Summary
The hereditary disorders chorea acanthocytosis and Cohen syndrome are caused by mutations in different members of a family of genes that are orthologs of yeast VPS13. In vegetatively growing yeast, VPS13 is involved in the delivery of proteins to the vacuole. During sporulation, VPS13 is important for formation of the prospore membrane that encapsulates the daughter nuclei to give rise to spores. We report that VPS13 is required for multiple aspects of prospore membrane morphogenesis. VPS13 (1) promotes expansion of the prospore membrane through regulation of phosphatidylinositol phosphates, which in turn activate the phospholipase D, Spo14; (2) is required for a late step in cytokinesis that gives rise to spores; and (3) regulates a membrane-bending activity that generates intralumenal vesicles. These results demonstrate that Vps13 plays a broader role in membrane biology than previously known, which could have important implications for the functions of VPS13 orthologs in humans.
Key words: Cytokinesis, Phospholipase D, Prospore membrane
Introduction
Spore formation in Saccharomyces cerevisiae, the equivalent of gametogenesis in metazoans, is a differentiation program in which the four haploid chromosome sets produced by meiosis are packaged into daughter cells (Neiman, 2011). Packaging requires the generation of new membrane compartments within the cytoplasm, termed prospore membranes. At the start of the second meiotic division, secretory vesicles coalesce at each of the four spindle poles to form small double-membrane caps. As meiosis II progresses, each prospore membrane expands so that after nuclear division each daughter nucleus is engulfed by a prospore membrane. The closure of a prospore membrane around a nucleus is a cytokinetic event, separating the cytoplasm of the daughter cell (the spore) from that of the mother cell (now called the ascus). After closure, the prospore membrane serves as the plasma membrane of the newly formed spore (Neiman, 2011).
Proper growth and closure of the prospore membrane require a number of different factors. The phospholipase D Spo14 is essential for membrane formation (Rudge et al., 1998). Spo14 catalyzes the hydrolysis of phosphatidylcholine to phosphatidic acid (PA) and enzymatic activity is essential for the fusion of vesicles during prospore membrane formation (Rudge et al., 1998; Nakanishi et al., 2006). Activation of Spo14 during sporulation requires both the translocation of Spo14 to the prospore membrane from its endosomal and/or cytosolic localization in vegetative cells, and the presence, in the prospore membrane, of sufficient levels phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), which stimulates activity of the enzyme (Rudge et al., 1998; Sciorra et al., 1999; Rudge et al., 2004). Closure of the membrane requires the removal of the leading edge protein complex (LEP), which is localized to the mouth of the prospore membrane (Maier et al., 2007; Diamond et al., 2008). The LEP is composed of at least three subunits Don1, Ady3 and Ssp1, of which Ssp1 is the most crucial. Degradation of Ssp1, triggered at the end of meiosis II by the anaphase-promoting complex and its regulator Ama1, is necessary to allow the subsequent closure of the prospore membrane (Diamond et al., 2008). In addition, several genes have been identified that are essential for proper growth of the prospore membrane including VPS13 (Rabitsch et al., 2001; Nakanishi et al., 2007).
VPS13 was first identified in mutant cells defective in the delivery of carboxypeptidase Y to the vacuole and in the regulation of Kex2 cycling between the Golgi and late endosomes, indicating a role in trafficking between the late Golgi and the endosome (Bankaitis et al., 1986; Brickner and Fuller, 1997). More than 50 VPS genes affecting vacuolar delivery have been identified in S. cerevisiae (Bonangelino et al., 2002; Burston et al., 2008). Mutations in approximately 20 of these VPS genes have been found to reduce or block sporulation prior to prospore membrane formation, probably as a result of defects in autophagy (Enyenihi and Saunders, 2003; Neiman, 2005; Nakanishi et al., 2007; Piekarska et al., 2010). VPS13 appears unique in that mutants are defective in growth of the prospore membrane (Nakanishi et al., 2007). Thus, VPS13 seems to have a function in prospore membrane growth that is distinct from its role in vacuolar sorting.
VPS13 is highly conserved, with orthologs in all eukaryotic genomes that have been sequenced. In humans there are four VPS13 orthologs (Velayos-Baeza et al., 2004). The function(s) of their different proteins in human cells has not been defined, but mutations in two of the orthologs, VPS13A (also known as CHAC) and VPS13B (COH1) give rise to the hereditary diseases chorea acanthocytosis and Cohen syndrome, respectively (Rampoldi et al., 2001; Kolehmainen et al., 2003). Because of the established role of yeast VPS13 in vacuolar transport, it has been inferred that the human phenotypes result from comparable defects in membrane transport. We report here new functions for yeast VPS13 during prospore membrane formation. The role of VPS13 in membrane expansion is mediated through effects on the phospholipase D, Spo14. Separately, VPS13 is required for closure of the prospore membrane, possibly through control of a membrane-bending activity.
Results
VPS13 is required for cytokinesis during spore formation
Fluorescence loss in photobleaching (FLIP) can be used to monitor prospore membrane closure, because of the ability of a cytosolic green fluorescent protein (GFP) to diffuse between the presumptive ascal and spore cytoplasms (Diamond et al., 2008). In this assay, a spot in the cytoplasm outside of the prospore membrane is repeatedly photobleached and the cytoplasmic GFP fluorescence inside of the prospore membrane is monitored. Loss of GFP fluorescence inside the prospore membrane indicates that the cytoplasms are connected and the prospore membrane is open. Whereas 100% of late stage prospore membranes were closed in wild-type cells, vps13Δ mutants displayed a severe closure defect, with over 90% of membranes open (Table 1; supplementary material Fig. S1). This closure defect was not simply a consequence of the smaller prospore membranes in vps13Δ cells, because another mutant that forms smaller prospore membranes, gip1Δ, displayed efficient closure (76%) (Ishihara et al., 2009). Thus, vps13Δ mutants have a strong cytokinesis defect during sporulation.
Table 1.
vps13 mutants fail to undergo cytokinesis during sporulation
| Percentage of prospore membranes | |||
| Relevant genotypea | Closed | Open | Ind.b |
| Wild typec | 100 | 0 | 0 |
| vps13Δ | 0 | 94 | 6 |
| gip1Δ | 76 | 16 | 7 |
| vps13Δ 2µ-Myr-SPO14 | 11 | 71 | 18 |
| snf7Δ | 69 | 10 | 21 |
The closure of at least 50 late stage prospore membranes was examined by FLIP assay in AN390 (wild type), JSP26 (gip1Δ), JSP164 (vps13Δ) and JSP287 (snf7Δ). 2µ-Myr-SPO14 expresses the myristoylated form of SPO14 from a high copy vector.
Indeterminate: fluorescence loss inside of the prospore membrane was intermediate between open and closed.
Data for wild-type cells is from Diamond et al. (Diamond et al., 2008).
