Abstract
Astrocytes play an active role in the modulation of synaptic transmission by releasing cell-cell signaling molecules in response to various stimuli that evoke a Ca2+ increase. We expand on recent studies of astrocyte intracellular and secreted proteins by examining the astrocyte peptidome in mouse astrocytic cell lines and rat primary cultured astrocytes, as well as those peptides secreted from mouse astrocytic cell lines in response to Ca2+-dependent stimulations. We identified 57 peptides derived from 24 proteins with LC–MS/MS and CE–MS/MS in the astrocytes. Among the secreted peptides, four peptides derived from elongation factor 1, macrophage migration inhibitory factor, peroxiredoxin-5, and galectin-1, were putatively identified by mass-matching to peptides confirmed to be found in astrocytes. Other peptides in the secretion study were mass-matched to those found in prior peptidomics analyses on mouse brain tissue. Complex peptide profiles were observed after stimulation, suggesting that astrocytes are actively involved in peptide secretion. Twenty-six peptides were observed in multiple stimulation experiments but not in controls and thus appear to be released in a Ca2+-dependent manner. These results can be used in future investigations to better understand stimulus-dependent mechanisms of astrocyte peptide secretion.
Keywords: glia, astrocytes, secreted peptides, peptidomics, LC–MS, CE–MS, Ca2+-dependent stimulation
INTRODUCTION
Neurons and glia are the two most common cell types found in the central nervous system (CNS). Neurons play important roles in coordinating CNS activities and function.1 Glia, although present in higher numbers than neurons,2 have been considered as passive companions to neurons with respect to cell-to-cell signaling. Recently, however, more active roles for astrocytes—a common subtype of glia in the CNS—have been reported. They are involved in modulating nervous system circuits by integrating neuronal signals, exhibiting Ca2+-induced excitability, and processing signaling information.3-6 From these landmark studies has emerged the concept of the tripartite synapse, which refers to chemical communication between the pre- and post-synaptic terminals of neurons and surrounding astrocytes.7, 8
Like other cells, glia contain many small molecules, peptides, and proteins. Using a traditional bottom-up approach, proteomic analyses of primary cultured astrocytes have identified hundreds of proteins9, 10 and generated a protein database to aid further studies of astrocyte function and physiology. Astrocytes have also been reported to express membrane ion channels,11, 12 neurotransmitters,4 and receptors13 related to important signaling pathways in the CNS, providing the foundation for astrocyte-astrocyte and astrocyte-neuron communication. The active role of astrocytes that are intimately associated with synapses may be mediated by the release of chemical substances. For instance, recent studies have shown that astrocytes modulate synaptic transmission by releasing small signaling molecules such as glutamate,14 ATP,15 and D-serine16 in response to various stimuli that evoke a Ca2+ increase.17 A proteomics study on astrocyte-conditioned medium also revealed more than 100 proteins released from astrocytes,18, 19 and some of these may be released through a classic vesicular mechanism.20 In contrast to small molecules and large proteins, knowledge about peptide content in astrocytes is limited.21-23 Traditional peptidome measurements have identified a number of endogenous peptides from brain regions that include astrocytes and neurons,24-28 with only a few studies measuring a defined and specific cell type.29-32 Collectively, the body of literature suggests a need for global and systematic investigations of peptides and their secretion from a defined population of astrocytes.
We used two sources of astrocytes in order to enlarge our astrocyte peptide database. More specifically, we identified peptides in mouse astrocytic cell lines and in rat primary cultured astrocytes using liquid chromatography (LC) coupled with electrospray ionization (ESI) ion-trap (IT) tandem mass spectrometry (MS/MS), and capillary electrophoresis (CE) coupled with ESI quadrupole (Qq) time-of-flight (TOF) MS/MS. The identified astrocyte peptides were then compared to the previous peptidomics analyses of mouse brain tissue33 and human cell lines32 to highlight the peptides in common to both sample types.
