Abstract
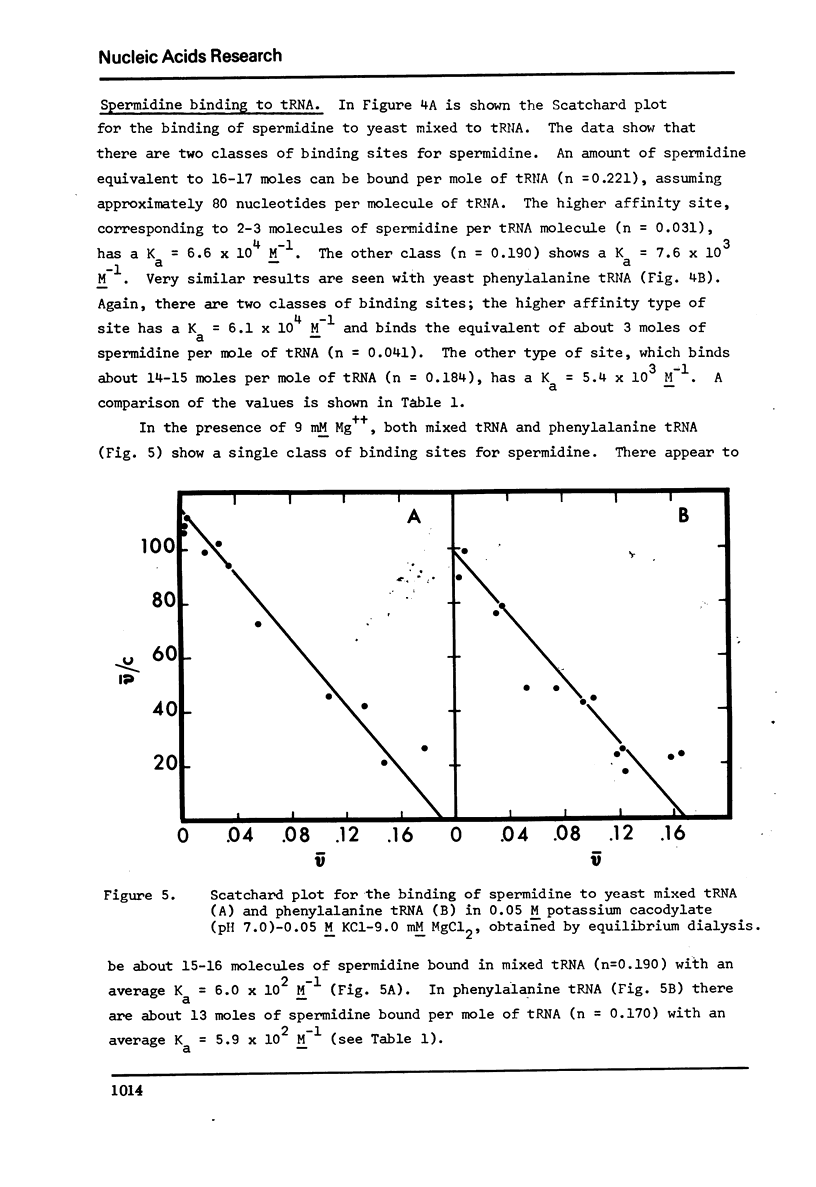
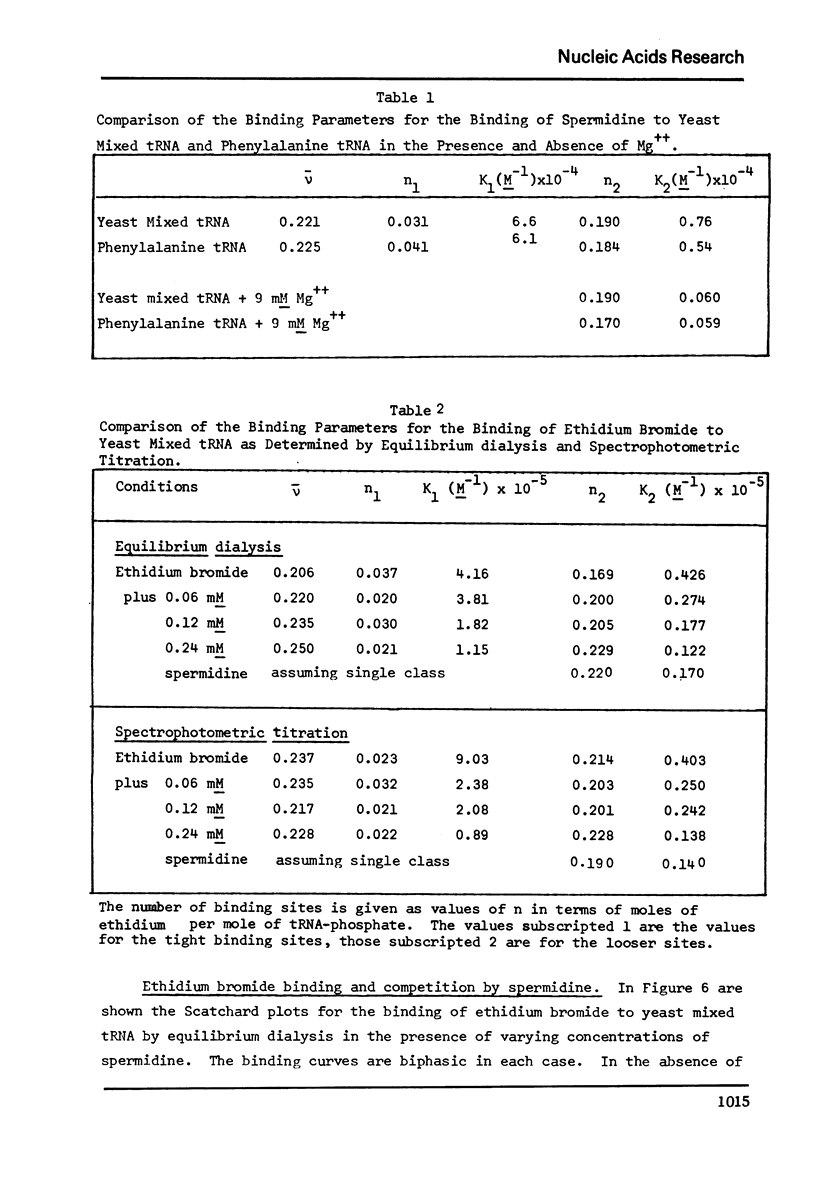
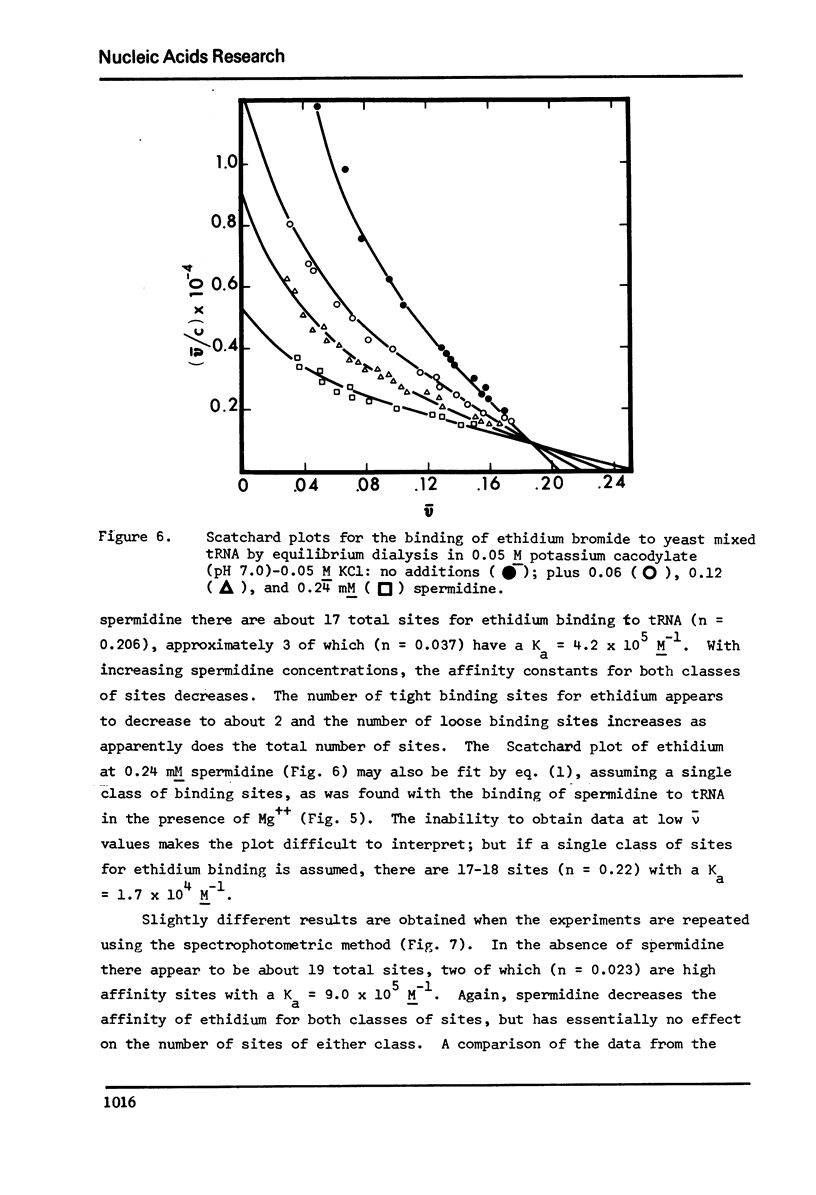
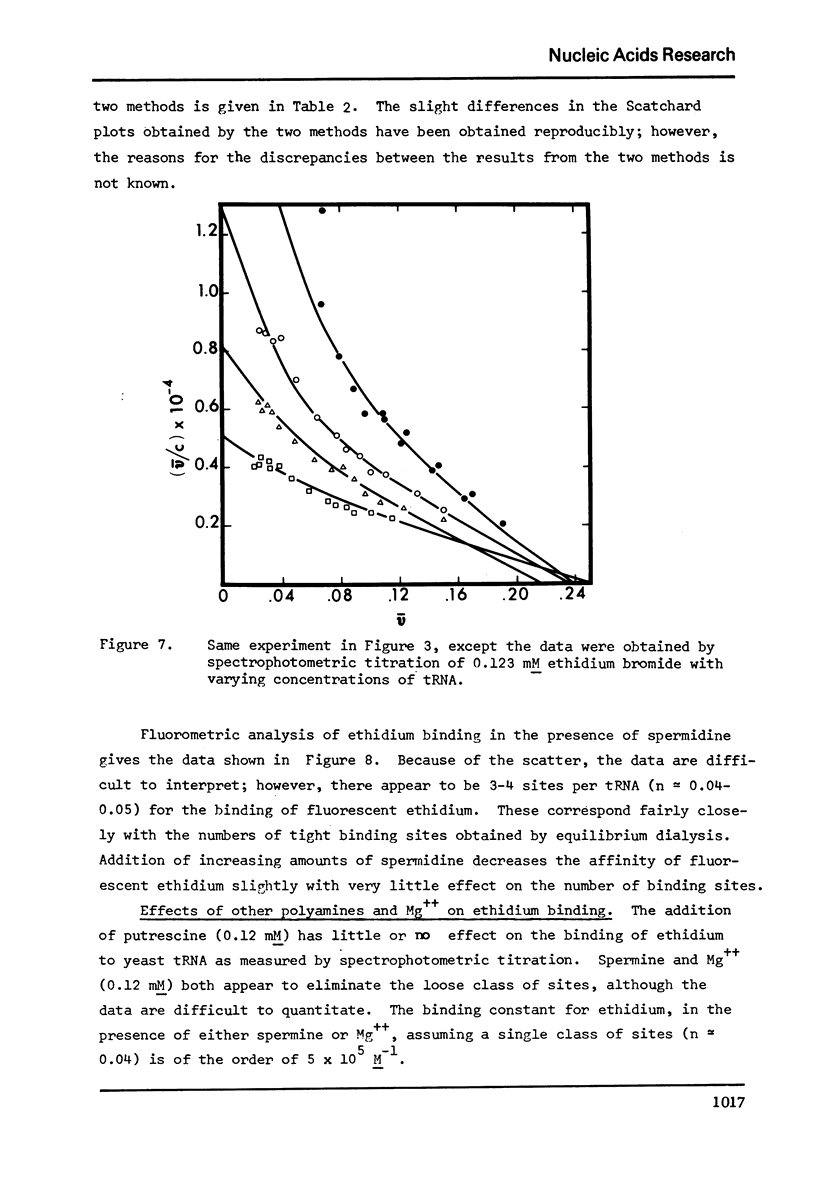
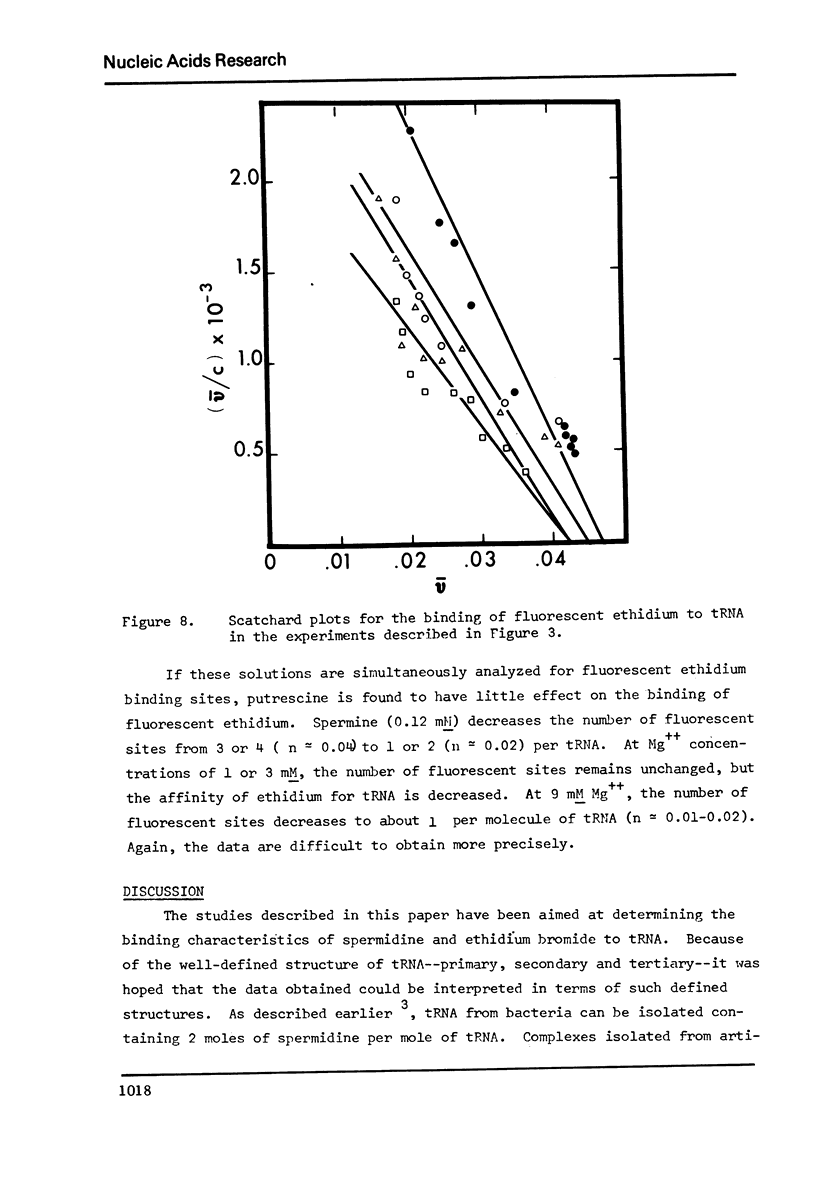
The binding of spermidine and ethidium bromide to mixed tRNA and phenylalanine tRNA has been studied under equilibrium conditions. The numbers and classes of binding sites obtained have been compared to those found in complexes isolated by gel filtration a low ionic strength. The latter complexes contain 10-11 moles of either spermidine or ethidium per mole of tRNA; either cation is completely displaceable by the