Abstract
Three tumor suppressor genes at the small (<50 kb) INK4-ARF (CDKN2A/B) locus on human chromosome 9p21 coordinate a signalling network that depends on the activities of the retinoblastoma protein (RB) and the p53 transcription factor. Disruption of this circuitry, frequently by co-deletion of INK4-ARF, is a hallmark of cancer, begging the question of why the intimate genetic linkage of these tumor suppressor genes has been maintained in mammals despite the risk of their co-inactivation. The INK4-ARF locus is not highly expressed under normal physiologic conditions in young mammals, but its induction becomes more pronounced as animals age. Notably, INK4-ARF is actively silenced en bloc in embryonic, fetal, and adult stem cells but becomes poised to respond to oncogenic stress signals as stem cells lose their self-renewal capacity and differentiate, thereby providing a potent barrier to tumor formation. Epigenetic remodeling of the locus as a whole provides a mechanism for coordinating the activities of RB and p53. A hypothesis is that the INK4-ARF locus may have evolved to physiologically restrict the self-renewal capacities and numbers of stem and progenitor cells with the attendant consequence of limiting tissue regenerative capacity, particularly as animals age. Deletion of INK4-ARF contributes to the aberrant self-renewal capacity of tumor cells and occurs frequently in many forms of human cancer.
Introduction
Tumor suppressor genes (TSGs) counter deleterious actions of oncogenes by restricting the proliferation of incipient cancer cells. Prototypic TSGs are recessive, their bi-allelic inactivation being required for complete loss of function. RB1 (hereafter RB)and TP53 (hereafter p53), among the earliest discovered and canonical TSGs, regulate a signalling network that prevents aberrant cellular self-renewal. The INK4-ARF locus (formally designated CDKN2A and CDKN2B) includes three intimately linked TSGs (INK4A, INK4B, and ARF) that trigger the anti-proliferative activities of both RB and p53. Hence, INK4-ARF deletion or silencing mimics effects of RB and p53 co-inactivation, endowing cells with an aberrantly enhanced proliferative potential.1
RB and two other RB-family members [RBL1 (p107) and RBL2 (p130)]integrate extracellular signals that regulate progression through the cell division cycle. Growth factor signalling acts in part through the induction and stabilization of G1 cyclin D-dependent kinases (CDK4 and CDK6) that phosphorylate and inactivate the RB-family members, which, in turn, act as transcriptional corepressors. Principal among factors regulated by the RB-family are the E2Fs, which coordinate the expression of genes that enforce entry into S-phase and maintain cells in cycle (Figure 1). However, distinct molecular complexes containing RB-family proteins play additional roles in facilitating cellular quiescence, modulating differentiation decisions, maintaining chromosomal stability, dampening apoptosis, and enforcing senescence.2
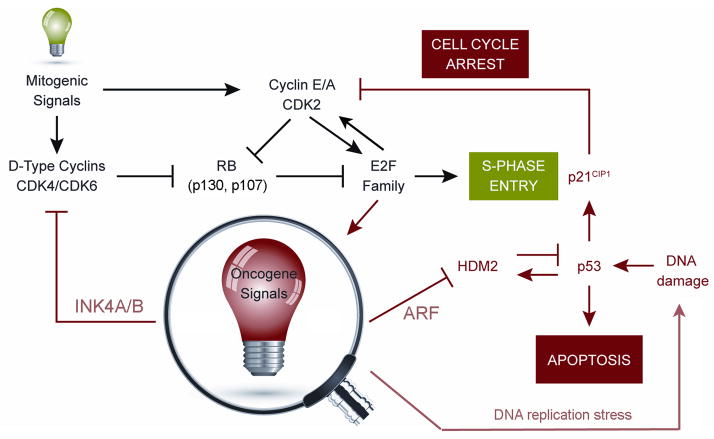
FIGURE 1.
The INK4-ARF signalling network. Physiologic mitogenic signals (green light, top left) stimulate the transcription of genes encoding D-type cyclins and facilitate their assembly into stable complexes with cyclin-dependent kinases (CDK4 and CDK6). These kinases promote the initial phosphorylation of RB and other RB family members (p130 and p107), cancelling their negative regulation of E2F transcription factors and triggering an E2F-dependent program that stimulates entry into the DNA synthetic (S) phase of the cell division cycle. E2F-responsive genes include those encoding cyclins E and A, which assemble with CDK2 to enforce RB-family protein phosphorylation and drive S-phase entry. Aberrant thresholds of hyperproliferative signals emanating from constitutively active oncogenes (magnified red light, bottom) activate INK4-ARF gene expression to inhibit the activities of cyclin-dependent kinases and HDM2. ARF-mediated inhibition of HDM2 E3 ubiquitin ligase activates the p53 transcriptional program, leading either to apoptosis or cell cycle arrest. Apart from the INK4 proteins, another key mediator of cell cycle arrest is the p53-responsive CDK2 inhibitor, p21Cip1. Multiple types of DNA damage activate p53, including DNA replication errors triggered by oncogenes (bottom right). Many feedback loops regulate the network. Inactivation of p53 leads to increased ARF expression; loss of RB leads to increased p16INK4A levels (not shown). At least one of the transcription factors activating the ARF gene is E2F.
