Abstract
Intestinal epithelial cells and antigen-presenting cells orchestrate mucosal innate immunity. This study investigated the role of bacterial DNA in modulating epithelial and bone marrow-derived antigen-presenting cells (BM-APCs) and subsequent T-lymphocyte responses. Murine MODE-K epithelial cells and BM-APCs were treated with DNA from either Bifidobacterium breve or Salmonella enterica serovar Dublin directly and under coculture conditions with CD4+ T cells. Apical stimulation of MODE-K cells with S. Dublin DNA enhanced secretion of cytokines from underlying BM-APCs and induced interleukin-17 (IL-17) and gamma interferon (IFN-γ) secretion from CD4+ T cells. Bacterial DNA isolated from either strain induced maturation and increased cytokine secretion from BM-APCs. Conditioned medium from S. Dublin-treated MODE-K cells elicited an increase in cytokine secretion similar to that seen for S. Dublin DNA. Treatment of conditioned medium from MODE-K cells with RNase and protease prevented the S. Dublin-induced increased cytokine secretion. Oral feeding of mice with B. breve DNA resulted in enhanced levels of colonic IL-10 and transforming growth factor β (TGFβ) compared with what was seen for mice treated with S. Dublin DNA. In contrast, feeding mice with S. Dublin DNA increased levels of colonic IL-17 and IL-12p70. T cells from S. Dublin DNA-treated mice secreted high levels of IL-12 and IFN-γ compared to controls and B. breve DNA-treated mice. These results demonstrate that intestinal epithelial cells are able to modulate subsequent antigen-presenting and T-cell responses to bacterial DNA with pathogenic but not commensal bacterial DNA inducing effector CD4+ T lymphocytes.
INTRODUCTION
The apical surface of the intestinal epithelium is in close contact with luminal microbes and their products, while the basolateral surface is closely associated with underlying immune cells. Recognition of luminal microbes and the ability to discriminate between harmful and commensal organisms is a seminal role of the innate immune system. Recent studies suggest that epithelial cells in collusion with dendritic cells and macrophages act together to orchestrate appropriate mucosal innate immune responses and maintain gut homeostasis (21). It has been shown that epithelial cells release soluble factors such as thymic stromal lymphopoietin (TSLP) that condition underlying dendritic cells to prime CD4+ T helper cells to undergo type-2 (Th2) differentiation and secrete interleukin-4 (IL-4) and IL-10 in response to live and irradiated bacteria (22, 30). In addition, Rimoldi et al. (22) have shown that gut dendritic cells are conditioned by factors derived from epithelial cells to become “noninflammatory.” These dendritic cells are unable to release IL-12, highlighting the role of epithelial cells in maintaining homeostasis.
Epithelial-microbial interactions involve innate immune receptors such as Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD) molecules (1). TLRs play critical roles in early innate recognition and inflammatory responses against pathogenic microbes. Gut epithelial cells express TLR1 to TLR9, as well as Nod1 and Nod2 (19). TLR9 is activated by unmethylated CpG-containing DNA found in high concentrations in microbes, some double-stranded DNA viruses, and synthetic CpG oligodeoxynucleotides (ODN) (1, 9). In epithelial cells, TLR9 is expressed on the cell surface and is upregulated in response to the presence of DNA from pathogenic strains (5, 15). The epithelial response to bacterial DNA is dependent upon whether TLR9 is activated on the apical or the basolateral surface, and also upon the specific bacterial strain (5, 12, 15). TLR9 signaling is associated with gut homeostasis, where loss of TLR9 signaling results in increased susceptibility to colonic inflammation (17). Further, TLR9 has been shown to be involved in the anti-inflammatory effects of probiotics (13, 20), demonstrating a role for probiotic bacteria DNA as an active factor that imparts a beneficial effect. More recently, Hall et al. (8) have shown a role for TLR9 signaling and gut flora DNA in modulating the levels of T-regulatory and T-effector cells in the gut.
Based on these findings, we hypothesized that the differential response of epithelial cells to DNA isolated from commensal and pathogenic bacterial strains would be communicated through dendritic cell signaling and result in altered T-cell differentiation. Here we demonstrate that in vitro treatment of intestinal epithelial cells (IECs) with bacterial DNA from Salmonella enterica serovar Dublin, a pathogenic strain, results in the release of soluble mediators which act to increase cytokine secretion from underlying bone marrow-derived antigen-presenting cells and enhance IL-17 secretion from CD4+ T cells. In contrast, epithelial cells treated with bacterial DNA isolated from Bifidobacterium breve did not release any mediators that altered cytokine secretion from underlying bone marrow-derived antigen-presenting cells. In vivo experiments showed B. breve DNA to have an anti-inflammatory effect on colonic tissues, while S. Dublin DNA induced IL-12p70, IL-17, and gamma interferon (IFN-γ). These findings highlight the role of bacterial DNA and epithelial cells in modulating gut innate immune responses through antigen-presenting cells and CD4+ T-cell responses. Further, these findings provide a mechanism by which the intestinal epithelium can distinguish between commensal/probiotic organisms and pathogens, thus providing a robust immune response or eliciting tolerance.
MATERIALS AND METHODS
Bacterial strains and preparation of DNA.
Salmonella enterica serovar Dublin strain Lane (ATCC 15480) was selected for these studies as a pathogenic bacterium due to the ability of its DNA to induce IL-8 secretion from intestinal epithelial cells (12). Bifidobacterium breve Y8 (VSL Pharmaceutics) was selected as a commensal bacterium due to the ability of its DNA to reduce basal IL-8 secretion (12). Bacteria were inoculated at 0.18% (vol/vol) into 25 ml of Mann-Rogosa Sharpe broth (Difco 0370-17-3) and grown statically overnight (18 to 20 h) at 37°C. For DNA isolation, cells were centrifuged at 11,700 × g for 10 min, washed with SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and resuspended in 0.01 sodium phosphate buffer with 20% sucrose and 2.5 mg/ml lysozyme for 45 min at 37°C followed by lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 500 mg pronase B, 1% SDS, pH 8) for 30 min at 37°C. DNA was extracted by adding an equal volume of 1:1 buffer-saturated phenol and chloroform to the bacterial solution. The mixture was spun for 5 min at 4,000 × g and the aqueous layer removed. The extraction was repeated until no interface was visible. Traces of phenol were removed with chloroform, and the salt concentration was adjusted by the addition of a 1/10 volume of sodium acetate (3 M), pH 5.2. DNA was precipitated with cold 100% ethanol, washed with 70% ethanol, and resuspended in sterile Tris-EDTA buffer. A postisolation treatment to remove endotoxin was performed with a 5% volume of Tx114 (Promega) at 4°C for 30 min. Further isopropanol precipitation was done to isolate endotoxin-free DNA. Only preparations with endotoxin levels not exceeding 0.1 ng/μg DNA were used. The concentration and purity of DNA preparations were confirmed by measuring optical density at 260 nm (OD260) absorbance and OD260/280 ratio and by running agarose gel electrophoresis as previously described (12). DNA preparations were assayed for endotoxin using the Pyrochrom amebocyte assay (DP0704; Associates of Cape Cod Inc., East Falmouth, MA). DNase-treated preparations were used as controls in all experiments. DNA preparations were incubated overnight at 37° C with 5 μg/ml DNase I (Sigma-Aldrich) in the presence of 5 mM MgCl2. DNA depletion was confirmed by agarose gel electrophoresis and ethidium bromide staining as previously described (12). In some experiments, DNA methylation was carried out using 37.5 U/ml SssI DNA methylase (New England BioLabs) at 37°C for 1 h. Reaction mixtures were inactivated by heating to 85°C for 10 min prior to application onto BM-APCs.
Isolation of bone marrow-derived antigen-presenting cells.
Antigen-presenting cells were isolated from the long bones of male 129/SvEv mice using a modified protocol (16). Briefly, long bones of mice were isolated and cleaned and then placed in 70% ethyl alcohol (EtOH) for 5 min and transferred to calcium and magnesium-free phosphate-buffered saline (CMF-PBS). The ends of bones were clipped and the marrow was rinsed with 10 ml CMF-PBS. Cells were spun at 2,000 × g for 5 min, resuspended in CMF-PBS, and plated at 2 × 106 in 10 ml of medium. Cells were maintained at 37°C with 5% CO2 in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (1,000 units/ml), streptomycin (1 μg/ml), l-glutamine (2 mM) (Invitrogen), and 1,000 U/ml mouse recombinant granulocyte-macrophage colony-stimulating factor (mrGM-CSF; R&D Systems). After 10 days of incubation, GM-CSF was removed from the medium. Prior to experimentation, cells were analyzed by flow cytometry. The majority of the cells were CD11c+++ (80%) with ∼20% of cells being CD11b+. A small population of cells (<2%) was positive for CD19, indicating the presence of a small population of contaminating B cells. Following 24 h of incubation, flow cytometry analysis demonstrated a low percentage of CD11bdull CD8α+ cells, with half the population of cells being major histocompatibility complex class II positive (MHC-II+), and of those, the major proportion (>90%) being CD11c+ CD11b+ and CCR6+. This population was defined as bone marrow-derived antigen-presenting cells (BM-APCs) and was used in all experiments. For experimental studies, BM-APCs were plated at a concentration of 1 × 106 cells/ml without mrGM-CSF and incubated for 24 h with either B. breve or S. Dublin DNA (2 to 50 μg/ml) or conditioned medium (CM) derived from MODE-K cells.