In order to ensure that the secreted peptides were from astrocytes and not other cell types that inevitably end up in primary astrocyte cultures, we measured secretion only from the well-defined mouse astrocytic cell lines. As only a small fraction of the peptides from these cells is secreted, there is not enough sample to perform MS/MS on the secreted peptides; hence the peptides secreted from the mouse cell lines upon chemical stimulation were identified by mass matching to the peptides identified in the same brain regions or from the same cells, similar to other recent studies.27, 34 In addition, sensitive capillary LC coupled off-line with matrix-assisted laser desorption/ionization (MALDI) TOF MS was used to detect peptides released from the astrocytic cell lines.25, 35 These peptides were then compared with the identified peptides from the rat cell culture homogenates in this work, and with previous peptidomics analyses of mouse brain tissue.33 Unlike proteomics analyses of astrocyte secretion,18, 20 no external enzymes were used in these experiments to produce the peptides.
To determine the range of peptides secreted from the astrocytes, different chemical stimuli were used to induce peptide secretion. These included ionomycin,22, 36, 37 KCl,38-40 serotonin,41-43 and bradykinin,21, 23, 44 all of which have been reported to evoke a Ca2+ increase in astrocytes. Complex peptide profiles were observed after stimulation, suggesting that astrocytes are actively involved in peptide secretion. Twenty-six peptides observed in multiple stimulation experiments but not in controls are more likely to be the subset released in a Ca2+-dependent manner. These peptides can be used to guide future efforts to better understand stimulus-dependent mechanisms of astrocyte peptide secretion.
EXPERIMENTAL PROCEDURES
Cell Cultures
Astrocyte type I cell lines (C8-D1A) derived from the cerebella of C57BL/6 mice were purchased from American Type Cell Collection (Manassas, VA) and maintained in standard Dulbecco’s modified Eagle media (DMEM) with 10% fetal bovine serum (FBS) in a humidified 5% CO2/95% air atmosphere according to the supplier’s protocol.45 This specific astrocyte cell line was used for targeting secreted peptides upon various chemical stimulations and identifying the peptide content in these astrocytes. A primary culture was also used to help enlarge the database of astrocyte peptides for identifying the secreted peptides. Primary cultured astrocytes from rat cortical tissue were prepared as described previously.23 Briefly, 2- to 4-d old Long-Evans/BluGill rat pups (University of Illinois at Urbana-Champaign) were sacrificed by rapid decapitation in compliance with protocols established by the University of Illinois Institutional Animal Care and Use Committee and in accordance with all state and federal regulations. Astrocytes were isolated and maintained in tissue culture flasks in standard DMEM astrocyte media with 10% FBS and 1% penicillin/streptomycin in a humidified 5% CO2/95% air atmosphere. After reaching confluence in ~7–9 d, the cells were shaken at ~1.9 × g at 37 °C for four consecutive periods of 18 h to remove loosely adherent non-astrocyte cells, such as neurons and microglia, interspersed by ~30 h periods of overnight recovery. Staining with a glial fibrillary acidic protein antibody (Millipore, Billerica, MA) revealed that ~70% of the cells were astrocytes, and staining with a CD11b antibody (Abcam, Cambridge, MA) revealed that microglia were not present after shaking.