Unlike RB which is largely controlled by physiologic cues, p53 is induced by stress, typically involving genomic damage incurred in response to DNA replication errors, irradiation and genotoxic drugs, failure of the mitotic spindle checkpoint, telomere attrition, hypoxia, reactive oxygen species, and oncogene activation. Genes induced by p53 include those encoding CDK inhibitors (CDKN1A, p21Cip1), pro-apoptotic proteins, and MDM2 which inhibits p53-induced transcription and ubiquitinates p53 to target its degradation and terminate the p53 response. Activation of p53 initiates a program of gene expression that leads to cell cycle arrest or apoptosis, thereby eliminating incipient cancer cells.3,4
Two genes within the INK4-ARF locus – INK4A and INK4B – encode polypeptide inhibitors (p16INK4A and p15INK4B) of CDK4and CDK6, preventing the initial G1-phase phosphorylation of Rb-family proteins to maintain them in their growth-suppressive state. The third gene (ARF) specifies a distinct protein (p14ARF in humans and p19Arf in the mouse) encoded in part by an alternative reading frame within the second of three exons that comprise the INK4A gene (Figure 2). The ARF protein antagonizes the E3 ligase activity of MDM2to activate the p53-mediated transcriptional program. Because the INK4A and INK4B genes were defined before ARF was discovered, they were named CDKN (CDK inhibitor)-2A and -2B that, in retrospect, designate only the RB-dependent activities of two of three genes in the cluster. Given the role of INK4-ARF in modulating activities of RB and p53, it is not surprising that deletion of the locus is frequently detected in many distinct tumor types. But, given its compact size and apparent ease of inactivation, why has this TSG cluster been evolutionarily conserved in mammals?
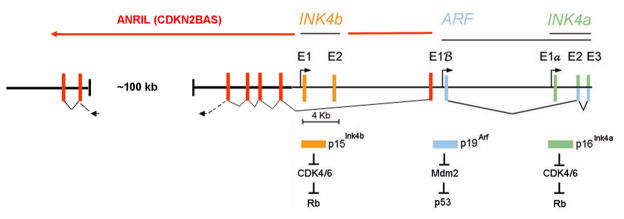
FIGURE 2.
Expanded view of the INK4-ARF locus. The two INK4 genes and ARF are schematically drawn to scale. Rectangles indicate coding exons of the three genes separated by intronic sequences (black horizontal line). Exons 2 (E2) and three (E3) of the INK4A gene (far right) are translated in alternative reading frames to generate the p16Ink4a protein (green exons and bar) and p19Arf protein (blue exons andbar, p14ARF in human cells). Promoters 5′ to INK4b exon 1 (E1) and to the alternative ARF and INK4a 5′ exons (E1β and E1α, respectively) are noted by arrows. The INK4 proteins are shown to inhibit CDK4/6 to maintain Rb in its growth-suppressive mode. By inhibiting Mdm2, p19Arf activates p53. A long intragenic noncoding RNA (designated ANRIL or CDKN2BAS) is transcribed from the ARF promoter (or from an unidentified promoter element close to exon E1β) in an antisense direction with respect to the primary INK4 and ARF transcripts. ANRIL transcripts are spliced, and putative exons are indicated by red rectangles. A ~60 kb segment within the 100 kb gap region illustrated in the schematic includes single nucleotide polymorphisms on human chromosome 9p21 in proximityto CDKN2A/B that are associated with susceptibility to coronary artery disease, aortic aneurysm, stroke, and type II diabetes.
INK4-ARF MEDIATED TUMOR SUPPRESSION
Barriers to cellular self-renewal
Unlike immortal cancer cells, cultured primary cells exhibit a limited proliferative capacity and eventually undergo replicative senescence. Early investigations of DNA tumor viruses led to the realization that co-inactivation of RB and p53 greatly extend cellular lifespan.5 The T antigen of SV40, the E6 and E7 oncoproteins of human papilloma viruses, and theE1A and E1B oncoproteins of adenoviruses disrupt both RB and p53 function, preventing exit from the cell cycle and allowing further population doublings. Notably, many primary rodent somatic cells can bypass senescence and be established as continuously growing cell lines either following DNA tumor virus infection, co-inactivation of RB and p53, or Ink4-Arf deletion. In contrast, human fibroblasts fail to generate established cell lines in response to inactivation of the RB-p53 signaling network.