MODE-K cells.
The murine intestinal epithelial cell line MODE-K used for these studies was kindly provided by K. Croitoru (University of Toronto, Toronto, ON). This cell line was derived from C3H/He mice, exhibits morphological and phenotypic characteristics similar to those of normal enterocytes, and expresses MHC-II molecules and TLR9 (7, 26). The cells were maintained at 37°C and 5% CO2 in Dulbecco minimal essential medium (DMEM) high glucose with HEPES and l-glutamine (Invitrogen) supplemented with 10% heat-inactivated FBS. To prepare conditioned medium, MODE-K cells were rinsed with Mg+- and Ca2+-free PBS and then incubated with HEPES and l-glutamine DMEM media (Invitrogen) containing either B. breve or S. Dublin DNA for 1 h. For experiments, cells were cultured in Transwells (Costar) and allowed to reach confluence. Monolayers were rinsed with Mg+- and Ca2+-free PBS, and medium was replaced with fresh warm high-glucose medium with HEPES and l-glutamine DMEM for 6 h. Medium was removed from the basolateral compartment and used in subsequent experiments to stimulate BM-APCs. To investigate the mechanism of signal communication, basolateral conditioned medium from DNA-stimulated or nonstimulated MODE-K cells was added to BM-APCs after enzymatic treatment (RNA digestion, protein digestion) or added to BM-APCs in conjunction with antibodies (anti-IL-6 and/or anti-TSLP; R&D Biosystems). Enzymatic digestions prior to BM-APC treatment were as follows: RNase digestion with 10 μg/ml RNase A (Qiagen) at 37°C for 1 h, protease digestion with 0.5 μg/ml Proteinase K (Ambion) at 37°C for 2 h. All reaction mixtures were inactivated by heating to 85°C for 10 min (RNase) or 20 min (Proteinase K) prior to their application onto BM-APCs. For the antibody treatments, BM-APCs were pretreated with 100 ng/ml anti-IL-6 or 100 ng/ml anti-TSLP for 30 min prior to the addition of MODE-K conditioned media containing the respective antibody. DMEM high glucose with 1× nonessential amino acids (NEAA), 5% FBS, 15 mM HEPES, and 100 U/ml penicillin and 100 μg/ml streptomycin was used to create the conditioned medium and as a control on the APCs. All medium treatments were applied to the BM-APCs for 24 h prior to supernatant collection for secreted cytokine analysis.
Coculture experiments.
MODE-K cells were plated on the apical surface of Transwells coated with mouse type IV collagen (Sigma-Aldrich) at a concentration of 7 × 105 cells per Transwell and cultured for 2 to 4 days until confluent monolayers were formed. MODE-K monolayers were treated apically with either S. Dublin or B. breve (50 μg/ml) with or without BM-APCs (1 × 106 cells/ml) in the basolateral compartment.
Analysis of cytokines and cell surface molecules.
BM-APCs were scraped from the bottom of the well and the underside of the apical Transwell surface and rinsed with CMF-PBS, and the suspension was spun at 2,000 × g for 5 min. The supernatant was collected and frozen at −70°C for measurement of cytokine release by the MesoScale platform. Levels of cytokines were analyzed by the MesoScale platform. The remaining cell pellets were split into two samples for measurement of cell viability and expression of surface markers. Briefly, cells were washed twice in PBS and fixed in 0.5% paraformaldehyde until analysis by flow cytometry. Cell surface expression of CD11c, MHC-II IA/IE, CD11b, CD8α, and CCR6 (eBioscience) was analyzed using a laser flow cytometer (FACSCanto). Data were analyzed using FCS Express software (De Novo Software). For measurement of cell viability, cells were incubated with 7-amino actinomycin D (7-AAD) and the proportions of positive and negative labeled cells determined by flow cytometry. There was no difference in viability between the groups (data not shown).
Measurement of epithelial resistance.
Barrier function was measured in MODE-K cells grown to confluence on Transwells by measuring electrical conductance and the spontaneous potential across the monolayer with an Evom Volt-Ohm meter and an STX-2 electrode set (World Precision Instruments, Sarasota, FL).
CD4+ T-cell isolation.
Male 129/SvEv mice between 8 and 12 weeks old were euthanized and the spleens harvested under sterile conditions. The organs were homogenized between sterile frosted glass slides and rinsed with 5 ml of iMag buffer (CMF-PBS with bovine serum albumin [BSA] and 0.09% sodium azide) (BD Bioscience, Mississauga, ON). Cell isolates were strained through a sterile 100-μm-pore-size strainer and centrifuged at 200 × g for 5 min. Cell pellets were resuspended in iMag buffer (BD Bioscience) and red blood cells lysed. Splenocytes were counted and diluted to 20 × 106 cells/ml in iMag buffer in preparation for negative selection. Cells were selected using the BD CD4+ T-cell enrichment protocol as per the manufacturer's instructions (BD Bioscience, Mississauga, ON). Briefly, cells were incubated with biotinylated CD4+ T-cell negative selection enrichment cocktail for 30 min on ice and then centrifuged, and supernatant was removed. Biotin-coated magnetic beads were added to the cells and placed in the BD iMagnet for 5 min. Cells in the fraction not bound to the magnet were harvested as CD4+ T cells and were ∼90 to 95% pure. T lymphocytes were cultured at 37°C with 5% CO2 at a concentration of 5 × 105 cells/ml with a 1:1 concentration of anti-CD3- and CD28-coated Dynabeads (Invitrogen).
T-cell coculture systems.
BM-APCs were plated at 5 × 104 cells/ml in medium and incubated for 24 h with either S. Dublin or B. breve DNA (50 μg/ml). CD4+ T lymphocytes were then added to the culture in a 1:10 ratio of T lymphocyte/BM-APC with 5 × 105 cells/ml αCD3- and CD28-coated Dynabeads (Invitrogen). Cells were incubated for 48 h and supernatants harvested for analysis. For triculture systems, MODE-K cells were plated on Transwell supports (Costar) coated with mouse type IV collagen (Sigma-Aldrich) at a concentration of 7 × 105 cells per Transwell and cultured for 2-4 days until complete monolayers were formed. MODE-K monolayers were then cocultured with BM-APCs (5 × 104 cells/ml) in the basolateral compartment, and S. Dublin or B. breve DNA (50 μg/ml) was applied to the apical surface. After 24 h, the MODE-K cells and the apical medium were removed to prevent any contact between T lymphocytes and MODE-K cells. CD4+ T lymphocytes were added to the culture in a 1:10 ratio of T lymphocyte/BM-APC with 5 × 105 cells/ml anti-CD3- and CD28-coated Dynabeads (Invitrogen). Cells were incubated for 48 h and supernatants harvested for analysis.
In vivo studies with bacterial DNA.
Male 129/SvEv mice aged between 8 and 12 weeks were raised in conventional housing. The mice were gavaged daily with a total of 50 μg S. Dublin or B. breve DNA dissolved in 30 μl water/day or vehicle (saline) for 7 days. On day 8, the mice were euthanized and the colons, spleens, and mesenteric lymph nodes (MLNs) harvested. The colons were snap-frozen in liquid nitrogen for subsequent homogenization for cytokine analysis, while the spleens and MLNs were processed to isolate CD4+ T cells as described above (see “CD4+ T-cell isolation” above). Frozen colonic tissue was homogenized in PBS containing 0.05% Tween 20 and protease inhibitor cocktail (P8340; Sigma-Aldrich). The homogenate was centrifuged at 10,000 × g for 10 min at 4°C. Supernatants were collected and frozen for enzyme-linked immunosorbent assay (ELISA), while the remaining homogenized tissue was air dried for dry weight measurements. ELISA was performed for the following cytokines (using Duo Set from R&D Systems): IL-10, IL-12p70, IL-17, IFN-γ, transforming growth factor β (TGFβ), and tumor necrosis factor alpha (TNF-α).
Characterization and culturing of CD4+ cells.
Freshly isolated CD4+ T-cell isolates were fixed with FoxP3 fixatives for 30 min on ice (BD Bioscience). CD4+ T cells were washed with iMag buffer, centrifuged at 200 × g for 5 min, and resuspended in freezing medium (90% FCS and 10% dimethyl sulfoxide [DMSO]). Cells were placed in a −80°C freezer until further use for flow cytometry characterization. Thawed CD4+ T-cell isolates were washed with fluorescence-activated cell sorter (FACS) buffer (1% BSA, 0.01% sodium azide in PBS) and centrifuged at 200 × g for 5 min. They were resuspended in prewarmed FoxP3 permeabilizing buffer (BD Bioscience) and incubated for 30 min in the dark at 37°C. Cells were washed twice in FACS buffer. Resuspended cells were incubated with either CD4-peridinin chlorophyll protein (PerCP)/Cy5.5, IFN-γ-phychoerythrin (PE), IL-10-fluorescein isothiocyanate (FITC), or CD4-PerCP/Cy5.5, IL-17A-PE, FoxP3-Alexa 647, or appropriate antibody isotypes (eBiosciences). Cells were incubated for 30 min in the dark at room temperature, washed twice with FACS buffer, and resuspended for FACS analysis using BD FACSCanto (BD BioSciences). Negatively selected CD4+ T cells were obtained from mice as described above. Freshly isolated CD4+ T cells were cultured in RPMI 1640 medium (Gibco) supplemented with 5% FBS and incubated on CD3-coated plates for 4 days. Supernatants were collected and cytokines measured via ELISA.
