. Cress WD, Seto E. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184: 1–16 [DOI] [PubMed] [Google Scholar]
- 7. Cunha GR, Hayward SW, Wang YZ, Ricke WA. 2003. Role of the stromal microenvironment in carcinogenesis of the prostate. Int. J. Cancer 107: 1–10 [DOI] [PubMed] [Google Scholar]
- 8. Dyson N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12: 2245–2262 [DOI] [PubMed] [Google Scholar]
- 9. Hodgson MC, et al. 2005. The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J. Biol. Chem. 280: 6511–6519 [DOI] [PubMed] [Google Scholar]
- 10. Horlein AJ, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377: 397–404 [DOI] [PubMed] [Google Scholar]
- 11. Humbert PO, et al. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14: 690–703 [PMC free article] [PubMed] [Google Scholar]
- 12. Iaquinta PJ, Lees JA. 2007. Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 19: 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Innocente SA, Lee JM. 2005. p53 is a NF-Y- and p21-independent, Sp1-dependent repressor of cyclin B1 transcription. FEBS Lett. 579: 1001–1007 [DOI] [PubMed] [Google Scholar]
- 14. Innocente SA, Lee JM. 2005. p73 is a p53-independent, Sp1-dependent repressor of cyclin B1 transcription. Biochem. Biophys. Res. Commun. 329: 713–718 [DOI] [PubMed] [Google Scholar]
- 15. Kooistra A, Romijn JC, Schroder FH. 1997. Stromal inhibition of epithelial cell growth in the prostate; overview of an experimental study. Urol. Res. 25(Suppl. 2):S97–S105 [DOI] [PubMed] [Google Scholar]
- 16. Li Y, et al. 2008. Decrease in stromal androgen receptor associates with androgen-independent disease and promotes prostate cancer cell proliferation and invasion. J. Cell Mol. Med. 12: 2790–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liao G, et al. 2003. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem. 278: 5052–5061 [DOI] [PubMed] [Google Scholar]
- 18. Linja MJ, Visakorpi T. 2004. Alterations of androgen receptor in prostate cancer. J. Steroid Biochem. Mol. Biol. 92: 255–264 [DOI] [PubMed] [Google Scholar]
- 19. Liu M, et al. 2009. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc. Natl. Acad. Sci. U. S. A. 106: 19035–19039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu S, Liu M, Epner DE, Tsai SY, Tsai MJ. 1999. Androgen regulation of the cyclin-dependent kinase inhibitor p21 gene through an androgen response element in the proximal promoter. Mol. Endocrinol. 13: 376–384 [DOI] [PubMed] [Google Scholar]
- 21. Lu Z, et al. 2006. E2F-HDAC complexes negatively regulate the tumor suppressor gene ARHI in breast cancer. Oncogene 25: 230–239 [DOI] [PubMed] [Google Scholar]
- 22. Moras D, Gronemeyer H. 1998. The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol. 10: 384–391 [DOI] [PubMed] [Google Scholar]
- 23. Nantermet PV, et al. 2004. Identification of genetic pathways activated by the androgen receptor during the induction of proliferation in the ventral prostate gland. J. Biol. Chem. 279: 1310–1322 [DOI] [PubMed] [Google Scholar]
- 24. Nesslinger NJ, Shi XB, DeVere White RW. 2003. Androgen-independent growth of LNCaP prostate cancer cells is mediated by gain-of-function mutant p53. Cancer Res. 63: 2228–2233 [PubMed] [Google Scholar]
- 25. Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- 26. Peng Y, et al. 2008. Distinct nuclear and cytoplasmic functions of androgen receptor cofactor p44 and association with androgen-independent prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 105: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rayman JB, et al. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rowland BD, Bernards R. 2006. Re-evaluating cell-cycle regulation by E2Fs. Cell 127: 871–874 [DOI] [PubMed] [Google Scholar]
- 29. Shand RL, Gelmann EP. 2006. Molecular biology of prostate-cancer pathogenesis. Curr. Opin. Urol. 16: 123–131 [DOI] [PubMed] [Google Scholar]
- 30. Shenk JL, et al. 2001. p53 represses androgen-induced transactivation of prostate-specific antigen by disrupting hAR amino- to carboxyl-terminal interaction. J. Biol. Chem. 276: 38472–38479 [DOI] [PubMed] [Google Scholar]
- 31. Takahashi Y, Rayman JB, Dynlacht BD. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14: 804–816 [PMC free article] [PubMed] [Google Scholar]
- 32. Trapman J, Brinkmann AO. 1996. The androgen receptor in prostate cancer. Pathol. Res. Pract. 192: 752–760 [DOI] [PubMed] [Google Scholar]
- 33. Trimarchi JM, et al. 1998. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc. Natl. Acad. Sci. U. S. A. 95: 2850–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trimarchi JM, Lees JA. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3: 11–20 [DOI] [PubMed] [Google Scholar]
- 35. Wang D, et al. 2011. Protein signatures for classification and prognosis of gastric cancer a signaling pathway-based approach. Am. J. Pathol. 179: 1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H, et al. 2011. Crocidolite asbestos-induced signal pathway dysregulation in mesothelial cells. Mutat. Res. 723: 171–176 [DOI] [PubMed] [Google Scholar]
- 37. Wang Y, et al. 2001. A human prostatic epithelial model of hormonal carcinogenesis. Cancer Res. 61: 6064–6072 [PubMed] [Google Scholar]
- 38. Wu L, et al. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414: 457–462 [DOI] [PubMed] [Google Scholar]
- 39. Ye F, et al. 2009. The effect of Scutellaria baicalensis on the signaling network in hepatocellular carcinoma cells. Nutr. Cancer 61: 530–537 [DOI] [PubMed] [Google Scholar]
- 40. Zalmas LP, et al. 2008. DNA-damage response control of E2F7 and E2F8. EMBO Rep. 9: 252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang DY, et al. 2009. Proteomics, pathway array and signaling network-based medicine in cancer. Cell Div. 4: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu W, Giangrande PH, Nevins JR. 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23: 4615–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.