Although RB-p53 co-inactivation extends the lifespan of cultured primary human cells, they eventually enter a second phase of “crisis” from which only few immortalized clones emerge.6 These species-specific differences between rodent and human cells depend upon the respective lengths of chromosomal telomeres which are much longer in laboratory mouse strains than in human cells, and which, when critically shortened after repeated cell divisions, trigger chromosomal end-to-end joining, fusion-bridge-breakage cycles, mitotic catastrophe and cell death.6–8 Continuously proliferating human cells are more precipitously called upon to solve “the telomere end-replication problem”, usually by reactivating telomerase or using alternative recombinational mechanisms to protect the ends of chromosomes.6 Inactivating telomerase in the germ line of laboratory mice leads to symptoms of DNA damage and organismal aging only after the deficient strains are interbred for multiple sequential generations, which eventually shortens their telomeres and “humanizes” them.7–8 Strikingly, these deficiencies in late generation telomerase-deficient mice are reversed by reinstating telomerase activity.9 Thus, unlike more permissive rodent cells, the immortalization of human cell strains following inactivation of RB-p53 or INK4-ARF is highly subject to the additional restraints imposed by telomere attrition.
TSG activities of INK4 and ARF genes in humans and mice
Deletion of INK4-ARF in cancers provoked attempts to ascertain which product(s) of the locus limit the formation or progression of different tumor types. INK4A undergoes inactivating point mutations in various human cancers,10–12 and mutations that target exon1α (Figure 2) leave ARF coding sequences unaffected, underscoring the primacy of INK4A in these tumors. In mice, the germline deletion of Ink4a alone predisposes animals to spontaneous tumorigenesis and accelerates the appearance of carcinogen-induced cancers.13 Few spontaneous tumors arise in a strain carrying an Ink4a point mutation that is frequently detected in human tumors, although interbreeding these animals to Arf heterozygotes sensitizes them to carcinogen-induced melanomagesis.14 Mutations that disable p16INK4A perturb the structure of tandem ankyrin repeats that make up the body of the protein, resulting in its inability to bind and inhibit CDK4 and CDK6.15 For reasons that remain unclear, mutations targeting INK4B in human tumors are far less common.10–12 In mice, disruption of Ink4b alone has only modest phenotypic effects, but Ink4b compensates for Ink4a inactivation and plays a more crucial role when p16Ink4a is absent.16 The presence of two other chromosomally unlinked INK4 family members, CDKN2C (INK4C)and CDKN2D (INK4D), that encode proteins with CDK inhibitory activities biochemically indistinguishable from those of INK4A and INK4B, further complicate the picture.10 INK4C has TSG activity and can compensate for INK4A loss in some cancers,17 but INK4D lacks documented TSG activity, and its disruption in mice is well tolerated.
The ARF protein bears no structural relationship to INK4 proteins. It is unusually basic and composed of approximately 20% arginine residues; the polypeptide exhibits no recognizable motifs, is natively unstructured, and acquires activity and stability only when bound to targets, such as MDM2.18 ARF can bind to many proteins, but the biological significance of most such interactions remains controversial. At least one other associating protein, nucleophosmin (NPM/B23), binds p19Arf with high stoichiometry, targets it to nucleoli, and stabilizes p19Arf by preventing its N-terminal ubiquitination and proteasomal degradation. NPM-Arf complexes do not contain Mdm2, and vice versa. Although most tumor suppressor activity of p19Arf is mediated through the Mdm2-p53 axis, p19Arf also has p53-independent activities, including an ability to induce sumoylation of other proteins, Mdm2 and NPM among them.18 The N-terminal domain of p19Arf, encoded in toto by exon-1β and comprising only a little more than 60 amino acid residues, is sufficient to bind to and inhibit Mdm2 E3 ligase activity, thereby inducing p53. Together, these features suggest that missense mutations that disable ARF function are improbable, and indeed, its specific inactivation by mutation is relatively rare.11
In murine cancers, targeted deletion of Arf alone (with retention of Ink4 gene function) is highly oncogenic,19 and results in comparatively greater spontaneous tumor penetrance than inactivation of Ink4a or Ink4b.13,14,16 Yet, all three genes manifest TSG activities, and their co-deletion leads to more dramatic effects than the inactivation of any one alone.16,20 In humans, the fact that INK4A is frequently mutated, whereas ARF alone is not has led to the general impression that alterations affecting p16INK4A play the prominent role in tumor suppression, whereas Arf figures more importantly in murine tumors. Nonetheless, given that frequent INK4-ARF deletions and p53 mutations occur as mutually exclusive events in certain human cancers, a parsimonious interpretation is that ARF interacts epistatically with p53 to suppress evolution of these tumor types.