Identification of Peptides in Astrocytes
Two sample preparation protocols were separately performed on the astrocytic cell line cultures and primary cultured astrocytes for the peptidomic analyses (Figure 1). In the protocol for astrocytic cell lines, after reaching 80% confluence, the cells (~108 cells in ~10 culture flasks) were rinsed with serum-free medium, boiled in water for 5 min, and extracted with iced 0.25% acetic acid in the culture flasks prior to being pooled for cell pellets by scraping.46 The homogenates were centrifuged and the supernatant was pre-concentrated with C18 spin columns (Pierce, Rockford, IL) before further analysis. For the primary culture protocol, we applied a previously published approach to study the peptidomics of cultured astrocytes from rat cortical tissue.23 About 30 culture flasks of cells from five rats were used for culturing, sampling and peptidome method optimization, and the results from the final culture flask, which contained the most complete identifications, are reported here. Specifically, in the optimized method, the cells were washed with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffered salt solution, extracted with 1–2 mL of 10 mM HCl, and then collected by scraping. Trifluoroacetic acid (TFA) (0.1% as a final concentration) was added and the samples were sonicated for 2–3 min before incubation at 70 °C for 20 min. The cell extracts were then centrifuged at 3300 × g for 10 min, and the supernatant was applied to a 10 kDa cutoff Centricon filter (Nanosep 10K Omega, Pall Corporation, Port Washington, NY). The flow-through was lyophilized and the sample extracts were resuspended in 20 μL of 0.1% TFA aqueous solution. The samples were further desalted with pipette tips (Perfect Pure C18, Eppendorf, Hauppauge, NY) before MS analysis.
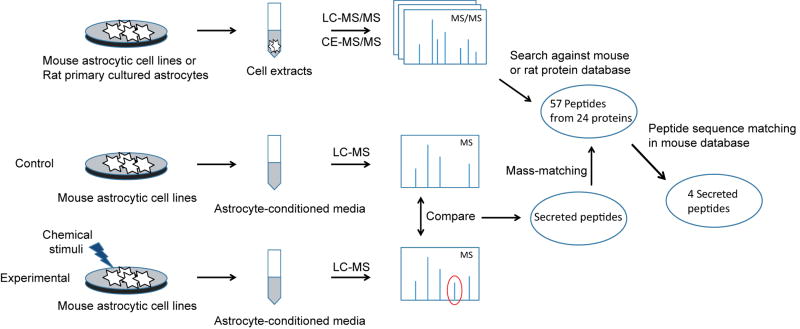
Figure 1.
Overview of the experimental design. Peptides in mouse astrocytic cell lines and rat primary cultured astrocytes were identified by analyzing the cell culture extracts with LC–MS/MS and CE–MS/MS and searching the data against the corresponding database. Peptides released from mouse astrocytic cell lines in response to various chemical stimuli were selected by comparing the control and experimental astrocyte-conditioned media. The secreted peptides were identified by mass-matching to the identified peptides in astrocytes, and then by sequence matching in the mouse database.
LC–ESI-IT MS/MS and CE–ESI-Qq-TOF MS/MS were then employed for peptide identification. The LC–ESI-IT MS/MS analysis was performed similarly to previously described protocols.27, 47, 48 Briefly, the samples were separated by a CapLC system (Micromass, Manchester, UK) using a capillary column (300 μm inner diameter × 15 cm, C18 PepMap100, 3 μm particle size, 100 Å pores, LC Packings,™ San Francisco, CA) at a flow rate of 2.5 μL/min in a gradient generated with Solvent A (95% water and 5% acetonitrile containing 0.1% formic acid (FA) and 0.01% TFA) and Solvent B (5% water and 95% acetonitrile containing 0.1% FA and 0.01% TFA). The gradient was 2–40% B in 55 min, ramping to 80% B in 20 min, staying at 80% B in 4 min, before coming down to 2% B. The ESI-IT mass spectrometer (HCTultra, Bruker Daltonics, Billerica, MA) was operated using EsquireControl software (Bruker Daltonics) in the mass range of m/z 50–2400 with a target mass of m/z 800. Data-dependent MS/MS was activated once the parent ion reached a specific intensity threshold (e.g., 104).