Prospore membrane closure requires the removal of the LEP, which is localized to the mouth of the prospore membrane (Maier et al., 2007; Diamond et al., 2008). Removal of this complex can be monitored by relocalization of the LEP component Don1 (as Don1–GFP) from the leading edge to the cytoplasm (Diamond et al., 2008). Don1–GFP fluorescence disappeared from the majority of prospore membranes in late stage vps13Δ cells, in contrast to ama1Δ mutants where Don1–GFP fluorescence persisted at the leading edge (Fig. 1). Though not as complete as in gip1Δ or wild-type cells, the disappearance of Don1–GFP from the leading edge of vps13Δ cells suggests that VPS13 acts after LEP removal, perhaps to directly promote membrane closure.
Fig. 1.
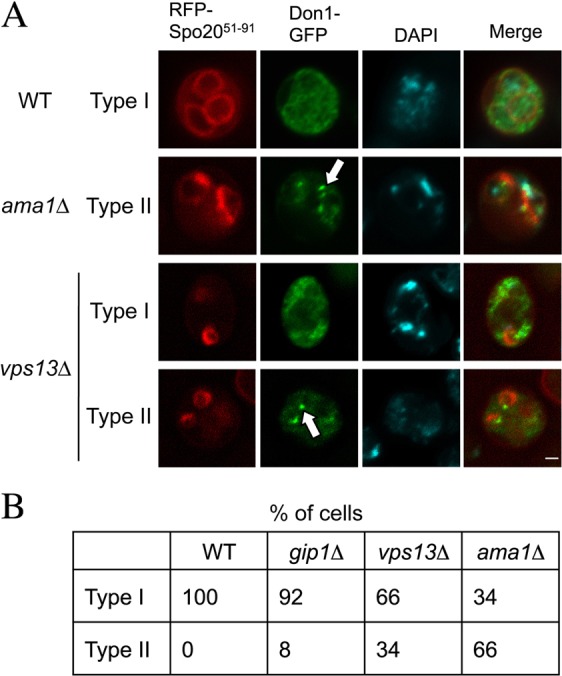
Don1 localization in vps13Δ cells. (A) Representative wild-type (AN120), gip1Δ (NY501), vps13Δ (HI29) and ama1Δ (ADY66) cells transformed with pRS426-RFP-Spo2051–91-DON1-GFP to visualize the prospore membranes and the LEP, respectively. Type I indicates that Don1–GFP was dispersed in the cytoplasm, whereas Type II indicates that Don1–GFP was concentrated on the prospore membrane. White arrows indicate LEPs. Scale bar: 1 µm. (B) Quantification of Don1–GFP distribution in the cells in A. More than 100 prospore membranes were examined for each strain.
Vps13 relocalizes to the prospore membrane during spore formation
In mitotic cells, Vps13 is localized to the endosome, raising the possibility that the effect of vps13Δ on prospore membrane closure is indirect (Huh et al., 2003). We found that, similar to mitotic cells, early in sporulation Vps13–GFP localizes diffusely in the cytoplasm with brighter cytoplasmic puncta (presumably endosomes). By contrast, later in meiosis it was still diffuse in the cytoplasm but also concentrated along the prospore membrane (Fig. 2). Thus, Vps13 relocalizes to the prospore membrane in sporulating cells, suggesting it has a more direct role in membrane expansion and closure.
Fig. 2.
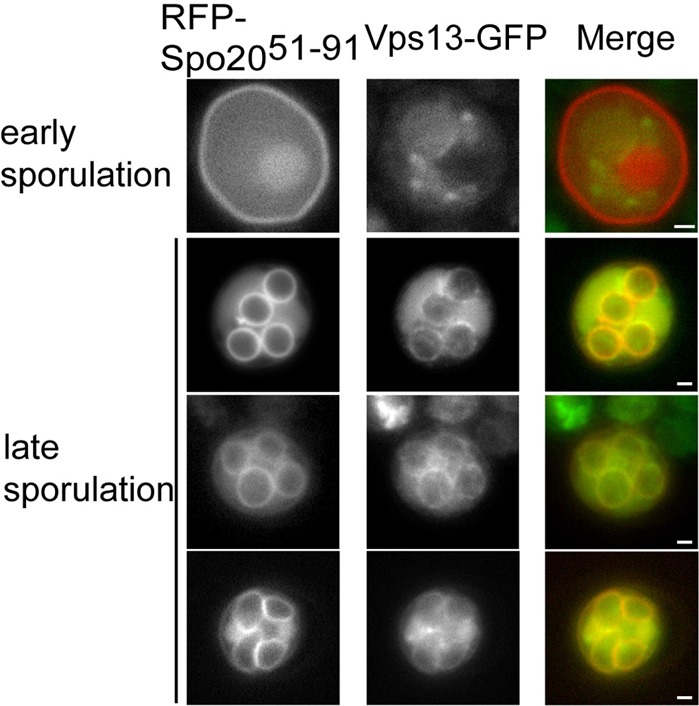
Localization of Vps13–GFP in sporulating cells. A Vps13–GFP strain (JSP257) carrying pRS426-RFP-Spo2051–91 as a prospore membrane marker was sporulated and GFP and RFP fluorescence was monitored during prospore membrane growth. Scale bars: 1 µm.
Prospore membranes in vps13Δ cells have reduced phosphatidic acid content
The relocalization of Vps13 is reminiscent of Spo14, which translocates from a cytosolic or endosomal localization in vegetative cells to the prospore membrane during sporulation (Rudge et al., 1998; Li et al., 2000). The soluble NSF attachment protein receptor (SNARE) complex, which mediates fusion at the prospore membrane, includes a sporulation-specific subunit, Spo20 that is recruited to the membrane through a 41-amino-acid PA-binding motif in the Spo20 N-terminus (Nakanishi et al., 2004). A fusion of GFP to this motif, GFP–Spo2051–91, acts as an in vivo reporter for PA and localizes to the prospore membrane in a Spo14-dependent fashion (Nakanishi et al., 2004; Nakanishi et al., 2006; Zeniou-Meyer et al., 2007). The intensity of the fluorescent signal of GFP–Spo2051–91 from the prospore membrane in vps13Δ cells appeared weaker than in wild-type cells, suggesting that the PA level in the prospore membranes of vps13Δ cells might be lower. To examine the relationship between the prospore membrane and PA, we compared the colocalization of RFP–Spo2051–91 to that of the integral membrane protein, Dtr1–GFP in wild-type and vps13Δ cells (Felder et al., 2002). Although colocalization of the markers was high in wild-type cells, in vps13Δ only 50% of prospore membranes identified by Dtr1–GFP displayed visible RFP–Spo2051–91 signal, indicating that the PA levels in these membranes are reduced (Fig. 3A,B). Note that PA must not be absent, just reduced below the level detectable by RFP–Spo2051–91, because complete loss of Spo14 activity blocks prospore membrane formation (Nakanishi et al., 2006). These results indicate that VPS13 is required for the full activity or localization of Spo14 during sporulation, resulting in lowered PA levels at the prospore membrane in vps13Δ cells.