Although the INK4 and ARF proteins separately target the RB and p53 “pathways” (Figure 2), there is significant crosstalk within the signalling network (Figure 1). For example, dismantling RB, either through INK4 mutation, cyclin D-CDK overexpression, or RB loss per se results in robust E2F activation of the ARF promoter,21 invoking ARF-MDM2-p53 signalling to protect cells from RB loss-of-function. In turn, p53-mediated induction of p21Cip1, a potent inhibitor of CDK2, leads to reduced RB phosphorylation, promoting cell cycle arrest. The fact that p16INK4A levels rise in response to RB inactivation, and that ARF expression is greatly increased when p53 function is abrogated, provide further examples of feedback control.22 Thus, the signalling circuits modulated by INK4-ARF are highly interconnected.
The configuration of the INK4-ARF genesis conserved in mammals, including the alternative exon-2 codon usage of INK4A and ARF. However, these features are not conserved in lower vertebrates.12 For example, only a rudimentary ARF gene encoded almost in its entirety by exon-1β is found in chickens, which lack exon-1α, do not encode p16INK4a, but retain an INK4B ortholog. In contrast, an INK4B-like gene is found in fish, but ARF is absent. Based on nucleotide sequence and species comparisons, it has been suggested that INK4A arose by duplication of INK4B, and that ARF exon-1β was acquired subsequently. In mammals, coordinate epigenetic regulation of the three closely spaced promoters (Figure 2) may confer an evolutionarily acquired advantage (see below).12
Stress-induced activation and mechanisms of INK4-ARF tumor suppression
Expression of INK4-ARF is not detected in most somatic tissues of young mammals, but it can be induced by various forms of oncogenic stress. Observations that an activated RAS oncoprotein induced both p16INK4A and p53 in primary fibroblasts provided the first evidence linking oncogenic signalling with INK4A induction.23 Expression of INK4A in response to constitutively active mutant forms of RAS has been well documented in cultured human and mouse primary cells, as well as in precancerous lesions from patients. In human cells, enforced RAS-RAF pathway activation has been reported to induce p16INK4A, but not p14ARF, whereas Ink4a and Arf tend to be co-regulated by mutant Ras in rodents.11,12
While remaining unresponsive to physiologic levels of mitogenic signalling, the entire murine Ink4-Arf locus is poised to respond to aberrant thresholds of hyperproliferative signals emanating from constitutively activated oncoproteins (Figure 1).1,4 Notably, oncogene-induced triggering of Ink4-Arf gene expression is an indolent process. This suggests that the locus is insulated in responding to acute stress signals, but undergoes chromatin remodelling and is progressively induced in the course of chronic oncogene stimulation. In mice, the general paradigm is that Arf loss of function, even when Ink4 genes are conserved, collaborates with oncogene activation to accelerate tumor progression and metastasis, closely mimicking effects of p53 inactivation. Functional collaboration between Arf inactivation and different up-regulated oncoproteins, including Ras, Myc, mutant EGF and PDGF receptors, Bcr-Abl, Wnt and others in mouse models, strongly supports the view that the Arf-Mdm2-p53 axis monitors mitogenic signal intensities, and that Arf loss, in turn, relieves oncogene-initiated cancer cells from such restraint.1,4
One outcome of oncogene-induced p53 and p16IINK4A activation is the induction of cellular senescence in vivo leading to durable cell cycle arrest and the eventual elimination of senescent cells.23 Senescence is characterized by lack of responsiveness to mitogenic growth factors, extensive chromatin remodeling (appearance of senescence-activated heterochromatic foci), and global gene expression changes associated with characteristic alterations in cellular morphology, production of particular inflammatory cytokines, and up-regulation of biomarkers, such as senescence-activated β-galactosidase. 24,25 These features distinguish senescent cells from quiescent (G0) populations that can re-enter the cell division cycle when stimulated with appropriate mitogens. Moreover, establishment of the senescent state seems to be a relatively slow process that requires the prolonged maintenance of p16INK4A-induced inhibition. Senescence provides a barrier to tumor formation and has been documented in pre-malignant tissues in rodents and humans.25–27 The host immune system is activated by cytokines released from senescent cells, triggering their elimination.25,27 In mice, both Ink4a and Arf can elicit oncogene-induced senescence; however, p16INK4A appears to play the central role in human cells.11,26
Apart from impinging on the INK4-ARF locus, oncogene activation in early premalignant tumors increases the frequency of DNA replication errors, resulting in a DNA damage response that activates p53 (Figure 1).28,29 Analysis of human precancerous lesions has indicated that markers of senescence and DNA damage are frequently concordant, implying that (ARF-independent) ATM/ATR kinase signalling strongly contributes to oncoprotein-mediated p53 activation. A frequently misunderstood distinction is that while p53 is activated by acute DNA damage, Arf is not.18,19 In turn, p53 activity is readily induced by DNA damage in Arf-null cells.19 In marked contrast, many chronic DNA damage signals have been reported to induce p16INK4A,11 further implying that various signalling inputs differentially engage INK4A and ARF.