The primary cultured astrocytes were also analyzed by CE–ESI-Qq-TOF-MS/MS, as previously described.48-50 Briefly, the CE separation was achieved by the application of 20 kV at the inlet while the outlet was at ground with 1% FA as the background electrolyte. Hydrodynamic injections of 6 nL were achieved via a 15 cm height difference with respect to the CE capillary outlet. A MicroTee (Idex Health & Science LLC, Oak Harbor, WA) was utilized for the CE–ESI sheath flow interface, equipped with a grounded stainless steel emitter (130 μm inner diameter, 260 μm outer diameter, part 21031A, Hamilton Company, Reno, NV). The sheath flow (50% (v/v) methanol with 0.1% (v/v) FA) was supplied through the emitter at 750 nL/min. Ions in the mass range of m/z 300–3000 were monitored and fragmented with a maXis ESI-Qq-TOF-MS/MS mass spectrometer (Bruker Daltonics).
DataAnalysis software 4.0 (Bruker Daltonics) was used to display the MS and MS/MS spectra and create a compound list with MS/MS after deconvolution. The data was then exported to the Biotools software program 3.1 (Bruker Daltonics) for searching with Mascot (Matrix Science, London, UK) against the mouse and rat Mass Spectrometry protein sequence DataBase (MSDB) and the International Protein Index (IPI) database, as well as an in-house database (http://neuroproteomics.scs.illinois.edu/neuropred.html) of neuropeptide prohormones, followed by manual verification. Within the Mascot search, no enzyme was specified; variable modifications such as N-terminal acetylation, C-terminal amidation, pyroglutamic acid, and methionine oxidation were considered; unsuspected cleavage sites were also allowed. For the Mascot searches, the tandem mass tolerances were set to 0.55 Da for the ESI-IT MS/MS data, and 0.05 Da for the ESI-Qq-TOF MS/MS data. Those peptides with ion scores higher than 30 were accepted. Manual verification of the Mascot results considered the parent mass matching, correct charge state assignment, and the major fragment ion matching. The peptides with lower ion scores were also manually examined for acceptance if they met the mass tolerances specified above and had more than three consecutive amino acids assigned.
Detection and Identification of Peptides Released from Mouse Astrocytic Cell Lines
The following individual chemical stimuli were used in this work: 5 μM ionomycin, 55 mM KCl, 1 μM serotonin, and bradykinin at different concentrations (100 nM, 1 μM, 100 μM, and 1 mM). Different experimental samples prepared at the same time (e.g., 100 μM and 1 mM bradykinin) shared one control sample, which consisted of the same cells, unstimulated. Therefore, a total of six controls were performed. The stimulus concentrations were selected according to previous studies on astrocyte secretion.21-23, 36-44 Bradykinin (m/z 1060.57) concentrations were varied to determine an optimal value large enough to induce a peptide profile change, while in sufficiently low concentrations to prevent interference with peptide detection. The stimulation experiments were performed when the cell line cultures reached 80% confluence. After the culture medium was removed, the cells were quickly rinsed twice with regular culture medium or serum-free medium to remove dead cells, and then subjected to the corresponding regular or serum-free medium with (experimental) or without (control) the chemical stimulus in a humidified 5% CO2/95% air atmosphere. The regular medium was used to minimize cell alteration for stronger and less-selective stimuli (e.g., ionomycin and KCl), and for exploring bradykinin at different concentrations. The serum-free medium was employed to minimize interference from serum during MS analysis for mild stimuli (e.g., serotonin and three replicates of bradykinin at the optimal concentration). After 15 min of stimulation, the astrocyte-conditioned media were respectively collected for control and experimental samples, and then centrifuged at 11,000 × g for 10 min. The supernatants were desalted and pre-concentrated using a C18 cartridge (Waters, Milford, MA). The solvent of the eluent was evaporated with a Savant SpeedVac Vacuum Concentrator (Thermo Scientific, Waltham, MA). The peptides were reconstituted into an aqueous solution containing 0.1% FA and 0.01% TFA.