Statistical analysis.
Data were tested for normality of distribution and analyses performed using the statistical software SigmaStat (Jandel Corporation, San Rafael, CA). Differences between means were evaluated using analysis of variance, paired t tests, or a nonparametric Mann-Whitney rank sum test where appropriate. Specific differences were tested using Dunn's or Bonferroni's post hoc analysis.
RESULTS
MODE-K cells differentiate between bacterial DNAs.
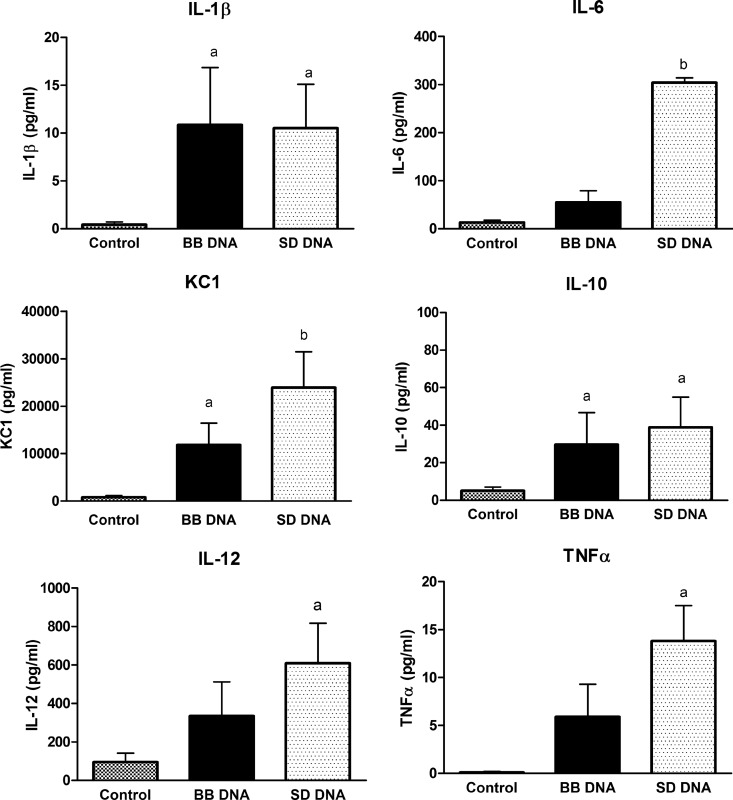
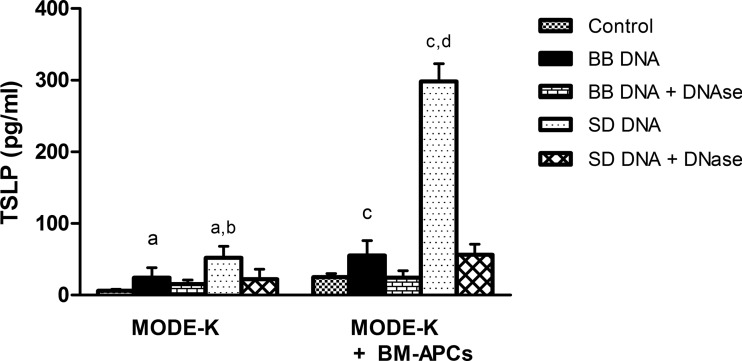
To confirm that mouse intestinal epithelial cells (MODE-K) were able to differentiate between bacterial DNAs in the same manner as we have previously shown in human epithelial cells (12), MODE-K cells were incubated with DNA from Salmonella Dublin or Bifidobacterium breve and cytokine secretion was measured. DNAs from these bacterial strains were selected for this study, as we have previously demonstrated clear differences in epithelial cell responses to S. Dublin, a pathogen, and B. breve, a commensal (12). MODE-K cells secreted IL-1β, IL-6, KC, and IL-10 when stimulated with S. Dublin or B. breve DNA, while S. Dublin DNA also induced IL-12 and TNF-α (Fig. 1). Similar to the differential response seen in human HT-29 cells (12), MODE-K cells released amounts of IL-6 and KC in response to S. Dublin DNA that were significantly higher than those released in response to B. breve DNA (Fig. 1). MODE-K cells also secreted higher amounts of TSLP (thymic stromal lymphopoietin) in response to S. Dublin DNA than in response to B. breve DNA (Fig. 2). DNase treatment prevented the S. Dublin and B. breve DNA-induced increase in TSLP secretion (Fig. 2). MODE-K cells did not secrete either IL-18 or TGFβ in response to either bacterial DNA (data not shown).
Fig 1.
Cytokine secretion from MODE-K cells in response to Salmonella Dublin (SD) DNA and Bifidobacterium breve (BB) DNA. MODE-K cells were treated apically for 24 h with either S. Dublin or B. breve DNA (50 μg/ml). MODE-K cells responded to S. Dublin and B. breve DNA with increased IL-1β, KC, and IL-10 secretion. S. Dublin DNA elicited significantly more IL-6 and KC secretion than B. breve DNA. Bars are mean ± standard error of the mean (SEM) of triplicate measurements made in two separate experiments. a, P < 0.05 compared with control; b, P < 0.05 compared with B. breve DNA and control.
Fig 2.
TSLP secretion from MODE-K cells and MODE-K cells coincubated with BM-APCs. MODE-K cells were treated apically for 24 h with either S. Dublin (SD) or B. breve (BB) DNA (50 μg/ml). MODE-K cells secreted barely detectable levels of TSLP in the absence of microbial stimuli. S. Dublin DNA induced more TSLP secretion than B. breve DNA. The presence of underlying BM-APCs resulted in enhanced TSLP secretion. Treatment with DNase prevented both B. breve and S. Dublin DNA-induced TSLP secretion. Bars are mean ± SEM of triplicate measurements made in two separate experiments. a, P < 0.05 compared with MODE-K control, B. breve DNA plus DNase, and S. Dublin plus DNase; b, P < 0.05 compared with B. breve DNA; c, P < 0.05 compared with MODE-K and BM-APC control, B. breve DNA plus DNase; d, P < 0.05 compared with B. breve DNA and S. Dublin DNA plus DNase.
Epithelial cells modulate BM-APC cytokine secretion in response to apical DNA.
In the gut, bacterial DNA would normally interact with an intact epithelial cell layer which would then release signals to underlying dendritic cells or macrophages (21). In subsequent experiments, we examined the response of underlying BM-APCs when epithelial cells were stimulated on the apical surface with bacterial DNA. Intestinal MODE-K cells were cocultured with BM-APCs in the underlying basolateral compartment of Transwells and stimulated apically with B. breve or S. Dublin DNA. Cytokines secreted into the basolateral compartment were measured. Under these conditions, there would be limited direct contact between bacterial DNA and BM-APCs, but there would be direct contact between epithelial and immune cells due to the presence of a confluent epithelial monolayer. When S. Dublin DNA was added to the apical compartment of intestinal MODE-K cells, enhanced secretion of IL-1β, IL-6, TNF-α, and IL-10 was seen (Table 1). In contrast, the presence of B. breve DNA in the apical compartment did not elicit any response from underlying BM-APCs. Interestingly, the presence of BM-APCs resulted in a significantly larger amount of TSLP secretion from MODE-K cells in response to S. Dublin DNA (Fig. 2).
Table 1.
Cytokine release from MODE-K and BM-APCs in the presence and absence of bacterial DNAb
| Cell type | Condition | Concn (pg/ml) of indicated cytokine released |
||||
|---|---|---|---|---|---|---|
| IL-1β | IL-6 | IL-10 | IL-12 | TNF-α | ||
| BM-APCs + MODE-K | Medium | 14.5 ± 0.8 | 20.2 ± 5.3 | 18.6 ± 3.9 | 17.7 ± 4.2 | 7.0 ± 1.6 |
| B. breve DNA | 19.6 ± 4.8 | 73.5 ± 27.7 | 30.2 ± 6.7 | 17.8 ± 2.3 | 8.5 ± 2.2 | |
| S. Dublin DNA | 52.8 ± 16.6a | 6,090 ± 652a | 108.6 ± 28a | 16.8 ± 3.6 | 796.6 ± 277.5a | |
P < 0.05 compared with MODE-K-medium and B. breve DNA-treated MODE-K.
n = 3 biological replicates for all groups.
Response of BM-APCs to bacterial DNA.