Given that DNA damage stems from both oncogene-induced senescence,25–29 and telomere dysfunction,30,31 each of which has been associated with organismal aging,11,31,32 Arf would seem unlikely to be directly involved in these particular processes. Reinforcing this view, responses to telomere shortening, DNA damage, and degenerative aging that are reversed by p53 deletion in mice33 are not attenuated by disruption of Ink4a-Arf.34 If, in fact, DNA damage is central to malignant transformation, the p53-dependent (Arf-independent) DNA damage response should bear major responsibility for tumor suppression. This has been challenged by experiments in which a p53 allele that could be toggled between inactive and active states was functionally restored at different times after mice were exposed to ionizing radiation. In this setting, widespread p53-dependent apoptosis induced by DNA damage did not contribute to tumor suppression, whereas restoration of p53 after the DNA damage response had subsided was potently tumor suppressive and depended upon p19Arf.4
Key questions therefore concern the degree to which p53 is activated by oncogene-induced DNA damage, by ARF, or through both pathways in precancerous tissues, and whether there are significant species-specific differences in the manner by which the RB/p53/INK4-ARF tumor suppressor network responds to stress signals in humans and mice. Whatever the circuitry, oncogenic stress not only provides the force that induces INK4-ARF gene expression but also selects for the emergence of rare cells that delete the locus and subsequently escape tumor suppression. Thus, the finding that a cancer cell has sustained an INK4-ARF deletion may be taken as evidence that they locus was activated at some earlier stage of tumor development.
INK4-ARF AND STEM CELL SELF-RENEWAL
Silencing of INK4-ARF in stem cells
Self-renewal of dividing stem cells requires mechanisms that maintain both their pluripotent state and capacity to differentiate. In embryonic stem (ES) cells, pluripotency is promoted by an autoregulatory circuit of “core factors” (principally Oct4, Sox2, and Nanog) that repress cell differentiation.35 In turn, enforced expression of Oct4 and various combinations of other transcription factors, including Sox2 and Nanog, can reprogram somatic cells into induced pluripotent stem (iPS) cells, reversing the epigenetic landscape and resetting it to an ES-like state. ES cell division cycles are characterized by a short G1 phase marked by little or no hypophosphorylated RB, desensitizing them to cyclin D-dependent kinase regulation by mitogens or, conversely, to Ink4 inhibitors. ES cells also resist p53-dependent cell cycle arrest. Hence, they are relatively refractory to Ink4-Arf-mediated TSG activity (Box 1).
Ink4-Arfregulation and stage-specific tumor suppression
Cardinal features of cell cycle control and regulation of the Ink4-Arf/RB/p53 network are indicated. HSCs and pro-B cells exemplify several well defined differences between tissue stem cells and their more differentiated progeny.
Embryonic Stem Cells
G1 interval is short
D-type cyclin levels are low.
RB is constitutively hyperphosphorylated by cyclin E/A-CDK2.
p53 is insulated from activation by DNA damage.
Ink4-Arf locus is silenced.
Hematopoietic Stem Cells (fetal and young adult)
G1 interval is regulated by environmental cues.
D-type cyclins are essential.
RB is sequentially phosphorylated by cyclin D/E/A-CDKs
p53 is acutely activated by DNA damage (Atm/Atr pathways).
Ink4-Arf locus is silenced.
Pro-B cells (young adult)
G1 interval is regulated by environmental cues.
D-type cyclins are essential.
RB is sequentially phosphorylated by cyclin D/E/A-CDKs.
p53 is acutely activated by DNA damage (Atm/Atr pathways).
Ink4-Arf locus responds to hyperproliferative signals (Ink4a, Arf) and DNA damage (Ink4a). Deletion promotes cancer.
HSCs and Pro-B cells (aged adult)
Stress-induced DNA damage and cumulative mutations trigger apoptosis and cellular senescence.
Ink4-Arf expression increases with age.
Tissue regenerative capacity is diminished.
Cells resist oncogene-induced transformation, but disruption of Ink4-Arf/RB/p53 network predisposes to cancer.
ES cells can give rise to diverse tissue stem cells which, while multipotent, are generally restricted to forming cells from their tissues of origin. This hierarchical design connotes a loss of plasticity that accompanies tissue specification during development. Unlike ES cells, tissue-specific stem cells, such as hematopoietic stem cells (HSCs), neural stem cells (NSCs) and others, alter their cell cycles and become dependent upon extracellular mitogens that activate formation of cyclin D-CDK complexes during G1 phase.35 These enzymes not only promote G1 phase progression by phosphorylating and inactivating the Rb-family proteins, but they also sequester the p21Cip1 and p27Kip1 inhibitors of cyclin E/A-driven Cdk2 to facilitate S phase entry.36 Underscoring the differences between ES cells and HSCs, disruption of all three D-type cyclin genes prevents HSCs from undergoing normal cell division, resulting in their loss during fetal development.37 Presumably, the emergence of regulatory control by cyclin D-CDK and Rb-family proteins in fetal and adult tissue stem cells reflects an ability of extracellular cues to determine their proliferative capacity and Influence their differentiation.