Fig. 3.
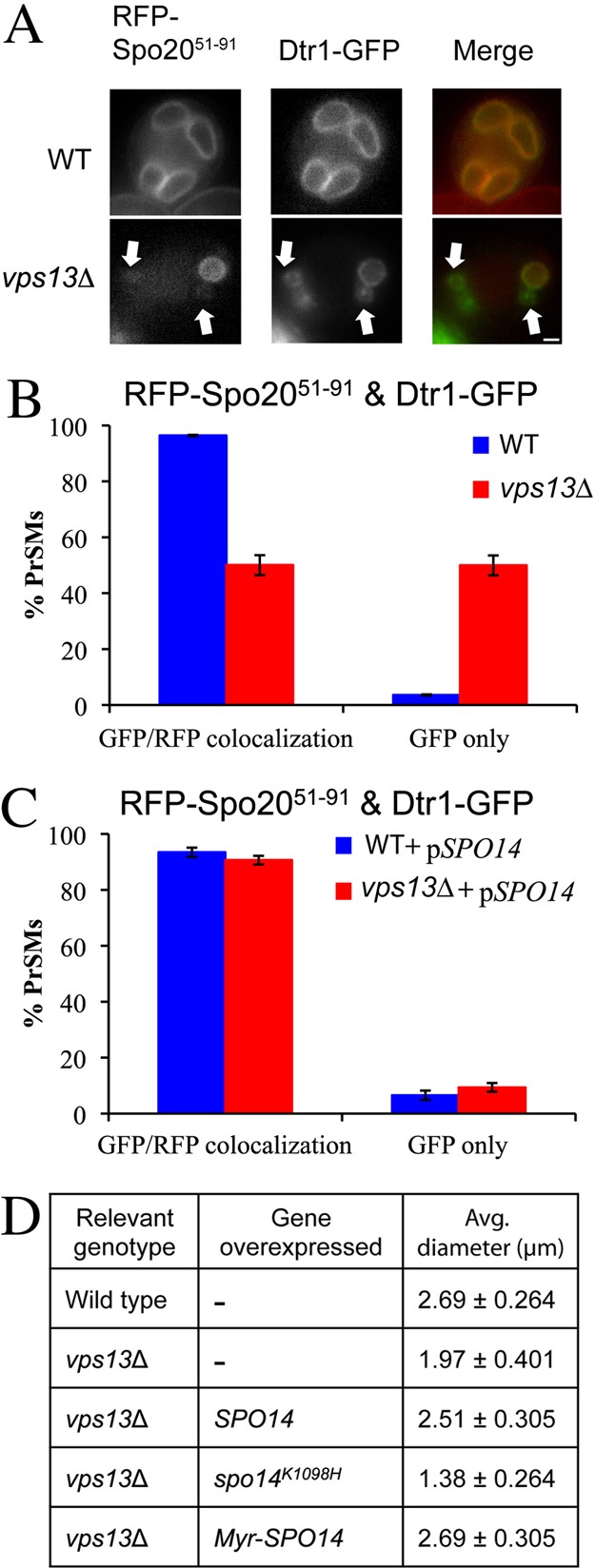
Effects of vps13Δ on phosphatidic acid in the prospore membrane. (A) Wild-type (AN120) or vps13Δ (HI29) cells expressing both an integral membrane marker for the prospore membrane, DTR1-GFP, and the PA sensor RFP-Spo2051–91 were examined during sporulation. Arrows indicate Dtr1–GFP marked prospore membranes that lack RFP fluorescence. (B) Quantification of Dtr1–GFP and RFP–Spo2051–91 colocalization in the strains in A. At least 110 prospore membranes were scored in two independent experiments. (C) Wild-type (AN120) or vps13Δ (HI29) cells transformed with both pRS424-DTR1-GFP and pRS426-RFP-Spo2051–91-SPO14 were analyzed as in B. (D) The diameter of the prospore membrane was measured in post-meiotic wild-type (AN120) or vps13Δ (HI29) cells overexpressing the indicated genes. More than 100 prospore membranes were measured for each strain. The numbers given are the average diameter ± one standard deviation. Scale bar: 1 µm.
Overexpression of SPO14 rescues the membrane size phenotype but not the cytokinesis defect of vps13Δ
To test whether reduced PA is responsible for the vps13Δ prospore membrane defects, colocalization of RFP–Spo2051–91 and Dtr1–GFP was examined in vps13Δ cells overexpressing SPO14. Overexpression of SPO14 rescued the colocalization of RFP–Spo2051–91 and Dtr1–GFP (Fig. 3C). Moreover, SPO14 overexpression also restored the prospore membranes in vps13Δ cells to sizes comparable with those of wild-type cells (Fig. 3D). This increase in size was significantly greater than in vps13Δ mutants without SPO14 overexpression (P<0.001). By contrast, overexpression of the gene for a catalytically inactive form of the enzyme, SPO14-K1098H (Rudge et al., 1998), did not rescue the membrane growth phenotype (Fig. 3D), indicating that PA production by the overexpressed SPO14 is necessary for the rescue. In fact, overexpression of SPO14-K1098H led to even smaller average membrane size in the vps13Δ cells (P<0.001).
VPS13 might influence PA levels by affecting the activity or the localization of Spo14. We were unable to directly assess the effect of vps13Δ on Spo14 localization because GFP–Spo14 could not be visualized at native expression levels. As an alternative approach, we tried to enhance Spo14 recruitment to the prospore membrane by fusing the N-terminal myristoylation signal from Gpa1 to the N-terminus of Spo14 (Stone et al., 1991). Myr–SPO14 slightly improved the rescue of the membrane growth phenotype of vps13Δ over that of wild-type SPO14 (Fig. 3D; P<0.01). Together, these results suggest that VPS13 promotes membrane expansion by enhancing the activity Spo14 at the prospore membrane with the consequent generation of PA, which in turn is necessary for efficient vesicle fusion.
To determine if the vps13Δ cytokinesis defect is also a consequence of lowered PA, the FLIP assay was performed using vps13Δ cells overexpressing Myr-SPO14. Though the prospore membrane growth defect is rescued in these cells, Myr-SPO14 overexpression only weakly suppressed the membrane closure defect (Table 1). Thus, the function of VPS13 in membrane expansion is separable from its role in cytokinesis.