other. In ethidium complexes, the first 2-3 moles are bound in fluorescent binding sites; the remaining 7-8 molecules bind in non-fluorescent form. At least one of the binding sites for spermidine appears similar to a binding site for fluorescent ethidium. Similar results are found with E. coli formylmethionine tRNA. Spermine, in excess of 18-20 moles per mole tRNA, causes precipitation of the complex. Putrescine does not form isolable complexes with yeast tRNA and displaces ethidium less readily from preformed ethidium-tRNA complexes. Under equilibrium conditions, in the absence of Mg++, there are 16-17 moles of spermidine bound per mole of tRNA as determined by equilibrium dialysis. Of these, 2-3 bind with a Ksence of 9 mM Mg++, the total number of binding sites is decreased slightly and there appears to be only one class of sites with a Ka = 600 M(-1). Quantitatively similar results are obtained for the binding of spermidine to yeast phenylalanine tRNA. When the interaction between ethidium bromide and mixed tRNA is studied by equilibrium dialysis or spectrophotometric titration, two classes of binding sites are obtained: 2-3 molecules bind with an average Ka = 6.6 x 10(5) M(-1) and 14-15 molecules bind with an average Ka = 4.1 x 10(4) M(-1). Spermidine, spermine, and Mg++ compete effectively for both classes of ethidium sites and have the effect of reducing the apparent binding constants for ethidium. When the binding of ethidium is studied by fluorometry, there are 3-4 highly fluorescent sites per tRNA. These sites are also affected by spermidine, spermine and Mg++. Putrescine has little effect on any of the classes of binding sites. These data are consistent with those found under non-equilibrium conditions. They suggest that polyamines bind to fairly specific regions of tRNA and may be involved in the maintenance of certain structural features of tRNA.