Despite their dependency on D-type cyclins, HSCs do not normally modulate Rb activity through engagement of the Ink4-Arf locus, which is kept in an epigenetically silenced state (Box 1). In young adult and fetal HSCs, active repression of Ink4a-Arf is maintained by different silencing complexes (Bmi1-containing Polycomb and Hmg2a-containing complexes, respectively).38,39 Fetal hematopoiesis proceeds in mice lacking Bmi1, but newborn animals die early in life from hematopoietic failure resulting from loss of bone marrow-derived HSCs. Defects in both HSCs and NSCs are significantly rescued in Bmi1-null mice that also lack Ink4a-Arf function,40,41 dramatically highlighting a requirement for Ink4-Arf repression in young adult stem cells. Conversely, co-deletion of Ink4a-Arf and p53 leads to rapid expansion of multipotent progenitors from HSCs.42 The fact that the Ink4-Arf locus is silenced in stem cells but becomes increasingly responsive to stress signals in more differentiated progenitors (such as pro-B cells, Box 1) reinforces the view that cells gain greater TSG potential as they lose “stemness”. Turning things on their head, expression of the Ink4-Arf locus may be incompatible with stem cell self-renewal. An attractive hypothesis is that the ability to superimpose these TSG-mediated restraints has been evolutionarily selected as one means of limiting the number of stem cells in various tissues.30,32,35,39
Stage-specific tumor suppression and “cancer stem cells”
Potential INK4-ARF-dependent responses to oncogene activation should also differ in stem cells versus their more differentiated progeny. This appears to be the case in chronic myelogenous leukemia (CML), a myeloproliferative disorder resulting from a chromosomal translocation (the Philadelphia chromosome, Ph+) in which elements of the cellular ABL1 gene on chromosome 9 are fused to a regulatory breakpoint cluster region (BCR) from chromosome 22. The product of the BCR-ABL fusion is a constitutively active tyrosine kinase that is expressed in HSCs and initiates the indolent “chronic phase” of CML in which only few leukemic blast cells reside in the bone marrow, and patients may have limited clinical symptoms. In this setting, INK4-ARF is neither expressed nor deleted,43 and drugs that inhibit the BCR-ABL kinase maintain patients in durable clinical remission. However, untreated cases of chronic phase CML routinely progress to myeloid or lymphoid blast crisis, both of which are aggressive, lethal leukemic syndromes. Deletion of INK4-ARF occurs in virtually all cases of lymphoid blast crisis, implying that differentiation in the B cell lineage facilitates INK4-ARF engagement and selects for its subsequent inactivation.43 (Mutations of p53 occur frequently only in myeloid blast crisis).
Constitutive activation of the BCR-ABL kinase can arise de novo in Ph+ acute lymphoblastic leukemia (ALL), a very aggressive disease with a poor prognosis in which lymphoid progenitors, but not HSCs, are affected. Approximately two-thirds of untreated Ph+ ALL patients at diagnosis already manifest bi-allelic INK4-ARF deletions.43 Mouse models of Ph+ ALL indicate that Arf inactivation conveys resistance to the suite of drugs that target the BCR-ABL kinase,44 and INK4-ARF deletion in human Ph+ ALL patients at diagnosis is an early predictor of relapse following conventional frontline treatment with both targeted and chemotherapeutic agents.45
The cancer stem cell hypothesis proposes that tumors mimic stem cells in retaining a capacity for cellular self-renewal as well as the ability to generate more differentiated cells that cannot propagate the tumor.46 This model is supported by observations that only a variable fraction of cells in a tumor population can reinitiate cancer following their transplantation into immunosuppressed mice, and that these so-called cancer stem cells again give rise to more differentiated, nontumorigenic progeny upon serial transplantation. Differentiated somatic cells that sustain particular genetic alterations can also be reprogrammed to a stem-like state in which they reacquire self-renewal capabilities. Hence, while all cancers share a capacity for unlimited proliferation, not all share the proclivity to differentiate into non-clonogenic progeny. As examples, chronic and acute myelogenous leukemias (CML and AML) conform to the cancer stem cell paradigm,46 whereas de novo Ph+ ALL does not.44,45 Importantly, these hypotheses are not mutually exclusive, and each has received experimental support in studies of different cancers.35 Focusing this argument only on functional collaboration between oncogene activation and INK4-ARF loss-of-function, INK4-ARF silencing in a stem cell or deletion of the locus in a more differentiated cell might equally allow cancer cells to arise.