Prospore membrane PtdIns-phosphate pools are reduced in the vps13Δ mutant
Spo14 requires PtdIns(4,5)P2 for its activity, and mutations in the PtdIns 4-kinase gene PIK1 or the PtdIns(4)P 5-kinase gene MSS4, which together generate PtdIns(4,5)P2, result in a failure to sporulate and to activate Spo14 during sporulation (Flanagan et al., 1993; Desrivières et al., 1998; Rudge et al., 2004). To investigate whether the effect of vps13Δ on Spo14 might be mediated through effects on PtdIns(4,5)P2 we used GFP fusions to the phosphatidylinositol 4-phosphate (PtdIns(4)P)-specific lipid-binding domain of Osh2 and the PtdIns(4,5)P2-specific lipid binding domain of PLCδ1 to examine the intracellular distribution of these lipids during sporulation (Stefan et al., 2002; Roy and Levine, 2004). In wild-type cells, the PtdIns(4)P reporter localized to prospore membranes in late meiotic cells (Fig. 4A,B). By contrast, in vps13Δ mutants the localization of the GFP fusion protein to the prospore membrane was reduced, similar to our results above with the PA reporter RFP–Spo2051–91 (Fig. 3). Very similar results were obtained with the PtdIns(4,5)P2 reporter (Fig. 4C,D). These results indicate that the levels of both PtdIns(4)P and, probably as a consequence, PtdIns(4,5)P2 are reduced in the prospore membrane in vps13Δ cells. PtdIns(4,5)P2 is necessary both for the activity and the recruitment of Spo14 (Sciorra et al., 2002) and the reduction of PtdIns(4,5)P2 levels probably accounts for the effects of vps13Δ on Spo14 activity.
Fig. 4.
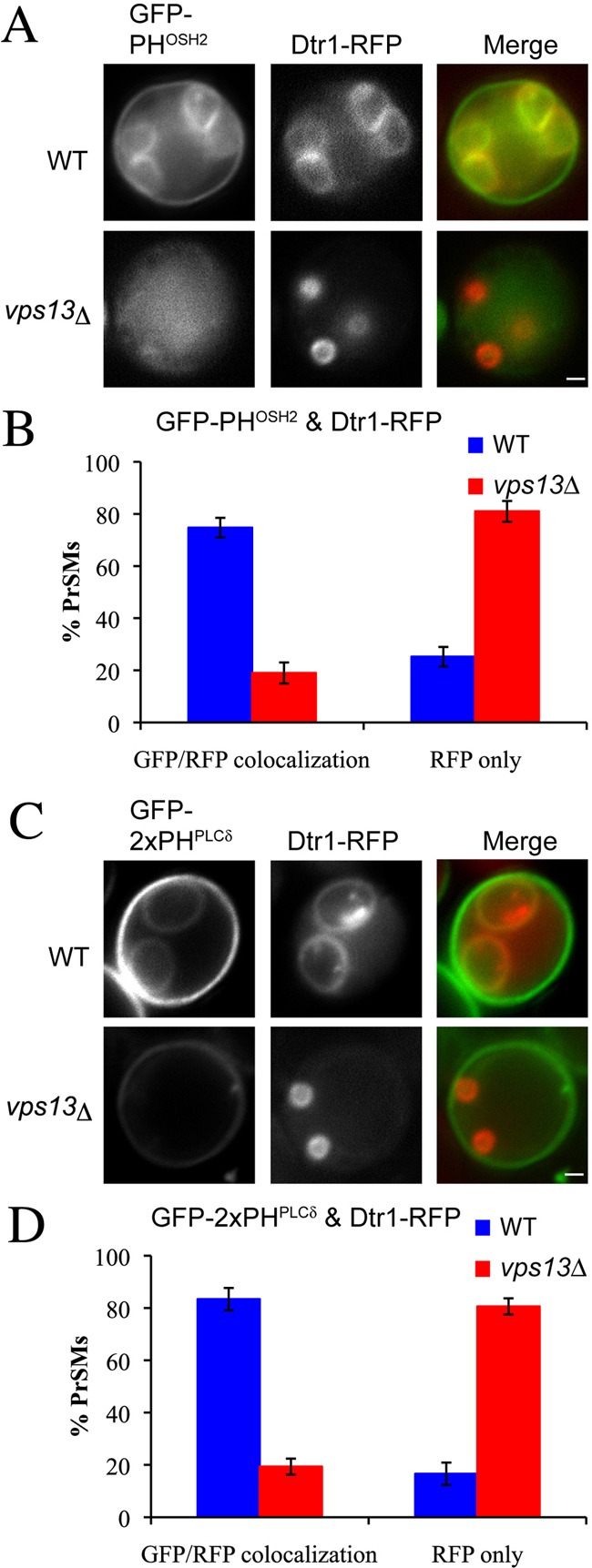
Effects of vps13Δ on prospore membrane PtdIns(4)P and PtdIns(4,5)P2 pools. (A) Wild-type (AN120) and vps13Δ (HI29) cells expressing the PtdIns(4)P sensor GFP–PHOSH2 and the prospore membrane marker Dtr1–RFP were examined during sporulation. (B) Quantification of GFP–PHOSH2 and Dtr1–RFP colocalization in A. (C) Wild-type (AN120) and vps13Δ (HI29) cells expressing the PtdIns(4,5)P2 sensor GFP–2XPHPLCδ and the prospore membrane marker Dtr1–RFP were examined during sporulation. (D) Quantification of GFP–2XPHPLCδ and Dtr1–RFP colocalization in C. More than 100 prospore membranes were observed for each strain. Scale bars: 1 µm.
Prospore membranes accumulate intralumenal vesicles in vps13Δ
Transmission electron microscopy (TEM) studies of vps13Δ cells revealed an unusual phenotype in sporulating cells. In wild-type cells, the width of the lumen between the inner and outer bilayers of the prospore membrane is uniform along the entire length of the membrane (Fig. 5A). By contrast, membrane-bound inclusions within the prospore membrane lumen were frequently seen in vps13Δ cells (visible in 75% of prospore membrane profiles; Fig. 5B,C,D). These intralumenal vesicles appear to form by an inbudding or invagination of the prospore membrane into the lumen (Fig. 5E,F). Overexpression of SPO14 did not suppress the appearance of intralumenal vesicles (68% of profiles; Fig. 5G), suggesting that reduced PA in the prospore membrane is not the basis for this phenotype.
Fig. 5.