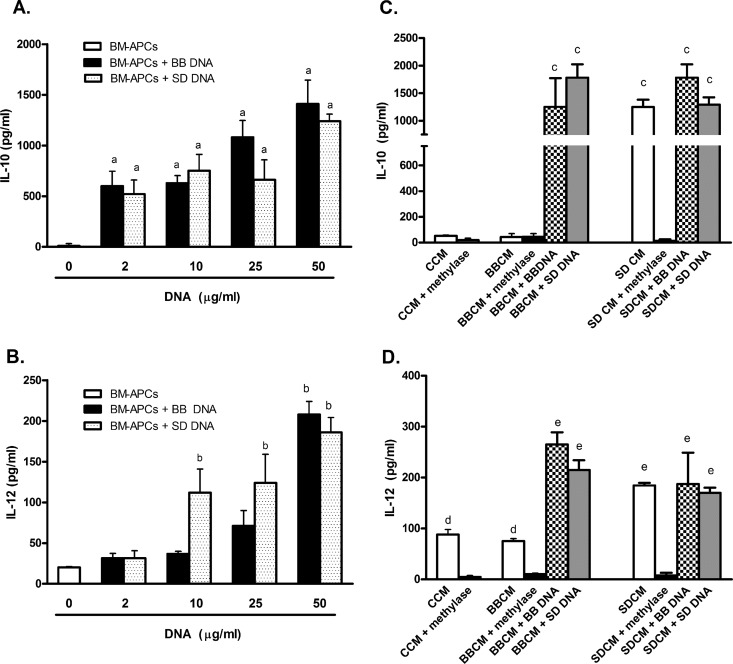
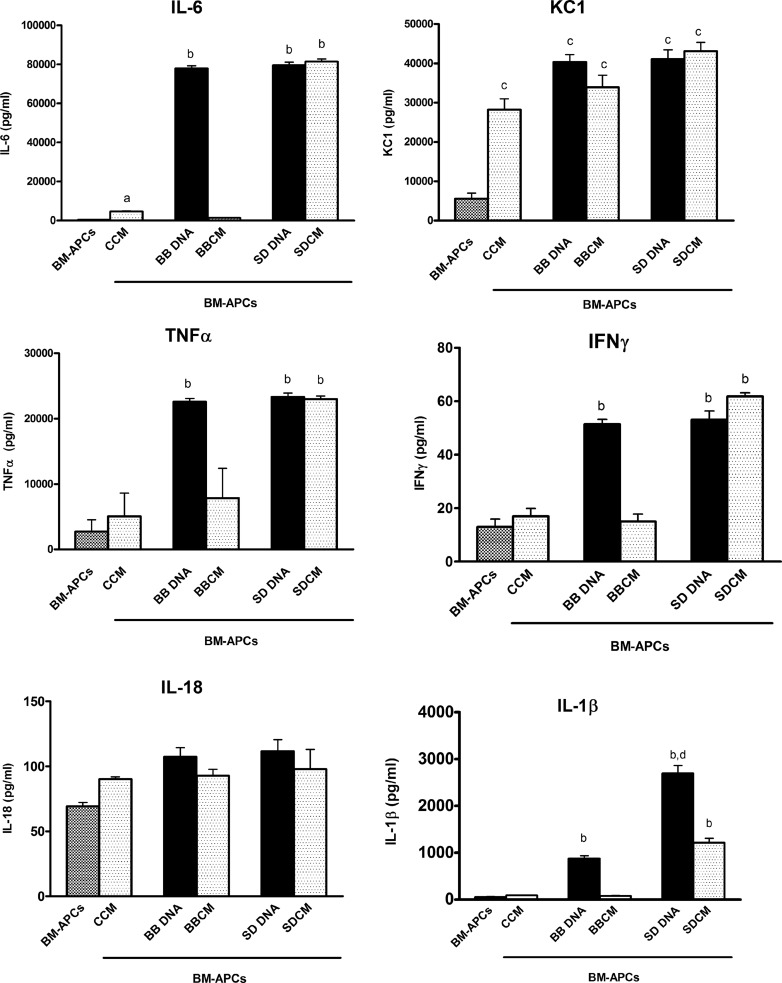
As diffusion of bacterial DNA to the basolateral compartment over the course of the incubation could have occurred, resulting in a stimulation of BM-APCs by S. Dublin DNA, direct effects of bacterial DNA on BM-APCs were determined. BM-APCs were incubated with increasing amounts of either B. breve or S. Dublin DNA, and the release of IL-10 and IL-12 was measured. Following 24 h of incubation with bacterial DNA, all BM-APCs were CD11c+, CD11b+, MHC++, and CCR6+. A similar increase in IL-10 secretion occurred in response to both B. breve and S. Dublin DNA at all concentrations tested (2 to 50 μg/ml) (Fig. 3A). For IL-12, a concentration dependence was seen, with S. Dublin DNA, but not B. breve DNA, eliciting increased IL-12 secretion at 10 and 25 μg/ml (Fig. 3B). At the highest concentration tested (50 μg/ml), S. Dublin and B. breve DNA elicited similar levels of IL-12 secretion. In the presence of both B. breve and S. Dublin DNA, the secretion of IL-10 exceeded the secretion of IL-12p70. In order to determine if B. breve and S. Dublin DNA elicited different levels of secretion of other cytokines, further experiments were carried out using the highest concentration of bacterial DNA (50 μg/ml). Similar to what has been reported previously (29), BM-APCs responded to both B. breve and S. Dublin DNA with an increased secretion of IL-6, KC, TNF-α, and IFN-γ (Fig. 4). However, S. Dublin DNA induced a high level of IL-1β secretion compared with B. breve DNA (Fig. 4). These results indicated that in the absence of epithelial cells, BM-APCs reacted to both B. breve and S. Dublin DNA with increased cytokine secretion.
Fig 3.
IL-10 (A) and IL-12p70 (B) secretion from BM-APCs in response to S. Dublin and B. breve DNA (2 to 50 μg/ml) and conditioned medium from S. Dublin (SD)- and B. breve (BB)-treated MODE-K cells (C and D, respectively) after 24 h of incubation. (A) B. breve and S. Dublin DNA stimulated IL-10 secretion from BM-APCs at all concentrations tested. (B) S. Dublin DNA stimulated IL-12 secretion at low concentrations (10 μg/ml) compared with B. breve DNA. Both S. Dublin and B. breve DNA stimulated similar amounts of IL-12 secretion at a concentration of 50 μg/ml. (C) Conditioned medium from nontreated (CCM) and B. breve-treated MODE-K (BBCM) cells did not induce IL-10 secretion from BM-APCs. Conditioned medium from S. Dublin-treated MODE-K (SDCM) cells induced similar amounts of IL-10 secretion as did direct stimulation with S. Dublin DNA. BBCM did not have any effect on B. breve DNA- or S. Dublin DNA-induced IL-10 secretion. SDCM also did not have any effect on B. breve- or S. Dublin-induced IL-10 secretion. Methylation of S. Dublin DNA prevented the SDCM-induced increase in IL-10 secretion. (D) CCM and BBCM induced similar amounts of IL-12 secretion, while SDCM induced higher levels of secretion. Methylation of S. Dublin and B. breve DNA prevented the increase in IL-12 secretion. BBCM did not affect IL-12 secretion in response to either B. breve DNA or S. Dublin DNA. SDCM cells stimulated similar amounts of IL-12 as did direct stimulation with S. Dublin DNA. SDCM did not affect IL-12 secretion in response to either B. breve DNA or S. Dublin DNA. Bars are mean ± SEM of triplicate measurements made in two separate experiments. a, P < 0.05 compared with nonstimulated BM-APCs; b, P < 0.05 compared with nonstimulated BM-APCs, B. breve and S. Dublin DNA (2 μg/ml), and B. breve DNA (10 μg/ml); c, P < 0.02 compared with CCM, CCM plus methylase, BBCM, BBCM plus methylase, and SDCM plus methylase; d, P < 0.05 compared with CCM plus methylase, BBCM plus methylase, and SDCM plus methylase; e, P < 0.02 compared with CCM, CCM plus methylase, BBCM, BBCM plus methylase, and SDCM plus methylase.
Fig 4.
Cytokine secretion from BM-APCs treated with S. Dublin and B. breve DNA (50 μg/ml) and conditioned medium from nonstimulated (CCM), B. breve-treated (BBCM), and S. Dublin-treated (SDCM) MODE-K cells. B. breve and S. Dublin DNA stimulated IL-6, KC, TNF-α, IFN-γ, and IL-1β secretion from BM-APCs. S. Dublin DNA induced high amounts of IL-1β secretion compared with B. breve DNA. SDCM induced the same response as S. Dublin DNA, with increased IL-6, KC, TNF-α, IFN-γ, and IL-1β. BBCM induced only KC secretion. CCM induced IL-6 and KC secretion. Bars are mean ± SEM of triplicate measurements made in two separate experiments. a, P < 0.05 compared with BM-APCs alone and BBCM-treated BM-APCs; b, P < 0.01 compared with BM-APCs, CCM-treated BM-APCs, and BBCM-treated BM-APCs; c, P < 0.05 compared with BM-APCs; d, P < 0.02 compared with B. breve DNA-treated BM-APCs.
Effects of DNA on MODE-K cell viability and resistance.
In that BM-APCs responded to direct stimulation with B. breve and S. Dublin DNA but apical stimulation of MODE-K cells with only S. Dublin DNA resulted in cytokine secretion from underlying BM-APCs, it was possible that S. Dublin DNA induced alterations in monolayer permeability, allowing a greater diffusion of DNA into the basolateral compartment. To test this, the effects of S. Dublin and B. breve DNA on cellular integrity and epithelial resistance were measured with 7-AAD and transepithelial electrical resistance, respectively. We found no effects of either S. Dublin or B. breve DNA on MODE-K cellular viability as measured by 7-AAD (data not shown) or resistance as measured in Transwells (control, 67.7 ± 0.5; S. Dublin DNA, 68.1 ± 0.7; B. breve DNA, 69.9 ± 0.5 Ω/cm2) and equal amounts of DNA were detected in the basolateral compartment under all conditions (data not shown). These findings would suggest that while the presence of bacterial DNA in the basolateral compartment likely contributes to the stimulation of BM-APCs, an increased amount of S. Dublin DNA in the basolateral compartment was not likely the underlying mechanism by which apical stimulation of MODE-K cells with S. Dublin DNA induces cytokine secretion from BM-APCs.