INK4-ARF AND AGING
The idea that TSGs evolved to prevent the development of cancer is questionable, since in mammals, cancer is a disease that has its greatest impact in aging, post-reproductive populations. p53 is an ancestral gene, whose role in tumor suppression represents a recent evolutionary adaptation.4 Thus, while p53, RB, INK4A, and ARF are inactivated in various cancers, the evolutionary pressure to conserve them may instead reflect their ability to maintain normal tissue homeostasis, rather than to protect against cancer per se.
Although Ink4-Arf is actively silenced in stem cells, and while deletion of the Ink4-Arf locus has only minimal effects on HSC or NSC self-renewal in young mice,35 a decline in the capacity of various organs to respond to physiologic demands in response to stress is a defining characteristic of aging tissues in which expression of p16Ink4a, p15Ink4b and p19Arf increases47 and is associated with reduced stem and progenitor cell function, and with the more limited regenerative capacity of aging tissues (Box 1).48–50 In humans, p16INK4A levels have been used as a biomarker of aging,51 although the role of ARF remains uncertain. In mouse tissues that require cyclin D-dependent kinases for their proliferation (HSCs, NSCs, and pancreatic islet cells) and in which p16Ink4a levels increase markedly with aging, germline p16Ink4a deficiency partially abrogates their age-dependent decline in proliferative capacity, although p16-independent mechanisms continue to play a role.48–50 It has proven more difficult to oncogenically transform aged B-lymphoid cells that have a more limited proliferative capacity than their younger progenitors, but this restrictive behaviour was lost upon Ink4-Arf deletion.52 Somatic deletion of p16Ink4a in murine T or B lymphocytes rescues many aging phenotypes, but, unsurprisingly, promotes B-cell neoplasia.53 Overexpression of the Ink4-Arf locus in mice harboring these genes on a bacterial artificial chromosome limits spontaneous tumorigenesis; these “super Ink4-Arf” mice manifest no evidence of increased aging and exhibit normal lifespan, but when crossed with analogous “super p53” mice, show extended longevity.54 Possibly, age-related increases in Ink4-Arfexpression, while limiting tumor occurrence and regenerative capacity, play a salutary role in restricting deleterious effects of proliferative damage, such as atherosclerosis.
Sahin and DePinho31 highlighted the importance of four pathways that influence tissue aging in response to genotoxic stress. These include (i) downstream components of the phosphatidylinositol 3′-kinase signaling pathway (Pten, Tsc1/2, FoxO and mTOR) that regulate stem cell numbers; (ii) DNA repair pathways, including those responding to telomere attrition, that prevent loss of stem cell reserves; (iii) mitochondrial functions crucial for stem cell maintenance; and (iv) genetic regulators of cellular mortality, including Ink4a-Arf. There is significant cross-talk between these systems. For example, Bmi1, an Ink4a-Arf repressor, facilitates physiological mitochondrial function and prevents overproduction of reactive oxygen species that can activate the DNA damage response.55
Genome-wide association studies across numerous patient samples have pinpointed a group of single nucleotide polymorphisms (SNPs) within 120 kb of the INK4-ARF locus on chromosome 9p21 that are associated with increased risk of development of coronary artery disease (CAD), aortic aneurysm, stroke, and type-2 diabetes, all of which are degenerative disorders associated with aging. SNP variants associated with significantly increased disease risk did not correlate with hyperlipidemia, hyperglycemia, or increased body mass. The region containing these SNPs is transcribed within a primary long intragenic noncoding (LINC) RNA (designated ANRIL or CDKN2BAS) that originates near the ARF promoter, is transcribed in the opposite direction, and “anti-senses” the CDKN2B (INK4B) gene (Figure 2).56 Altered expression of INK4-ARF transcripts is found in individuals harboring a SNP associated with increased risk of atherosclerosis.57 Targeted deletion of the syntenic chromosomal region in mice resulted in downregulation of Ink4-Arf gene expression and an increased propensity of tumor formation.58 Enhancer elements located within the human CAD interval may indirectly regulate ANRIL/CDKN2BAS expression,59 and, in humans, this LINC RNA binds the polycomb component CBX7 to methylate H3K27 and silence INK4A.60 Together, these findings provide additional evidence linking alterations in INK4-ARF gene expression with common age-related disorders.
Conclusion
As a general rule, the entire INK4-ARF locus is epigenetically silenced in embryonic, fetal, and adult stem cells, but in more differentiated cells, it is remodeled to become increasingly responsive to aberrant mitogenic signals that exceed normal physiologic thresholds. This process is reversed when somatic cells are induced to regain pluripotency through iPS reprogramming. Untoward expression of INK4-ARF limits stem cell self-renewal, suggesting that coordinated INK4-ARF expression may normally act to restrict stem cell numbers. As animals age and confront various forms of cellular stress, INK4-ARF expression increases and reflects an age-related decline in tissue regenerative capacity. That cancer is a disease of aging populations implies that cumulative mutational stress throughout life can eventually overcome barriers imposed by TSG activity. Herein lies the yin and yang of cancer and aging.