Full text
PDF
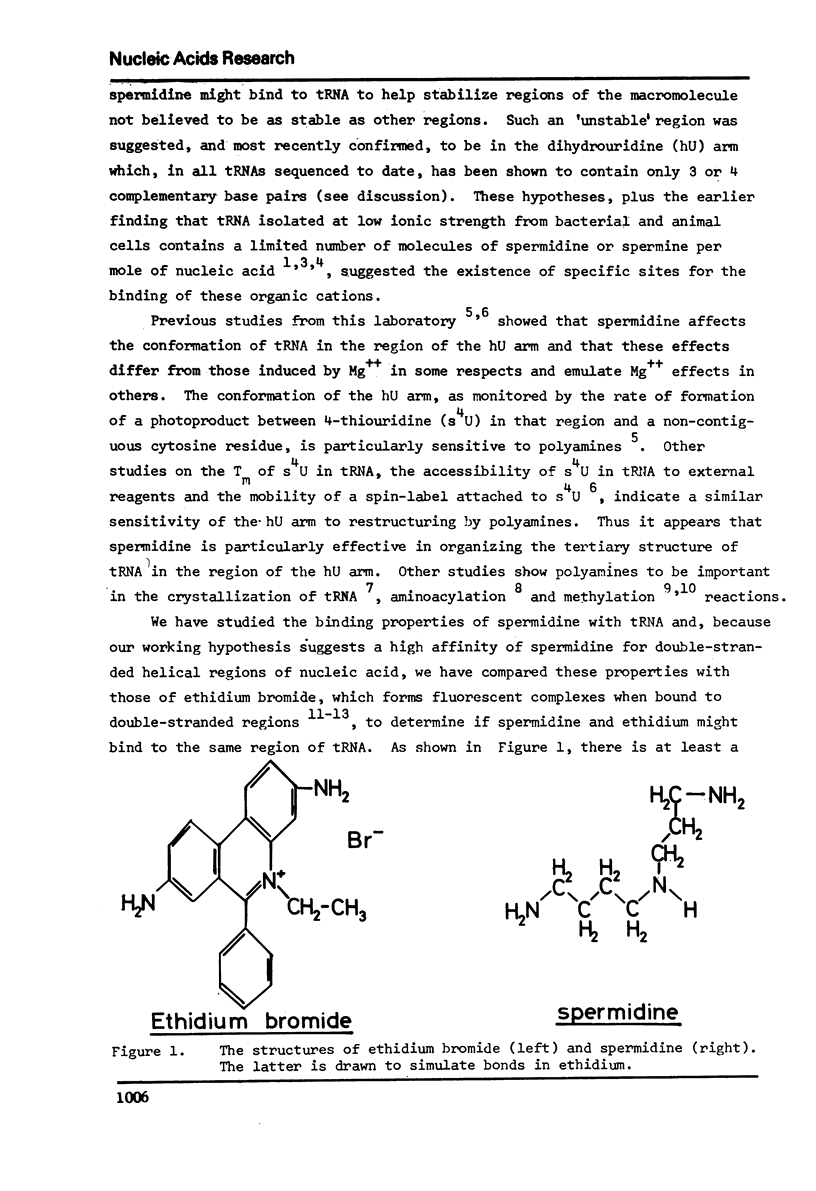



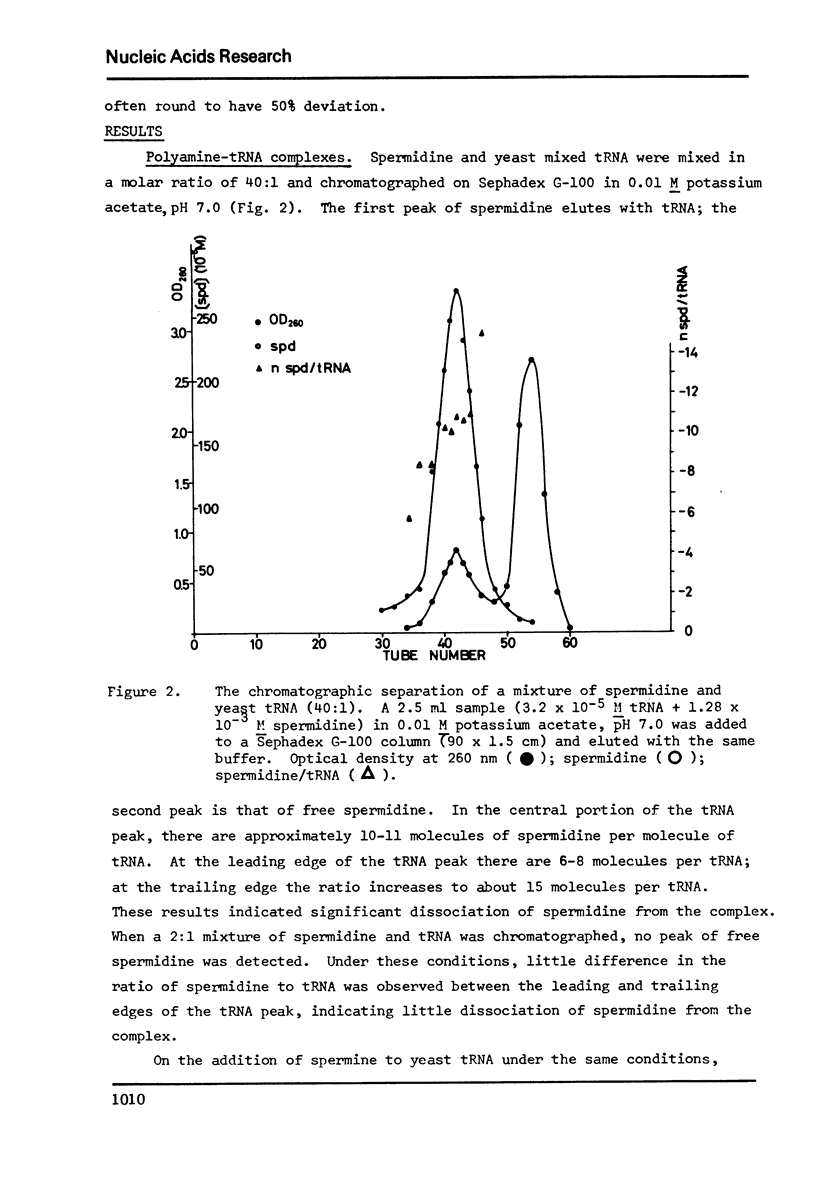
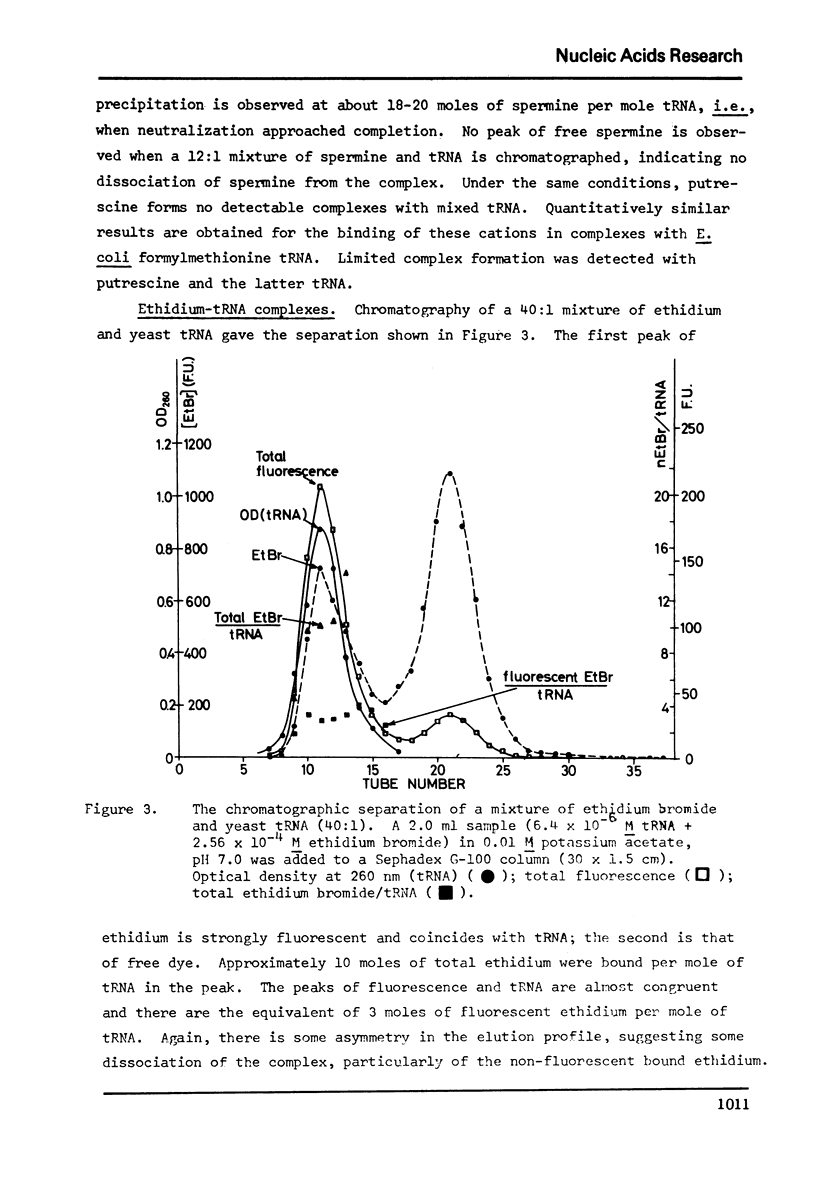

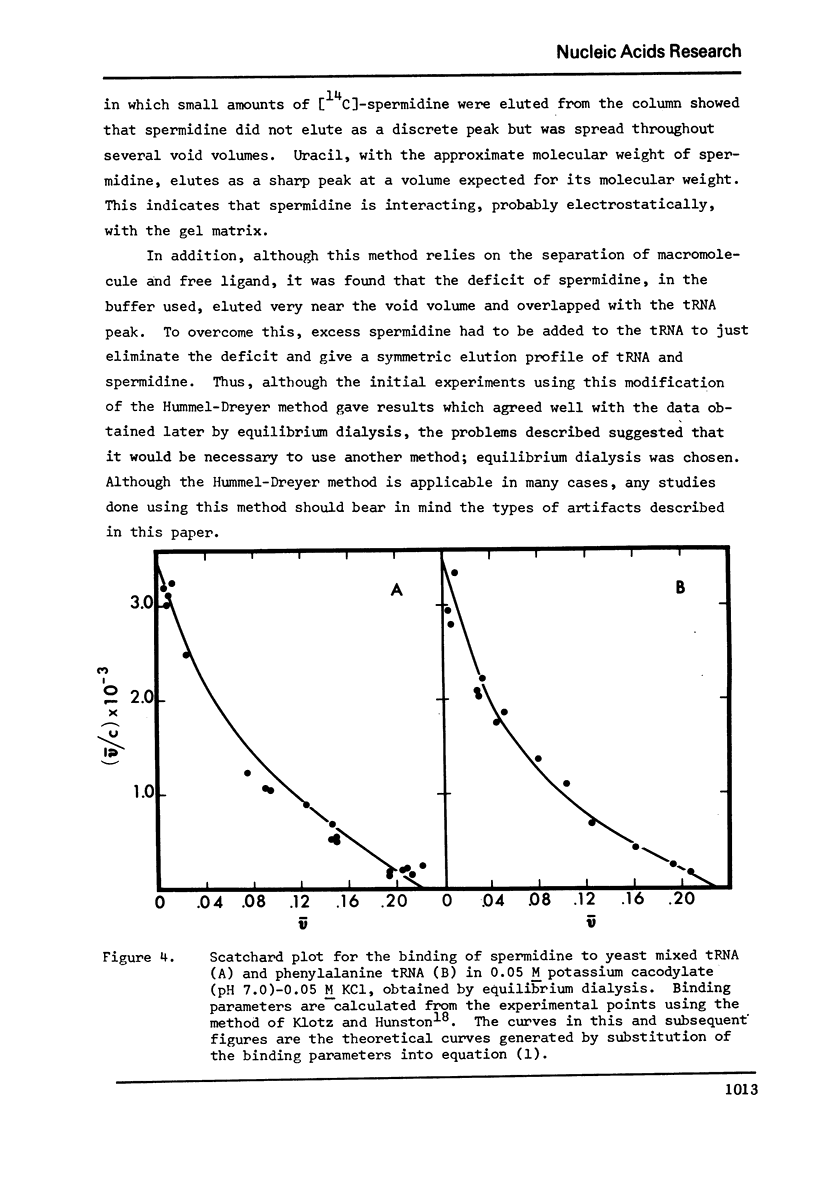




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cohen S. S., Morgan S., Streibel E. The polyamine content of the tRNA of E. coli. Proc Natl Acad Sci U S A. 1969 Oct;64(2):669–676. doi: 10.1073/pnas.64.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts S. M., Gangloff J., Dirheimer G. Conformational transitions in tRNA Asp (brewer's yeast). Thermodynamic, kinetic, and enzymatic measurements on oligonucleotide fragments and the intact molecule. Biochemistry. 1974 Sep 10;13(19):3938–3948. doi: 10.1021/bi00716a019. [DOI] [PubMed] [Google Scholar]
- HUMMEL J. P., DREYER W. J. Measurement of protein-binding phenomena by gel filtration. Biochim Biophys Acta. 1962 Oct 8;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- Hampel A., Bock R. General procedure for crystallization of transfer ribonucleic acid. Biochemistry. 1970 Apr 28;9(9):1873–1880. doi: 10.1021/bi00811a002. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Shulman R. G., Kim S. H. High resolution NMR study of the melting of yeast tRNA Phe. Biochem Biophys Res Commun. 1973 Dec 10;55(3):953–960. doi: 10.1016/0006-291x(73)91235-7. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Takeda Y. Polyamines and protein synthesis. VI. Role of spermine in aminoacyl-tRNA formation. Biochim Biophys Acta. 1970 Jul 16;213(1):240–243. doi: 10.1016/0005-2787(70)90029-8. [DOI] [PubMed] [Google Scholar]