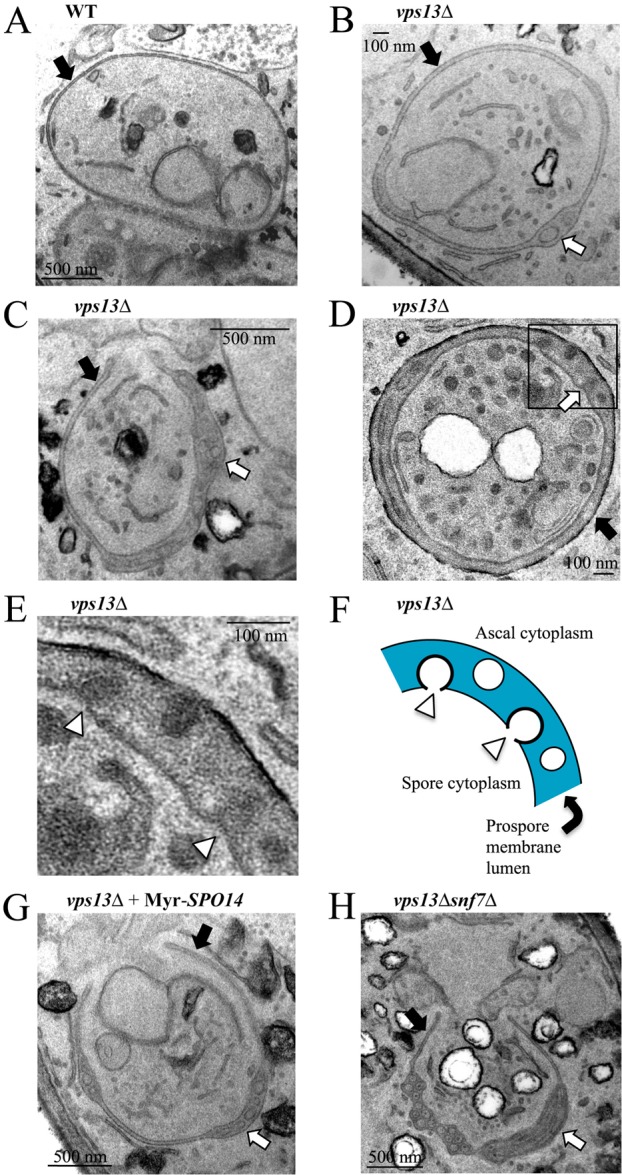
A previously unknown prospore membrane phenotype in vps13Δ cells. TEM images of prospore membranes in (A) wild-type (AN120) and (B–D) vps13 (HI29) cells. (E) Higher magnification image of the boxed region in D. White arrowheads indicate forming vesicles. (F) Cartoon of the prospore membrane region in E. (G) Prospore membrane in a vps13Δ cell overexpressing SPO14 (HI29 carrying pRS426-PSPS4-Myr-GFP-SPO14). (H) Prospore membrane in a vps13Δ snf7Δ cell (JSP286). In all panels, black arrows indicate prospore membranes, and white arrows point to regions where the lumen is expanded and contains vesicles. Lower magnification images of all the cells shown here are shown in supplementary material Fig. S2.
Intralumenal vesicle formation in vps13Δ is not dependent on the ESCRT complex
The lumenal vesicles seen in vps13Δ cells appear similar to multivesicular bodies (MVBs) (Piper and Katzmann, 2007). MVBs are carriers of endocytosed proteins to the vacuoles that are formed when a small region of the limiting membrane of the endosome pinches off into the endosomal lumen. Endosomal sorting complexes required for transport (ESCRT)-I, -II and -III complexes regulate the sorting of cargo into MVBs as well as their formation, scission and release into the lumen (Hurley and Hanson, 2010). ESCRT components have also been implicated in other topologically similar events in mammalian cells including cytokinesis and viral budding (Strack et al., 2003; Carlton and Martin-Serrano, 2007; Hurley and Hanson, 2010).
In particular, the ESCRT-III complex is required for membrane scission and the observation that mutants in different components of this complex display low sporulation efficiency suggests a potential role in prospore membrane biogenesis (Enyenihi and Saunders, 2003; Wollert et al., 2009). SNF7 encodes an essential component of the ESCRT-III machinery and although the majority of snf7Δ cells arrest before prospore membrane formation, a small fraction are able to form spores (Babst et al., 2002; Enyenihi and Saunders, 2003) (our unpublished observations). It was, therefore, possible to test the effects of mutation of SNF7 on later events in spore formation. Using the FLIP assay, we found that the prospore membranes that do form in snf7Δ cells efficiently undergo closure (Table 1; supplementary material Fig. S1), indicating that ESCRT is not involved in cytokinesis during sporulation. Moreover, in vps13Δ snf7Δ double mutants intralumenal vesicles were observed (79% of profiles) within the prospore membranes, indicating that ESCRT is not required for their formation (Fig. 5H). Thus, VPS13 might regulate a membrane bending and/or scission activity distinct from ESCRT.
Discussion
VPS13 was originally identified through its role in vesicular trafficking through the endosome to the vacuole (Bankaitis et al., 1986). Subsequent studies revealed that VPS13 is also required for sporulation (Enyenihi and Saunders, 2003; Nakanishi et al., 2007). This work demonstrates that VPS13 is required for many aspects of prospore membrane morphogenesis during spore formation: (1) it promotes membrane growth through the activity of the phospholipase D Spo14 at the prospore membrane; (2) it is required for cytokinesis at the end of prospore membrane growth; and (3) it regulates a membrane bending and/or scission activity capable of forming intralumenal vesicles.
The regulation of Spo14 by VPS13 appears to be mediated indirectly through effects on PtdIns-phosphate levels. Prospore membranes in vps13Δ cells exhibit reduced binding by reporters for both PtdIns(4)P and the Spo14-activating lipid PtdIns(4,5)P2. Notably, the effect of vps13Δ on the PtdIns-phosphate reporters was stronger than the effect on the PA reporter. This could reflect the relative affinities of Spo14 and the Osh2 and PLCδ1 reporters for PtdIns phosphates, as Spo14 binds to PtdIns(4,5)P2 at two independent sites on the protein (Sciorra et al., 2002). If it is recruited to the membrane more efficiently than the GFP reporters, then it can generate PA, leading to membranes that have detectable levels of PA even though they lack a signal for PtdIns phosphates.
The reduced PtdIns(4,5)P2 in vps13Δ cells probably reflects the lowered PtdIns(4)P levels, but how VPS13 influences PtdIns(4)P abundance remains to be determined. One possibility is that VPS13 regulates the activity of a PtdIns(4)-kinase. There are three PtdIns(4)-kinase enzymes in yeast, Pik1, Stt4 and Lsb6 (Flanagan et al., 1993; Yoshida et al., 1994; Han et al., 2002). PIK1 is reported to be required for sporulation (Rudge et al., 2004), suggesting it as a possible VPS13 target, but no interactions between any of these genes and VPS13 have been reported. Alternatively, VPS13 might negatively regulate one or more PtdIns-phosphate phosphatases. Increased activity of one or more phosphatases could also account for the vps13Δ mutant phenotype.
The effect of vps13Δ on Spo14 activity is distinct from the cytokinesis defect. However, it is not clear if these phenotypes of vps13Δ are truly independent or whether the cytokinesis defect might also be a consequence of reduced PtdIns-phosphate levels. Because the molecular function of VPS13 in vacuolar transport has not been determined, it is possible that dysregulation of PtdIns phosphates is also responsible for those defects. Interestingly, a recent synthetic genetic interaction screen found that mutants in VPS13 and in VPS30, a subunit of the PtdIns(3)-kinase complex, exhibit similar sets of genetic interactions (Hoppins et al., 2011), consistent with the idea that PtdIns phosphates are also dysregulated during vegetative growth in vps13Δ cells.