Bacterial DNA modulates BM-APC cytokine secretion through epithelial cell supernatants.
The next set of experiments was done to determine if MODE-K cells released soluble signals to underlying BM-APCs in response to bacterial DNA. Conditioned medium was removed from the basolateral compartment of Transwells underlying MODE-K cells that had been treated with bacterial DNA and added to BM-APCs. Conditioned medium from nonstimulated and B. breve-treated MODE-K cells induced similar levels of IL-12 (Fig. 3D) and KC (Fig. 4) secretion from BM-APCs, indicating that under basal conditions and in response to commensal bacteria, epithelial cell secretions can modulate cytokine release from BM-APCs. However, conditioned medium from S. Dublin-stimulated MODE-K cells resulted in an increase in IL-12 (Fig. 3D), IL-10 (Fig. 3C), IL-6, TNF-α, IFN-γ, and IL-1β (Fig. 4) that was significantly larger than the response seen to conditioned medium from either unstimulated MODE-K cells or MODE-K cells treated with B. breve DNA. Methylation of DNA in conditioned medium prevented the increase in IL-10 (Fig. 3C) and IL-12 (Fig. 3D), confirming a role for DNA in the stimulation.
Epithelial cell supernatants do not suppress cytokine secretion.
Our findings that B. breve DNA would induce IL-10 and IL-12 secretion when directly added to BM-APCs, yet when B. breve DNA diffused through MODE-K monolayers and contacted BM-APCs there was no response, would suggest that epithelial cells are able to suppress BM-APC responses. Thus, to determine if conditioned medium from B. breve DNA-treated MODE-K cells would suppress the response of BM-APCs to either S. Dublin or B. breve DNA, BM-APCs were incubated with S. Dublin or B. breve DNA with or without conditioned medium, and IL-10 and IL-12 secretion was measured. As seen in Fig. 3C and D, neither CM from B. breve-treated cells (BBCM) nor CM from S. Dublin-treated cells (SDCM) altered BM-APC responses when stimulated directly with bacterial DNA. These findings would suggest that epithelial cells are required to be present in order to suppress BM-APC responses to commensal bacterial DNA. However, the data also clearly indicate that epithelial cells appear to release soluble compounds in response to pathogenic bacterial DNA that can activate BM-APCs at least in part through cell contact-independent mechanisms.
Identification of soluble effectors.
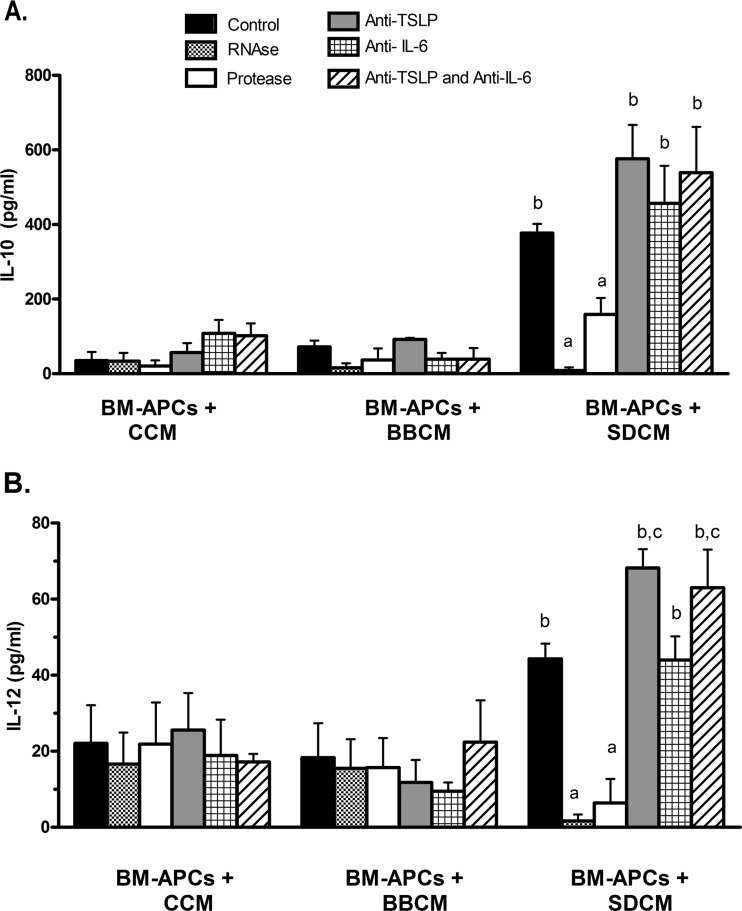
The major difference in cytokine secretion by MODE-K cells in response to S. Dublin and B. breve DNA was in the levels of IL-6 and TSLP. Thus, to determine if these were the factors in S. Dublin-conditioned medium that were responsible for inducing IL-10 and IL-12 secretion from BM-APCs, a series of experiments was carried out using antibodies to neutralize IL-6 and TSLP. As seen in Fig. 5, neutralization with specific antibodies against IL-6 had no effect on S. Dublin-induced IL-10 or IL-12 cytokine secretion. In contrast, the neutralization of TSLP resulted in enhanced IL-12 secretion in response to SDCM. A major stimulus of dendritic cell activation is through the recognition of RNA and DNA through TLR3, TLR7, and TLR9. Thus, to determine if RNA or other proteins were involved in the stimulation of BM-APCs, conditioned medium from nonstimulated and B. breve and S. Dublin DNA-treated MODE-K cells were incubated with RNase and then added to BM-APCs. As seen in Fig. 5, depletion of extracellular RNA from the conditioned medium completely prevented the SDCM induction of IL-10, IL-12, and IL-6 secretion from BM-APCs. Treatment of CM with Proteinase K protease also partially reduced IL-10 and IL-12 secretion.
Fig 5.
IL-10 (A) and IL-12p70 (B) secretion from BM-APCs incubated with conditioned medium from MODE-K cells. Treatment of SDCM with RNase or protease significantly reduced the SDCM-induced secretion of IL-10 and IL-12. Neutralization of TSLP had no effect on the SDCM-induced increase in IL-10 secretion but did enhance the IL-12 response. Neutralization of IL-6 had no effect on the SDCM-induced increase in either IL-10 or IL-12 secretion. Bars are means ± SEM. n = 4 for all conditions. a, P < 0.05 compared with BM-APCs plus SDCM, BM-APCs plus SDCM plus anti-TSLP, BM-APCs plus SDCM plus anti-IL-6, BM-APCs plus SDCM plus anti-TSLP and anti-IL-6; b: P < 0.01 compared with all BM-APCs plus CCM and all BM-APCs plus BBCM.
DNA from pathogenic bacteria induces T-effector cell differentiation.
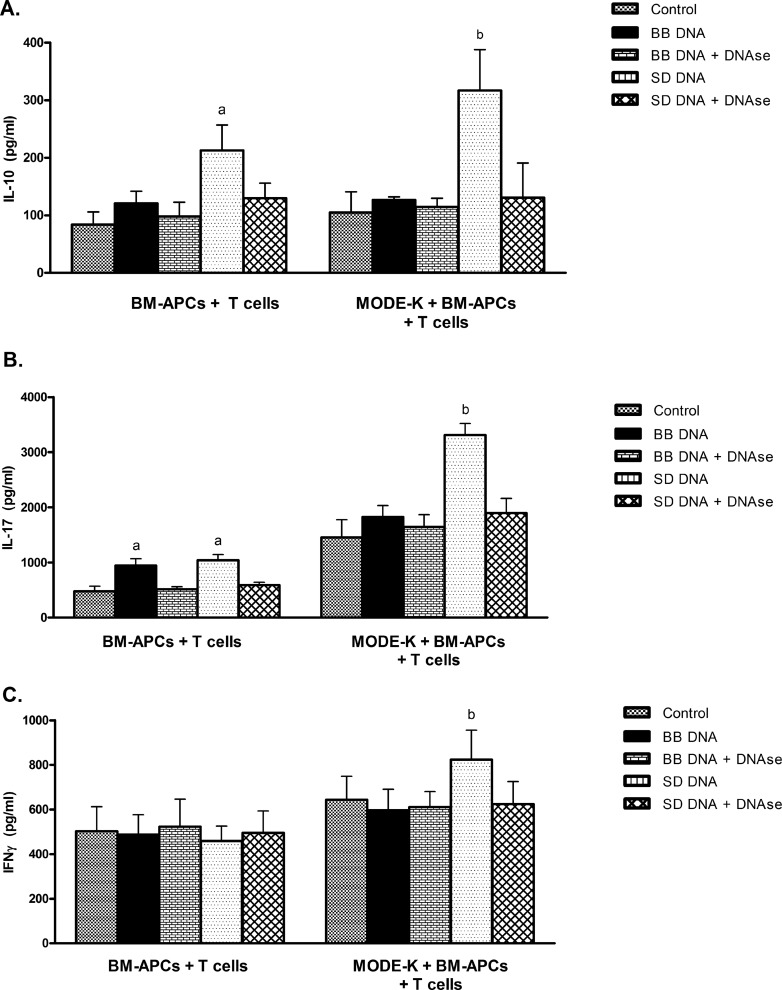
To examine whether stimulation of epithelial cells with bacterial DNA would be translated into an altered CD4+ T-cell response, a triculture system was used whereby MODE-K cells were incubated with apical bacterial DNA and then with BM-APCs and CD4+ T cells. In the absence of MODE-K cells, BM-APCs and CD4+ T cells incubated with S. Dublin DNA responded with enhanced IL-10 (Fig. 6A) and IL-17 (Fig. 6B) secretion and to B. breve DNA with enhanced IL-17 (Fig. 6B). IFN-γ secretion was not affected by either B. breve or S. Dublin DNA (Fig. 6C). However, in the presence of MODE-K cells, S. Dublin DNA stimulation of the epithelial cells resulted in an increase in secretion of IL-10, IL-17, and IFN-γ, while B. breve DNA treatment of MODE-K did not alter cytokine secretion. Treatment with DNase prevented the increased cytokine secretion in response to both S. Dublin and B. breve DNA.