- Jones C. R., Kearns D. R. Investigation of the structure of yeast tRNAphe by nuclear magnetic resonance: paramagnetic rare earth ion probes of structure. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4237–4240. doi: 10.1073/pnas.71.10.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne M. S., Cohn M. Enhancement of Tb(III) and Eu(III) fluorescence in complexes with Escherichia coli tRNA. Biochemistry. 1974 Sep 24;13(20):4159–4165. doi: 10.1021/bi00717a014. [DOI] [PubMed] [Google Scholar]
- Kearns D. R., Wong Y. P., Chang S. H., Hawkins E. Investigation of the structure of native and denatured conformations of tRNALeu3 by high-resolution nuclear magnetic resonance. Biochemistry. 1974 Nov 5;13(23):4736–4746. doi: 10.1021/bi00720a009. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Klotz I. M., Hunston D. L. Properties of graphical representations of multiple classes of binding sites. Biochemistry. 1971 Aug 3;10(16):3065–3069. doi: 10.1021/bi00792a013. [DOI] [PubMed] [Google Scholar]
- Klug A., Robertus J. D., Ladner J. E., Brown R. S., Finch J. T. Conservation of the molecular structure of yeast phenylalanine transfer RNA in two crystal forms. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3711–3715. doi: 10.1073/pnas.71.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEPECQ J. B., YOT P., PAOLETTI C. INTERACTION DU BROMHYDRATE D''ETHIDIUM (BET) AVEC LES ACIDES NUCL'EIQUES (A. N.). ETUDE SPECTROFLUORIM'ETRIQUE. C R Hebd Seances Acad Sci. 1964 Sep 7;259:1786–1789. [PubMed] [Google Scholar]