The relationship of the intralumenal vesicles that form in vps13Δ cells to the other mutant phenotypes also has yet to be established. It is noteworthy, however, that the membrane fission event that releases a vesicle into the lumen of the prospore membrane is topologically equivalent to the fission event that occurs at cytokinesis. Both events require the scission of a membrane oriented away from the cytoplasm. Indeed, in mammalian cells, ESCRT is important both for MVB formation and for cytokinesis (Raiborg et al., 2003; Carlton and Martin-Serrano, 2007). It seems probable, therefore, that the vps13Δ cytokinesis and intralumenal vesicle phenotypes are related. We favor a model in which Vps13 ordinarily restricts a membrane scission activity to the site of membrane closure, and loss of this Vps13-mediated restraint leads to ectopic ‘cytokinesis’ giving rise to intralumenal vesicles.
The formation of intralumenal vesicles in the vps13Δ cells is independent of the ESCRT complex. In mammalian cells, ESCRT-independent formation of intralumenal vesicles has been reported and these possibly involve the generation of specialized lipid species (Trajkovic et al., 2008; Babst, 2011). Our results provide the first evidence for a similar ESCRT-independent pathway in yeast.
Taken together, our findings suggest a much broader role for Vps13 in membrane biology than has been appreciated. Recent studies of Vps13 orthologs in other systems also suggest roles beyond vacuolar trafficking for this family of proteins. In Tetrahymena, the sole Vps13 ortholog has been localized to phagosomes, whereas in human cells, the VPS13B (COH1) protein, which is mutated in Cohen syndrome, localizes to the cis-Golgi (Samaranayake et al., 2011; Seifert et al., 2011).
Little is known about the molecular bases for Cohen syndrome or chorea acanthocytosis, caused by the mutation of VPS13B (COH1) and VPS13A (CHAC), respectively (Rampoldi et al., 2001; Kolehmainen et al., 2003). Our results identify new functions for VPS13 in yeast that could have direct implications for the functions of human orthologs. For example, a defining feature of chorea acanthocytosis is the presence of circulating ‘acanthocytes’; that is, misshapen red blood cells that have multiple protrusions of their plasma membranes (Delaunay et al., 1990). The shape of the erythrocyte is maintained by interactions of integral plasma membrane proteins with an underlying spectrin and actin cytoskeleton. The protein 4.1R plays an important role in linking the membrane proteins to the cytoskeleton (Takakuwa, 2000). 4.1R can bind to PtdIns(4,5)P2 and binding to the lipid modulates 4.1R interactions with other proteins (An et al., 2006). We speculate that VPS13A regulates PtdIns-phosphate levels in the red cell plasma membrane, as the yeast homolog does in the prospore membrane, and that alterations in plasma membrane PtdIns(4,5)P2 levels alters interaction of the plasma membrane with the cytoskeleton, giving rise to the misshapen red cells seen in chorea acanthocytosis patients. Because of the strong conservation of Vps13, further exploration of the roles of VPS13 in sporulation could provide significant insight into the molecular basis of these human diseases.
Materials and Methods
Yeast strains and media
Yeast strains used for this study are listed in supplementary material Table S1. Unless otherwise mentioned, standard yeast media and genetic techniques were used (Rose and Fink, 1990). To construct strain JSP141, SNF7 was deleted by PCR-based gene deletion (Longtine et al., 1998). The SNF7 deletion cassette was amplified with the primers JSO9 and JSO10; all oligonucleotide sequences are listed in supplementary material Table S2. To construct strain JSP101, the snf7Δ segregants from a cross of strains JSP141 and AN117-4B were mated. Similarly, strain JSP286 was created by mating snf7Δ vps13Δ segregants from a cross of JSP141 and HI27. Strain JSP257 was generated by the mating of segregants from a cross of AN117-4B to a VPS13::GFP-containing strain (Huh et al., 2003).
To construct strains for the FLIP assay, strain HI27, NY1 or JSP141 was mated with a strain containing a GFP-tagged TEF2 allele (Huh et al., 2003). After tetrad dissection, vps13Δ TEF2::GFP, gip1Δ TEF2::GFP or snf7Δ TEF2::GFP segregants were mated to generate strains JSP164, JSP26 and JSP287, respectively. These diploid cells were transformed with pRS424-RFP-SPO2051–91 or pRS426- RFP-SPO2051–91 prior to the assay.
Plasmids
Plasmids used in this study are listed in supplementary material Table S3. To construct pRS424-RFP-SPO2051–91-SPO14, pRS424-SPO14 was first constructed by homologous recombination in S. cerevisiae (Ma et al., 1987). Overlapping fragments of the SPO14 coding region including 500 bp of upstream and downstream sequence were amplified by PCR using the primers, JSO127 and JSO128-r and JSO128 and JSO129, respectively. The 5′ ends of the primers JSO127 and JSO129 carry ∼50 nucleotides that allow recombination with the plasmid pRS424 (Christianson et al., 1992). spo14Δ mutant diploid cells were transformed with the two PCR products and KpnI-linearized pRS424 and pRS424-SPO14 was assembled by homologous recombination in the cell. The plasmid was extracted from spo14Δ cells whose sporulation defect was restored by successfully constructed pRS424-SPO14. The KpnI fragment from pRS424-SPO14 carrying SPO14 was ligated into KpnI-digested pRS424-RFP-SPO2051–91, giving pRS424-RFP-SPO2051–91-SPO14. To construct pRS426-PSPS4-Myr-GFP-SPO14, the SPS4 promoter (from −1 to −1000) was first amplified using primers HJO166 and HJO167 and then cloned into a pRS426 as a SacI–SpeI fragment. A sequence encoding the first seven amino acids of GPA1, which constitute an N-myristoylation motif (Stone et al., 1991), were included in the primer nAC01 used along with AC06 to amplify GFP–SPO14 from the plasmid YEp351-GFP-SPO14 (Rudge et al., 1998). This PCR product, digested with SpeI and XhoI, was ligated into pRS426-PSPS4 to create pRS426-PSPS4-Myr-GFP-SPO14. Plasmid pRS426-GFP-spo14K1098H was constructed by moving GFP-spo14K1098H as a NotI–XhoI fragment from pRS424-GFP-spo14K1098H (Nakanishi et al., 2006) into pRS426. To construct pRS426-RFP-SPO2051–91-DON1-GFP, a DON1-GFP fragment with KpnI sites at both ends was created by PCR amplification with primers HT19Kpn1F and HT66Kpn1R. This fragment was ligated into the KpnI site of pRS426-RFP-SPO2051–91. To construct pRS426-DTR1-GFP a BamHI–XhoI fragment from pRS424-DTR1-mRFP (Mathieson et al., 2010) carrying DTR1-mRFP was ligated into similarly digested pRS426. To construct pRS424-DTR1-RFP-SPO14, the KpnI fragment from pRS424-SPO14 carrying SPO14 was ligated into KpnI-digested pRS424-DTR1-RFP.