Fig 6.
Cytokine secretion from B. breve (BB)- and S. Dublin (SD)-treated cocultures of BM-APCs incubated with CD4+ T cells with or without MODE-K cells. MODE-K cells were treated with B. breve or S. Dublin DNA followed by the addition of BM-APCs to the basolateral compartment. MODE-K cells were then removed and CD4+ T cells added. In the absence of MODE-K cells, BM-APCs were treated with B. breve or S. Dublin DNA for 24 h and then CD4+ T cells were added in a 1:10 ratio of T cells to BM-APCs with αCD3- and CD28-coated Dynabeads. Cells were incubated for 48 h and supernatants harvested for analysis. (A) IL-10 secretion was increased in response to S. Dublin DNA both in the presence and absence of MODE-K cells. (B) IL-17 secretion from CD4+ T cells in coculture with BM-APCs was increased in response to both bacterial DNAs. In the presence of MODE-K cells, S. Dublin DNA enhanced IL-17 secretion while B. breve DNA had no effect. (C) IFN-γ secretion was not altered by incubation of BM-APCs with bacterial DNA. In the presence of MODE-K cells, IFN-γ secretion was enhanced with S. Dublin DNA, but not in the presence of B. breve DNA (C). DNase treatment prevented the B. breve- and S. Dublin DNA-induced changes in cytokine secretion. Bars are means ± SEM. n = 4 for all conditions. a, P < 0.05 compared with BM-APCs plus T cells plus medium, BM-APCs plus T cells plus B. breve DNA, BM-APCs plus T cells plus B. breve DNA plus DNase, BM-APCs plus T cells plus S. Dublin DNA plus DNase; b, P < 0.05 compared with MODE-K plus BM-APCs plus T cells plus medium, MODE-K plus BM-APCs plus T cells plus B. breve DNA, MODE-K plus BM-APCs plus T cells plus B. breve DNA plus DNase, MODE-K plus BM-APCs plus T cells plus S. Dublin DNA plus DNase.
Salmonella Dublin promotes an inflammatory environment in the colon.
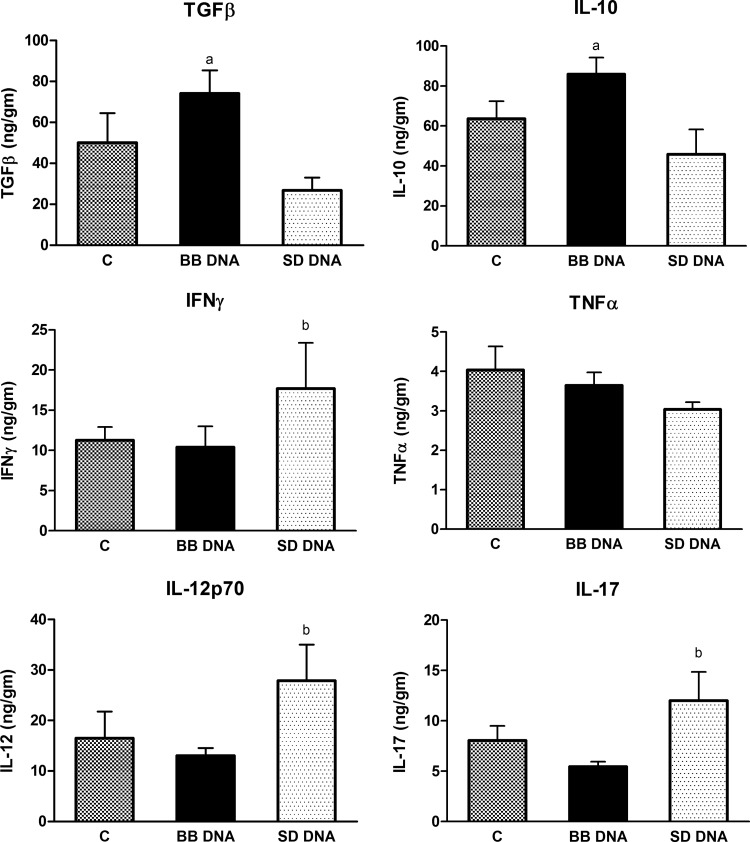
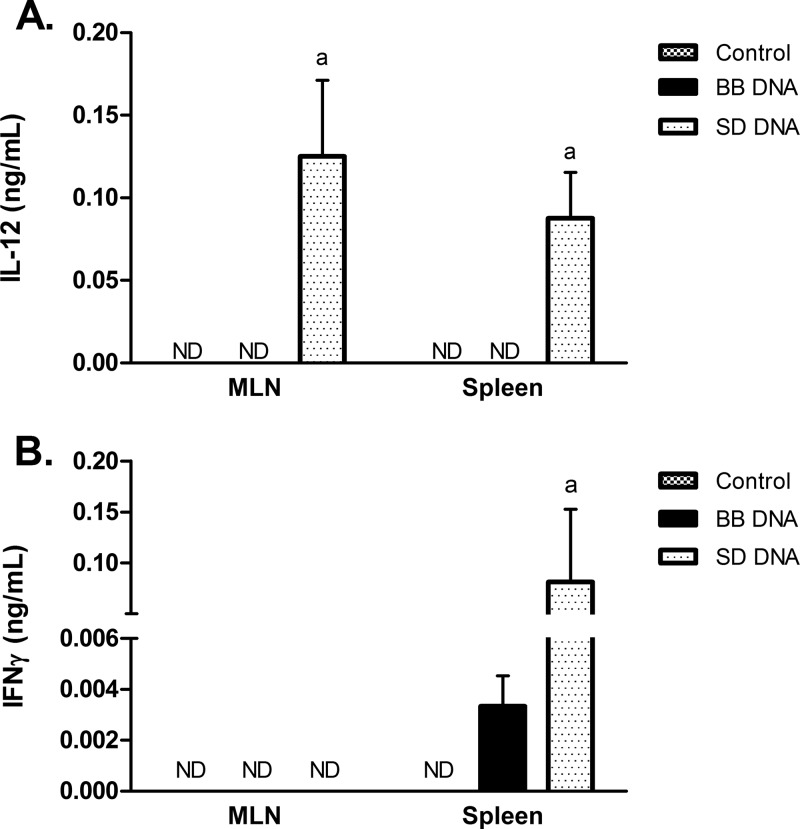
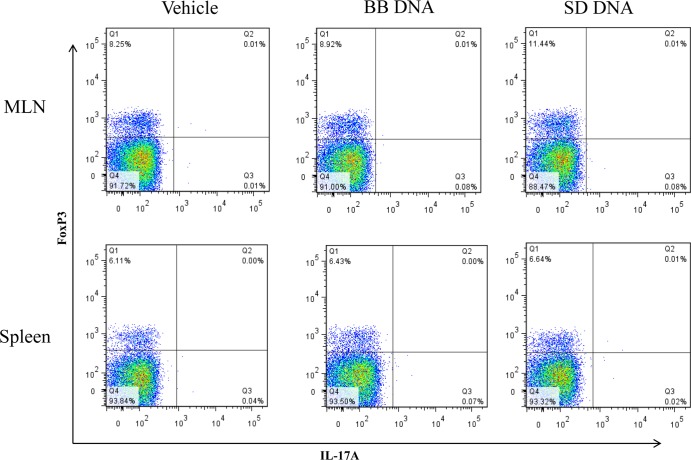
To determine if bacterial DNA had similar differential effects in vivo, wild-type 129/SvEv mice were treated with B. breve or S. Dublin DNA for 7 days. Colonic tissue from mice treated with B. breve DNA had high levels of IL-10 and TGFβ compared to what was seen for mice treated with S. Dublin DNA (Fig. 7). In contrast, S. Dublin DNA treatment increased the levels of IFN-γ, IL-12p70, and IL-17 in colonic tissues, while B. breve DNA-treated mice had similar levels as vehicle-treated mice (Fig. 7). There were no significant differences in the levels of TNF-α found among the three treatment groups. CD4+ T cells isolated from the mesenteric lymph nodes (MLNs) and spleens of mice treated with S. Dublin DNA secreted more IL-12 than controls and B. breve DNA-treated mice, whereas CD4+ T cells isolated from the spleens of S. Dublin DNA-treated mice secreted more IFN-γ than vehicle- or B. breve DNA-treated mice (Fig. 8). Analysis of CD4+ T cells isolated from MLNs showed increased FoxP3+ and Th17+ T cells in MLNs in the S. Dublin DNA-treated mice compared to what was seen for B. breve DNA-treated mice (Table 2; Fig. 9).