Acknowledgments
I thank Douglas Green and Sean Morrison for helpful suggestions about the manuscript. Many investigators worldwide have contributed to the studies reviewed in this article, and an overwhelming number of papers extant in the literature now describe functions of what I have called the RB-p53-INK4-ARF tumor suppressive network. Given limitations in allowed citation numbers, I have chosen to highlight excellent and much more detailed review articles written by many prominent experts in the field whose laboratories have made seminal contributions to our understanding of the subject. I sincerely apologize to the many others whose primary contributions could not be cited.
References
- 1.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 2.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Juntilla MR, Evan GI. P53 – a jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–829. doi: 10.1038/nrc2778. [DOI] [PubMed] [Google Scholar]
- 5.Howley PM, Livingston DM. Small DNA tumor viruses: large contributors to biomedical sciences. Virology. 2009;384:256–259. doi: 10.1016/j.virol.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright WE, Shay JW. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
- 7.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp P, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/S0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 8.Artandi SE, Chang S, Lee SL, Alson S, Gottleib GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406: 641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 9.Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, Cadiñanos J, et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2011;469:102–106. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roussel MF. The INK4 family of cell cycle inhibitors in cancer. Oncogene. 1999;18:5311–5317. doi: 10.1038/sj.onc.1202998. [DOI] [PubMed] [Google Scholar]
- 11.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127: 265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 13.Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguiree AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 14.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 15.Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- 16.Krimpenfort P, Ijpenberg A, Song JY, vand der Valk M, Nawjin M, Zevenhoven J, Berns A. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 17.Wiedemeyer R, Brennan C, Hefferman TP, Xiao Y, Mahoney J, Protopopov A, Zheng H, Bignell G, Furnari F, Cavenee WK, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherr CJ. Divorcing Arf and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 19.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame protein p19ARF. Cell. 1997:649–659. doi: 10.1016/S0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 20.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/S0092-8674(00)81079-X. [DOI] [PubMed] [Google Scholar]
- 21.Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. P14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 22.Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras promotes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 88:593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 24.Collado M, Serrano M. The power and promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- 25.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krizhanovsky V, Xue W, Zender L, Yon M, Hernando E, Lowe SW. Implications of cellular senescence in tissue damage response, tumor suppression, and stem cell biology. Cold Spr Hbr Symp Quant Biol. 2008;73:513–522. doi: 10.1101/sqb.2008.73.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halazonetis TD, Gorgoulis VG, Bartek J. Anoncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 29.Campisi J, D’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs JJ, de Lange T. Significant role for p16INK4a in p53-dependent telomere-directed senescence. Curr Biol. 2004;14:2302–2308. doi: 10.1016/j.cub.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 31.Sahin E, DePinho RA. Linking functional decline of telomeres, mitochondria, and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collado M, Blasco MA, Serrano M. Cellular Senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/S0092-8674(00)80762-X. [DOI] [PubMed] [Google Scholar]
- 34.Khoo CM, Carrasco DR, Bosenberg MW, Paik JH, DePinho RA. Ink4a/Arf tumor suppressor does not modulate the degenerative conditions or tumor spectrum of the telomerase-deficient mouse. Proc Natl Acad Sci USA. 2007;104:3931–3936. doi: 10.1073/pnas.0700093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Dev Cell Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 36.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 37.Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 38.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 39.Levi BP, Morrison SJ. Stem cells use distinct self-renewal programs at different ages. Cold Spr Harb Symp Quant Biol. 2008;73:539–553. doi: 10.1101/sqb.2008.73.049. [DOI] [PubMed] [Google Scholar]
- 40.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 41.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akala OO, Park IK, Qian D, Pihalja M, Becker MF, Clarke MF. Long-term haematopoietic reconstitution by Trp53−/− p16Ink4a−/− Arf−/− multipotent progenitors. Nature. 2008;453:228–232. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- 43.Mullighan CG, Williams RT, Downing JR, Sherr CJ. Failure of CDKN2A/B (INK4A/B-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes Dev. 2008;22:1411–1415. doi: 10.1101/gad.1673908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev. 2007;21:2283–2287. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 46.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4A family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 48.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;28;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 49.Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 50.Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev. 2008;22:3115–3120. doi: 10.1101/gad.1715808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Johnson SM, Fedoriw Y, Rogers AB, Yuan H, Krishnamurthy J, Sharpless NE. Expression of p16INK4a prevents cancer and promotes aging in lymphocytes. Blood. 2011;117:3257–3267. doi: 10.1182/blood-2010-09-304402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Via J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3369. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 57.Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, Ibrahim JG, Thomas NE, Sharpless NE. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu X-D, Topol E, Rosenfeld M, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signaling response. Nature. 2011;470:264–270. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]