Fluorescence microscopy
For detection of fluorescence, cells were fixed in 3.7% formaldehyde for 1 minute and washed once with PBS (137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4, pH 7.5). These cells were mounted with a mounting solution containing 4,6-diamidino-2-phenylindole (DAPI; Vectashield; Vector Laboratories, Peterborough, UK) to visualize the nuclei. Two fluorescence microscopes were used in this study; a Zeiss Axioplan2 microscope (Carl Zeiss, Thornwood, NY) with a Zeiss mRM Axiocam and a Zeiss Observer.Z1 microscope with an attached Orca II ERG camera (Hamamatsu, Bridgewater, NJ). Zeiss Axiovision 4.8 software was used to acquire images. Images in Fig. 1 were deconvolved using Zeiss Axiovision 4.8.
To measure prospore membranes (Fig. 3D), cells were first stained with DAPI so that post-meiotic cells could be identified. The longest diameter of the prospore membranes, visualized by Dtr1–GFP or RFP–Spo2051–91, was determined using the measurement tools in the Axiovision 4.8 software. The statistical significance of the differences between the distributions of membrane sizes in the different strains was analyzed using a Student's unpaired t-test with a two-tailed distribution.
For the FLIP assay, cells were first incubated either in liquid or on solid sporulation medium and their progression through sporulation was monitored by observing the prospore membrane morphology marked with RFP–Spo2051–91. When cells in the population began entering meiosis II, cells were transferred to agar pads and the FLIP assay was performed as previously described (Diamond et al., 2008), using a Zeiss LSM 510 META NLO inverted microscope coupled with a Coherent Chameleon XR Laser system.
Transmission electron microscopy
Sporulating cells were fixed in 3% glutaraldehyde in cacodylate buffer (100 mM sodium cacodylate, 5 mM CaCl2, pH 7.4) for 1 hour. Fixed cells were washed once with cacodylate buffer and left overnight at 25°C in 1 ml cacodylate buffer. The cells were collected, resuspended in 4% potassium permanganate in distilled water, and incubated at 25°C for 30 minutes. Cells were washed with distilled water until the supernatant became clear and resuspended in saturated uranyl acetate at 25°C for 2 hours. The samples were then dehydrated through a graded acetone series (30, 50, 70 and 95% acetone, two 10-minute incubations at each concentration) followed by four washes with 100% acetone. Dehydrated cells were left overnight in 100% acetone at 25°C. For embedding, the cells were first incubated 3×10 minutes in 100% acetonitrile and then resuspended in a 1∶1 acetonitrile: Epon mix (50% Epon 812, 15% dodecenyl succinic anhydride, 35% nadic methyl anhydride) for 4 hours at 25°C. These cells were collected and resuspended in 100% Epon mix for over 15 hours in vacuum. After two additional 1-hour incubations in 100% Epon mix under vacuum, the cells were transferred to Epon mix containing 1.5% 2,4,6-tris(dimethylaminomethyl)phenol (DMP-30) and were placed into 60°C under vacuum for 2 days. For transmission electron microscopy, the images were collected with an AMT XR-60 camera (AMT, Danvers, MA) attached to an FEI BioTwin G2 microscope (FEI, Hillsboro, OR). To quantify the ILV phenotype all prospore membranes visible in a given section were scored for the presence or absence of ILVs (regardless of number of vesicles). Thus, the percentage score indicates the fraction of prospore membranes with one or more ILVs in all the images of that strain.
Supplementary Material
Acknowledgments
The authors are grateful to Mark Lemmon (University of Pennsylvania) and Scott Emr (Cornell University) for plasmids, to Mike Frohman and Nancy Hollingsworth for comments on the manuscript, and members of the Neiman Lab for helpful discussions.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant number RO1 GM072540 to A.M.N.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.105114/-/DC1
References
- An X., Zhang X., Debnath G., Baines A. J., Mohandas N. (2006). Phosphatidylinositol-4,5-biphosphate (PIP2) differentially regulates the interaction of human erythrocyte protein 4.1 (4.1R) with membrane proteins. Biochemistry 45, 5725–5732 10.1021/bi060015v [DOI] [PubMed] [Google Scholar]
- Babst M. (2011). MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 23, 452–457 10.1016/j.ceb.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Estepa–Sabal E. J., Meerloo T., Emr S. D. (2002). Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3, 271–282 10.1016/S1534-5807(02)00220-4 [DOI] [PubMed] [Google Scholar]
- Bankaitis V. A., Johnson L. M., Emr S. D. (1986). Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA 83, 9075–9079 10.1073/pnas.83.23.9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino C. J., Chavez E. M., Bonifacino J. S. (2002). Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2486–2501 10.1091/mbc.02-01-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J. H., Fuller R. S. (1997). SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139, 23–36 10.1083/jcb.139.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burston H. E., Davey M., Conibear E. (2008). Genome-wide analysis of membrane transport using yeast knockout arrays. Methods Mol. Biol. 457, 29–39 10.1007/978-1-59745-261-8_3 [DOI] [PubMed] [Google Scholar]
- Carlton J. G., Martin–Serrano J. (2007). Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- Coluccio A. E., Rodriguez R. K., Kernan M. J., Neiman A. M. (2008). The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS ONE 3, e2873 10.1371/journal.pone.0002873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay J., Alloisio N., Morlé L., Pothier B. (1990). The red cell skeleton and its genetic disorders. Mol. Aspects Med. 11, 161–241 10.1016/0098-2997(90)90001-I [DOI] [PubMed] [Google Scholar]
- Desrivières S., Cooke F. T., Parker P. J., Hall M. N. (1998). MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 273, 15787–15793 10.1074/jbc.273.25.15787 [DOI] [PubMed] [Google Scholar]
- Diamond A., Park J. S., Inoue I., Tachikawa H., Neiman A. M. (2008). The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 134–145 10.1091/mbc.E08-06-0615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi A. H., Saunders W. S. (2003). Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder T., Bogengruber E., Tenreiro S., Ellinger A., Sá–Correia I., Briza P. (2002). Dtrlp, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 1, 799–810 10.1128/EC.1.5.799-810.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan C. A., Schnieders E. A., Emerick A. W., Kunisawa R., Admon A., Thorner J. (1993). Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science 262, 1444–1448 10.1126/science.8248783 [DOI] [PubMed] [Google Scholar]
- Han G. S., Audhya A., Markley D. J., Emr S. D., Carman G. M. (2002). The Saccharomyces cerevisiae LSB6 gene encodes phosphatidylinositol 4-kinase activity. J. Biol. Chem. 277, 47709–47718 10.1074/jbc.M207996200 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Collins S. R., Cassidy–Stone A., Hummel E., Devay R. M., Lackner L. L., Westermann B., Schuldiner M., Weissman J. S., Nunnari J. (2011). A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195, 323–340 10.1083/jcb.201107053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Hanson P. I. (2010). Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11, 556–566 10.1038/nrm2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara M., Suda Y., Inoue I., Tanaka T., Takahashi T., Gao X. D., Fukui Y., Ihara S., Neiman A. M., Tachikawa H. (2009). Protein phosphatase type 1-interacting protein Ysw1 is involved in proper septin organization and prospore membrane formation during sporulation. Eukaryot. Cell 8, 1027–1037 10.1128/EC.00095-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolehmainen J., Black G. C., Saarinen A., Chandler K., Clayton–Smith J., Träskelin A. L., Perveen R., Kivitie–Kallio S., Norio R., Warburg M., et al. (2003). Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 72, 1359–1369 10.1086/375454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Routt S. M., Xie Z., Cui X., Fang M., Kearns M. A., Bard M., Kirsch D. R., Bankaitis V. A. (2000). Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol. Biol. Cell 11, 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. (1987). Plasmid construction by homologous recombination in yeast. Gene 58, 201–216 10.1016/0378-1119(87)90376-3 [DOI] [PubMed] [Google Scholar]