Fig 7.
Cytokine measurements in colonic tissue homogenates. Mice were gavaged for 7 days with 50 μg/ml of either B. breve (BB) or S. Dublin (SD) DNA or with vehicle. ELISA measurements of colonic homogenates found production of TGFβ and IL-10 in mice gavaged with B. breve DNA that was significantly higher than that with mice gavaged with S. Dublin DNA. Increased concentrations of IFN-γ, IL-12p70, and IL-17 were seen in S. Dublin DNA-treated mice compared with B. breve DNA-treated mice. No difference in TNF-α levels was seen among the 3 treatment groups. Bars are means ± SEM. n = 6 or 7 for all groups. a, P < 0.05 compared with S. Dublin DNA-treated mice; b, P < 0.05 compared with B. breve DNA-treated mice.
Fig 8.
IL-12 (A) and IFN-γ (B) secretion from CD4+ T cells isolated from mesenteric lymph nodes and spleens of wild-type 129/SvEv mice treated with either B. breve (BB) or S. Dublin (SD) DNA. Following 4 days of culturing, there were significant levels of IL-12 released from CD4+ T cells isolated from mice that had been treated with S. Dublin DNA. No IL-12 was detected from CD4+ T cells isolated from mice treated with either vehicle or B. breve DNA. Splenocytes isolated from S. Dublin DNA-treated mice also demonstrated increased secretion of IFN-γ compared with B. breve DNA-treated or control mice. Bars are means ± SEM. n = 5 or 6 mice for all groups. a, P < 0.05 compared with vehicle- or B. breve DNA-treated mice. ND, not detected.
Table 2.
CD4+ T-cell population isolated from the MLNs and spleens of mice gavaged with B. breve or S. Dublin DNAc
| Group | Population of CD4+ T cells (% of total) |
|||
|---|---|---|---|---|
| FoxP3+ cells |
Th17+ cells (×10−2) |
|||
| MLN | Spleen | MLN | Spleen | |
| Vehicle | 8.1 ± 0.4 | 5.8 ± 0.6 | 3.8 ± 0.7 | 1.4 ± 0.7 |
| B. breve DNA | 8.9 ± 0.2 | 6.9 ± 0.3 | 5.0 ± 1 | 3.0 ± 1 |
| S. Dublin DNA | 11.5 ± 0.3a | 7.2 ± 0.8 | 7.2 ± 1b | 2.6 ± 0.6 |
P < 0.05 compared to B. breve DNA and vehicle treatment in MLNs.
P < 0.05 compared to vehicle treated in MLNs.
n = 3 replicates.
Fig 9.
CD4+ T-cell population isolated from the mesenteric lymph nodes (MLNs) and spleens of mice gavaged with either B. breve (BB) or S. Dublin (SD) DNA. Representative plots showing percentages of FoxP3 and IL-17A T cells in the CD4+ gate.
DISCUSSION
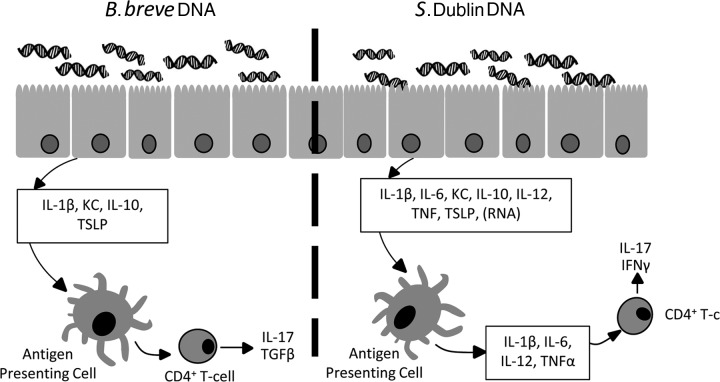
In this study, we show that apical stimulation of intestinal epithelial cells with bacterial DNA isolated from a pathogenic but not from a commensal bacterial strain results in the release of soluble mediators which effectively activate underlying BM-APCs. This activation of BM-APCs resulted in an increased production of proinflammatory cytokines and the induction of IL-17 and IFN-γ secretion from T cells. However, in the absence of intestinal epithelial cells, bone marrow-derived antigen-presenting cells (BM-APCs) did not discriminate between pathogenic and commensal bacterial DNA. Treatment of mice with pathogenic bacterial DNA resulted in an increase in tissue levels of IL-17 and IFN-γ along with increased percentages of CD4+ IL-17+ T cells in the mesenteric lymph nodes (MLNs). In contrast, in vitro stimulation of intestinal epithelial cells with bacterial DNA isolated from a commensal Bifidobacterium strain did not result in an increased IL-17 or IFN-γ secretion from BM-APCs, and treatment of mice with DNA from B. breve resulted in an enhanced tissue level of IL-10 and TGFβ. This highlights an important role for the ability of intestinal epithelial cells to recognize and respond appropriately to different bacterial DNAs in the modulation of mucosal immune responses (Fig. 10).
Fig 10.
Intestinal epithelial cells are constantly exposed to bacteria and its products such as DNA. Upon apical stimulation with bacterial DNA, intestinal epithelial cells (IECs) release IL-1β and neutrophil chemoattractant KC as well as anti-inflammatory cytokines such as thymic stromal lymphopoietin (TSLP) and IL-10. However, IECs distinguish among different strains of bacterial DNA by releasing a high amount of IL-6 and TNF-α in response to S. Dublin DNA stimulation compared with B. breve DNA stimulation. In addition to these cytokines, IECs also release other signaling molecules comprised of proteins as well as RNA. Upon S. Dublin DNA treatment, these signaling molecules affect underlining antigen-presenting cells (APCs), resulting in further release of IL-1β, IL-6, IL-12, and TNF-α. APCs treated with conditioned medium from IECs exposed to S. Dublin DNA stimulate naïve T cells to produce IL-17 and IFN-γ. In contrast, conditioned medium from IECs treated with BB DNA results in TGFβ-producing Th-17 cells.
Intestinal epithelial cells are constantly exposed to high numbers of both commensal and pathogenic microbes, and it is clear that intestinal epithelial cells respond differently to apical versus basolateral exposure and to different strains of bacterial DNA (5, 6, 12). Recognition of extracellular bacterial DNA by TLR9 or intracellular DNA through the inflammasome triggers signaling cascades in epithelial and immune cells that result in the activation of both innate and adaptive immune responses (18). The underlying mechanism by which epithelial cells respond differently to apical stimulation with pathogenic versus commensal DNA is not known but may involve the presence of inhibitory CpG motifs in commensal bacterial DNA such as seen in adenoviruses and synthetic oligonucleotides (14).
It is clear that epithelial cells are able to signal the presence of luminal pathogenic bacteria to underlying immune cells, and in this study we present evidence that this information can be found in isolated bacterial DNA. Studies using a similar coculture model as was used in our study demonstrated that apical stimulation of TLR9 and TLR4 on human epithelial cell lines enhanced the secretion of IL-12 in underlying peripheral blood mononuclear cells (PBMCs) via an epithelial cell-mediated mechanism (3, 24). In these studies, TLR4 ligation of epithelial cells drove an inflammatory response, while apical stimulation of TLR9 drove a regulatory Th1 effector immune response (3). Our results complement and extend these findings, clearly demonstrating that epithelial cells can, upon apical stimulation, drive underlying immune cells to either a tolerogenic or an inflammatory profile. In our in vitro studies, we found that S. Dublin DNA induced higher levels of secretion of TSLP, IL-6, and KC from MODE-K cells than B. breve DNA, with no differences in TGFβ secretion. We also found that while conditioned medium from MODE-K cells treated with B. breve DNA and nontreated MODE-K cells induced KC and IL-10 secretion from BM-APCs, conditioned medium from S. Dublin DNA-stimulated MODE-K cells induced IL-10, IL-12, IL-6, TNF-α, IFN-γ, KC, and IL-1β. Together, these findings stress that epithelial cells play a vital role in modulating mucosal immune responses.
Previous studies have shown that epithelial cells release soluble factors such as TGFβ, retinoic acid, and thymic stromal lymphopoietin (TSLP), which effectively stimulate the differentiation of dendritic cells into a tolerogenic profile and drive the development of CD4+ CD25+ Foxp3+ regulatory T cells (11, 21, 22). In addition, previous studies have shown that TSLP effectively suppresses the production of IL-12 and promotes a Th2 skewing response in dendritic cells (21, 22). We found that B. breve and S. Dublin DNA, when applied directly to BM-APCs, enhanced both IL-10 and IL-12 secretion. This is in agreement with previous studies showing bacterial DNA to induce cytokine secretion from bone marrow-derived dendritic cells (23, 29). Our results support the work of Yoshinaga et al. (29), where both Salmonella and Bifidobacterium DNAs effectively induced cytokine secretion from our bone marrow-derived dendritic cell population, as was shown for Escherichia coli DNA. However, when conditioned medium from MODE-K cells was applied to BM-APCs, only conditioned medium from Salmonella-treated MODE-K cells induced IL-12 secretion and, in the presence of antibodies against TSLP, IL-12 secretion increased compared to what was seen for controls. This would suggest that while TSLP did have a role in epithelial modulation of dendritic cell function, other molecules are also involved.