- Le Pecq J. B., Paoletti C. A new fluorometric method for RNA and DNA determination. Anal Biochem. 1966 Oct;17(1):100–107. doi: 10.1016/0003-2697(66)90012-1. [DOI] [PubMed] [Google Scholar]
- Le Pecq J. B., Paoletti C. Interaction du bromhydrate d'éthidium (BET) avec les polyribonucléotides. Applications à l'étude des réactions d'hybridation. C R Acad Sci Hebd Seances Acad Sci D. 1965 Jun 28;260(26):7033–7036. [PubMed] [Google Scholar]
- LePecq J. B., Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol. 1967 Jul 14;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- Leboy P. S. Influence of polyamines and salts on changing patterns of tRNA methylation. FEBS Lett. 1971 Aug 1;16(2):117–120. doi: 10.1016/0014-5793(71)80347-2. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Henley D. D., Fresco J. R. Molecular weight and molecular weight distribution of unfractionated yeast transfer ribonucleic acid. J Am Chem Soc. 1965 Nov 5;87(21):4961–4963. doi: 10.1021/ja00949a055. [DOI] [PubMed] [Google Scholar]
- Pochon F., Cohen S. S. 4-Thiouridine and the conformation of E. coli tRNA induced by spermidine. Biochem Biophys Res Commun. 1972 May 26;47(4):720–726. doi: 10.1016/0006-291x(72)90551-7. [DOI] [PubMed] [Google Scholar]
- Tritton T. R., Mohr S. C. Kinetics of ethidium bromide binding as a probe of transfer ribonucleic acid structure. Biochemistry. 1973 Feb 27;12(5):905–914. doi: 10.1021/bi00729a018. [DOI] [PubMed] [Google Scholar]
- Tritton T. R., Mohr S. C. Relaxation kinetics of the binding of ethidium bromide to unfractionated yeast tRNA at low dye-phosphate ratio. Biochem Biophys Res Commun. 1971 Dec 3;45(5):1240–1249. doi: 10.1016/0006-291x(71)90151-3. [DOI] [PubMed] [Google Scholar]