- Maier P., Rathfelder N., Finkbeiner M. G., Taxis C., Mazza M., Le Panse S., Haguenauer–Tsapis R., Knop M. (2007). Cytokinesis in yeast meiosis depends on the regulated removal of Ssp1p from the prospore membrane. EMBO J. 26, 1843–1852 10.1038/sj.emboj.7601621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson E. M., Schwartz C., Neiman A. M. (2010). Membrane assembly modulates the stability of the meiotic spindle-pole body. J. Cell Sci. 123, 2481–2490 10.1242/jcs.062794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., de los Santos P., Neiman A. M. (2004). Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15, 1802–1815 10.1091/mbc.E03-11-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Morishita M., Schwartz C. L., Coluccio A., Engebrecht J., Neiman A. M. (2006). Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 119, 1406–1415 10.1242/jcs.02841 [DOI] [PubMed] [Google Scholar]
- Nakanishi H., Suda Y., Neiman A. M. (2007). Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 120, 908–916 10.1242/jcs.03405 [DOI] [PubMed] [Google Scholar]
- Neiman A. M. (2005). Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 565–584 10.1128/MMBR.69.4.565-584.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M. (2011). Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189, 737–765 10.1534/genetics.111.127126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., Katz L., Brennwald P. J. (2000). Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155, 1643–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarska I., Kucharczyk R., Mickowska B., Rytka J., Rempola B. (2010). Mutants of the Saccharomyces cerevisiae VPS genes CCZ1 and YPT7 are blocked in different stages of sporulation. Eur. J. Cell Biol. 89, 780–787 10.1016/j.ejcb.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Piper R. C., Katzmann D. J. (2007). Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23, 519–547 10.1146/annurev.cellbio.23.090506.123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch K. P., Tóth A., Gálová M., Schleiffer A., Schaffner G., Aigner E., Rupp C., Penkner A. M., Moreno–Borchart A. C., Primig M., et al. (2001). A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11, 1001–1009 10.1016/S0960-9822(01)00274-3 [DOI] [PubMed] [Google Scholar]
- Raiborg C., Rusten T. E., Stenmark H. (2003). Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15, 446–455 10.1016/S0955-0674(03)00080-2 [DOI] [PubMed] [Google Scholar]
- Rampoldi L., Dobson–Stone C., Rubio J. P., Danek A., Chalmers R. M., Wood N. W., Verellen C., Ferrer X., Malandrini A., Fabrizi G. M., et al. (2001). A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 28, 119–120 10.1038/88821 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. (1990). Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Roy A., Levine T. P. (2004). Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 279, 44683–44689 10.1074/jbc.M401583200 [DOI] [PubMed] [Google Scholar]
- Rudge S. A., Morris A. J., Engebrecht J. (1998). Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140, 81–90 10.1083/jcb.140.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge S. A., Sciorra V. A., Iwamoto M., Zhou C., Strahl T., Morris A. J., Thorner J., Engebrecht J. (2004). Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol. Biol. Cell 15, 207–218 10.1091/mbc.E03-04-0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaranayake H. S., Cowan A. E., Klobutcher L. A. (2011). Vacuolar protein sorting protein 13A, TtVPS13A, localizes to the tetrahymena thermophila phagosome membrane and is required for efficient phagocytosis. Eukaryot. Cell 10, 1207–1218 10.1128/EC.05089-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra V. A., Rudge S. A., Prestwich G. D., Frohman M. A., Engebrecht J., Morris A. J. (1999). Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 18, 5911–5921 10.1093/emboj/18.21.5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra V. A., Rudge S. A., Wang J., McLaughlin S., Engebrecht J., Morris A. J. (2002). Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J. Cell Biol. 159, 1039–1049 10.1083/jcb.200205056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert W., Kühnisch J., Maritzen T., Horn D., Haucke V., Hennies H. C. (2011). Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J. Biol. Chem. 286, 37665–37675 10.1074/jbc.M111.267971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C. J., Audhya A., Emr S. D. (2002). The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell 13, 542–557 10.1091/mbc.01-10-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D. E., Cole G. M., de Barros Lopes M., Goebl M., Reed S. I. (1991). N-myristoylation is required for function of the pheromone-responsive G alpha protein of yeast: conditional activation of the pheromone response by a temperature-sensitive N-myristoyl transferase. Genes Dev. 5, 1969–1981 10.1101/gad.5.11.1969 [DOI] [PubMed] [Google Scholar]
- Strack B., Calistri A., Craig S., Popova E., Göttlinger H. G. (2003). AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114, 689–699 10.1016/S0092-8674(03)00653-6 [DOI] [PubMed] [Google Scholar]
- Suda Y., Nakanishi H., Mathieson E. M., Neiman A. M. (2007). Alternative modes of organellar segregation during sporulation in Saccharomyces cerevisiae. Eukaryot. Cell 6, 2009–2017 10.1128/EC.00238-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachikawa H., Bloecher A., Tatchell K., Neiman A. M. (2001). A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155, 797–808 10.1083/jcb.200107008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakuwa Y. (2000). Protein 4.1, a multifunctional protein of the erythrocyte membrane skeleton: structure and functions in erythrocytes and nonerythroid cells. Int. J. Hematol. 72, 298–309 [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brugger B., Simons M. (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- Velayos–Baeza A., Vettori A., Copley R. R., Dobson–Stone C., Monaco A. P. (2004). Analysis of the human VPS13 gene family. Genomics 84, 536–549 10.1016/j.ygeno.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott–Schwartz J., Hurley J. H. (2009). Membrane scission by the ESCRT-III complex. Nature 458, 172–177 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ohya Y., Goebl M., Nakano A., Anraku Y. (1994). A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J. Biol. Chem. 269, 1166–1172 [PubMed] [Google Scholar]
- Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. (2004). Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell 13, 677–688 10.1016/S1097-2765(04)00083-8 [DOI] [PubMed] [Google Scholar]
- Zeniou–Meyer M., Zabari N., Ashery U., Chasserot–Golaz S., Haeberlé A. M., Demais V., Bailly Y., Gottfried I., Nakanishi H., Neiman A. M., et al. (2007). Phospholipase D1 production of phosphatidic acid at the plasma membrane promotes exocytosis of large dense-core granules at a late stage. J. Biol. Chem. 282, 21746–21757 10.1074/jbc.M702968200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