Dendritic cells recognize and respond to RNA and DNA through TLR3, TLR7, and TLR9 receptors. It is known that intestinal epithelial cells secrete exosomes that contain major histocompatibility complex class I and class II molecules, numerous proteins, and functional RNA (17, 25). Bacterial components such as peptidoglycan can also be found in exosomes (2), and further, mRNA content within exosomes can be modulated based on host cell status (4). Our finding that treatment of conditioned medium with RNase prevented the induction of IL-12 in BM-APCs suggests a possible role for bacterial DNA-mediated epithelial cell-derived exosomes in the modulation of antigen-presenting cell function. Further studies will be required to confirm this possibility.
In vivo experiments demonstrated that treatment of mice with S. Dublin DNA resulted in a great secretion of IL-12p70 compared with B. breve treatment of mice. This higher level of IL-12 secretion in the S. Dublin DNA-treated group was associated with enhanced secretion of IL-17 and IFN-γ. In contrast, B. breve DNA treatment enhanced IL-10 and TGFβ release. This corroborates our previous findings where treatment with pathogenic DNA stimulated proinflammatory cytokines, while exposure to probiotic DNA suppressed proinflammatory and/or stimulated anti-inflammatory cytokines (12).
Our data support the hypothesis of an epithelial cell-driven spatially dependent and strain-dependent gut response to microbial DNA and that the cytokine milieu created by epithelial and dendritic cells drives the resulting T-cell responses. Hall and colleagues (8) showed that dendritic cells stimulated with gut flora DNA suppressed the conversion of Treg cells and increased both Th1 and Th17 cells. Our results also illustrate an increase in IL-17 and increased Th-17+ CD4+ T cells in the MLNs of mice treated with S. Dublin DNA. Our finding of increased levels of FoxP3+ CD4+ T cells in the MLNs of mice treated with S. Dublin is in agreement with other studies in which a gradual increase in FoxP3+ Treg cells has been shown to occur at inflamed sites (10, 27). In our model, we did not see a change in FoxP3+ Treg population in the MLNs of mice treated with B. breve or vehicle controls, similar to what has been shown previously (8). These findings support the idea of Treg cells having a role in suppressing disease progression by abolishing the inflammatory effector function of pathogenic T cells.
It must be noted that these studies were carried out using isolated bacterial DNA from Salmonella Dublin and not the live organism. Salmonella species can cause diseases ranging from mild gastroenteritis to systemic typhoid fever-like infections (28). Salmonella infection induces both mucosal and systemic inflammation by several different pathways, including direct cellular invasion, release of toxins, and stimulation of the innate immune system (28). Our findings that the presence of Salmonella DNA itself can be identified by intestinal epithelial cells suggest that this could serve as an early warning to the mucosal immune system of the presence of pathogens within the lumen, resulting in a priming of the immune system for possible future infection. At the same time, mechanisms are in place for a lack of response to normal commensal organisms such as bifidobacteria.
In conclusion, intestinal epithelial cells can distinguish between beneficial and pathogenic microbes based on their DNA. This information is transmitted to underlying immune cells, which in turn orchestrate critical responses; thus, intestinal epithelial cells play a crucial role in maintaining homeostatic balance or mounting host defense.
ACKNOWLEDGMENTS
These studies were supported by the Crohn's and Colitis Foundation of Canada, Canadian Institutes for Health Research, Alberta Innovates, and the Alberta IBD Consortium.
We thank Matt Emberg for his technical assistance with these studies.
Footnotes
Published ahead of print 21 May 2012
REFERENCES
- 1. Akira S, Hemmi H. 2003. Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 85:85–95 [DOI] [PubMed] [Google Scholar]
- 2. Bu HF, Wang X, Tang Y, Koti V, Tan XD. 2010. Toll-like receptor 2-mediated peptidoglycan uptake by immature intestinal epithelial cells from apical side and exosome-associated transcellular transcytosis. J. Cell. Physiol. 222:658–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Kivit S, van Hoffen E, Korthagen N, Garssen J, Willemsen LE. 2011. Apical TLR ligation of intestinal epithelial cells drives a Th1-polarized regulatory or inflammatory type effector response in vitro. Immunobiology 216:518–527 [DOI] [PubMed] [Google Scholar]
- 4. Eldh M, et al. 2010. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One 5:e15353 doi:10.1371/journal.pone.0015353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ewaschuk JB, et al. 2007. Surface expression of Toll-like receptor 9 is upregulated on intestinal epithelial cells in response to pathogenic bacterial DNA. Infect. Immun. 75:2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghadimi D, Vrese M, Heller KJ, Schrezenmeir J. 2010. Effect of natural commensal-origin DNA on toll-like receptor 9 (TLR9) signaling cascade, chemokine IL-8 expression, and barrier integrity of polarized intestinal epithelial cells. Inflamm. Bowel Dis. 16:410–427 [DOI] [PubMed] [Google Scholar]
- 7. Gopal R, Birdsell D, Monroy FP. 2008. Regulation of toll-like receptors in intestinal epithelial cells by stress and Toxoplasma gondii infection. Parasite Immunol. 30:563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hall J, et al. 2008. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29:637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hemmi H, et al. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740–745 (Erratum, 409:646, 2001.) [DOI] [PubMed] [Google Scholar]
- 10. Huehn J, Hamann A. 2005. Homing to suppress: address codes for Treg migration. Trends Immunol. 26:632–636 [DOI] [PubMed] [Google Scholar]
- 11. Iliev ID, et al. 2009. Human intestinal epithelial cells promote the differentiation of tolerogenic dendritic cells. Gut 58:1481–1489 [DOI] [PubMed] [Google Scholar]
- 12. Jijon H, et al. 2004. DNA from probiotic bacteria modulates murine and human epithelial and immune function. Gastroenterology 126:1358–1373 [DOI] [PubMed] [Google Scholar]
- 13. Kingma SD, et al. 2011. Lactobacillus johnsonii N6.2 stimulates the innate immune response through Toll-like receptor 9 in Caco-2 cells and increases intestinal crypt Paneth cell number in biobreeding diabetes-prone rats. J. Nutr. 141:1023–1028 [DOI] [PubMed] [Google Scholar]
- 14. Krieg AM. 2006. Therapeutic potential of Toll-like receptor 9 activation. Nat. Rev. Drug Discov. 5:471–484 [DOI] [PubMed] [Google Scholar]
- 15. Lee J, Rachmilewitz D, Raz E. 2006. Homeostatic effects of TLR9 signaling in experimental colitis. Ann. N. Y. Acad. Sci. 1072:351–355 [DOI] [PubMed] [Google Scholar]
- 16. Lutz MB, et al. 1999. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 223:77–92 [DOI] [PubMed] [Google Scholar]
- 17. Mathivanan S, et al. 2010. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteomics 9:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muller T, Hamm S, Bauer S. 2008. TLR9-mediated recognition of DNA. Handb. Exp. Pharmacol. 2008:51–70 [DOI] [PubMed] [Google Scholar]
- 19. Otte JM, Podolsky DK. 2004. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am. J. Physiol. Gastrointest. Liver Physiol. 286:G613–G626 [DOI] [PubMed] [Google Scholar]
- 20. Plantinga TS, et al. 2011. Differential Toll-like receptor recognition and induction of cytokine profile by Bifidobacterium breve and Lactobacillus strains of probiotics. Clin. Vaccine Immunol. 18:621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rescigno M. 2008. Intestinal epithelial cells control dendritic cell function. J. Pediatr. Gastroenterol. Nutr. 46(Suppl. 1):E17–E19 [DOI] [PubMed] [Google Scholar]
- 22. Rimoldi M, et al. 2005. Monocyte-derived dendritic cells activated by bacteria or by bacteria-stimulated epithelial cells are functionally different. Blood 106:2818–2826 [DOI] [PubMed] [Google Scholar]
- 23. Theiner G, et al. 2008. TLR9 cooperates with TLR4 to increase IL-12 release by murine dendritic cells. Mol. Immunol. 45:244–252 [DOI] [PubMed] [Google Scholar]
- 24. van Hoffen E, et al. 2010. Exposure of intestinal epithelial cells to UV-killed Lactobacillus GG but not Bifidobacterium breve enhances the effector immune response in vitro. Int. Arch. Allergy Immunol. 152:159–168 [DOI] [PubMed] [Google Scholar]
- 25. Van Niel G, et al. 2003. Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut 52:1690–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vidal K, Grosjean I, evillard JP, Gespach C, Kaiserlian D. 1993. Immortalization of mouse intestinal epithelial cells by the SV40-large T gene. Phenotypic and immune characterization of the MODE-K cell line. J. Immunol. Methods 166:63–73 [DOI] [PubMed] [Google Scholar]
- 27. Vignali DA, Collison LW, Workman CJ. 2008. How regulatory T cells work. Nat. Rev. Immunol. 8:523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wallis TS, Galyov EE. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997–1005 [DOI] [PubMed] [Google Scholar]
- 29. Yoshinaga T, Yasuda K, Ogawa Y, Nishikawa M, Takakura Y. 2007. DNA and its cationic lipid complexes induce CpG motif-dependent activation of murine dendritic cells. Immunology 120:295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ziegler SF, Liu Y-J. 2006. Thymic stromal lymphopoietin in normal and pathogenic T cell development and function. Nat. Immunol. 7:709–714 [DOI] [PubMed] [Google Scholar]