- Urbanke C., Römer R., Maass G. The binding of ethidium bromide to different conformations of tRNA. Unfolding of tertiary structure. Eur J Biochem. 1973 Mar 15;33(3):511–516. doi: 10.1111/j.1432-1033.1973.tb02710.x. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Complex formation between ethidium bromide and nucleic acids. J Mol Biol. 1965 Aug;13(1):269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Structural requirements for the binding of ethidium to nucleic acids. Biochim Biophys Acta. 1966 Feb 21;114(2):234–244. doi: 10.1016/0005-2787(66)90305-4. [DOI] [PubMed] [Google Scholar]
- Wildenauer D., Gross H. J., Riesner D. Enzymatic methylations: III. Cadaverine-induced conformational changes of E. coli tRNA fMet as evidenced by the availability of a specific adenosine and a specific cytidine residue for methylation. Nucleic Acids Res. 1974 Sep;1(9):1165–1182. doi: 10.1093/nar/1.9.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson J. M., Kearns D. R. Letter: Europium as a fluorescent probe of metal binding sites on transfer ribonucleic acid. I. Binding to Escherichia coli formylmethionine transfer ribonucleic acid. J Am Chem Soc. 1974 May 29;96(11):3653–3654. doi: 10.1021/ja00818a052. [DOI] [PubMed] [Google Scholar]
- Young D. V., Srinivasan P. R. The effect of polyamines on the methylation of Escherichia coli methyl-deficient transfer RNA by their homologous methylases. Biochim Biophys Acta. 1971 May 27;238(3):447–463. doi: 10.1016/0005-2787(71)90619-8. [DOI] [PubMed] [Google Scholar]