The six control and ten experimental samples were each sequentially subjected to LC–MALDI-TOF MS analysis (for a total of 16 LC–MS experiments). The samples were separated by the same capillary LC system mentioned above at a flow rate of 2 μL/min. The gradient was 3–5% B in 5 min, continuing to 30% B in 18 min, ramping up to 80% B in 3 min, and staying at 80% B for 2 min before ramping back to 3% B. Twenty-four 1 min fractions were collected between 16 and 45 min. A MALDI matrix, α-cyano-4-hydroxycinnamic acid (10 mg/mL), was used for measurements with the MALDI-TOF/TOF mass spectrometer (Ultraflex II, Bruker Daltonics) in the mass range of m/z 800–5000. FlexAnalysis software 3.0 (Bruker Daltonics) was used for data analysis. Peptides observed in each experimental sample were compared with the combined peptide list from all of the control samples. Peptide level changes between the control and experimental samples were not considered in this work. Therefore, the experimental peptides that were mass-matched to the control peptides within 0.30 Da were subtracted from the peptide list of experimental samples. The remaining peaks in the experimental samples were then manually verified based on peak intensity (e.g., >103), signal-to-noise ratio (e.g., >3), and isotopic distribution. Only the verified peaks in the experimental samples were mass-matched to the peptides identified above from the astrocyte cell lines/primary cultures within 0.50 Da and to the theoretical masses of the peptides in the previous peptidomics analysis of mouse brain tissue33 within 0.30 Da for peptide identities. If the mass of a secreted peptide from the mouse astrocytic cell lines matched the identified peptide in the rat primary cultured astrocytes, the sequence of this peptide had to be consistent between mouse and rat in order to be considered a positive identification.
RESULTS AND DISCUSSION
Peptidomics studies on the nervous system advance our understanding of the biochemical milieu of one of the most intricate, complex, and demanding organ systems. Traditional peptidomics approaches often sample the entire brain or specific regions to determine peptide content in various cell types, predominantly neurons and glia.24-28 More focused approaches investigate enriched cell species or individual cells.29-32 Here we modified traditional peptidomics approaches to analyze peptides present in astrocyte cell cultures, identifying a total of 57 peptides (Table 1 and Figure S1, Supporting Information). These peptides and their protein precursors were compared to the ones reported in previous peptidomics analyses of mouse brain tissue33 and human cell lines.32
Table 1.
Astrocyte peptides identified with LC–ESI-IT MS/MS and CE–ESI-Qq-TOF MS/MS.
| Protein | Peptide Sequence | (m/z)obs. | z | Mtheor. | ΔM (Da) |
|---|---|---|---|---|---|
| Actin alpha | *L.YGESDL.- | 683.57 | 1 | 682.28 | 0.28 |
| a #Actin beta | +D.ESGPSIVHRKCF.- | 680.35 | 2 | 1358.68 | 0.01 |
| +K.IIAPPERKYS.V | 587.60 | 2 | 1172.66 | 0.53 | |
| +L.RVAPEEHPVL.L | 574.03 | 2 | 1145.62 | 0.43 | |
| +L.YASGRTTGIVM.D | 578.50 | 2 | 1154.58 | 0.41 | |
| +W.IGGSILASLSTFQQ.M | 711.65 | 2 | 1420.76 | 0.53 | |
| +M.WISKQEYDESGPSIVHRKCF.- | 803.90 | 3 | 2408.16 | 0.52 | |
| +W.ISKQEYDESGPSIVHRK.C | 987.01 | 2 | 1972.00 | 0.00 | |
| +W.ISKQEYDESGPSIVHRKCF.- | 1112.18 | 2 | 2222.08 | 0.27 | |
| #Acyl-CoA-binding protein | +M.acetyl-SQADFDKAAEEVKRLK.T | 626.33 | 3 | 1875.97 | 0.00 |
| a #Calcium binding protein | +L.ALIYNEALK.- | 517.73 | 2 | 1033.58 | -0.14 |
| +F.LQTSQKRI.- | 973.58 | 1 | 972.57 | 0.00 | |
| +L.IGGLAIACHESF.L | 609.21 | 2 | 1216.59 | -0.19 | |
| +M.PTETERCIESL.I | 639.56 | 2 | 1276.60 | 0.51 | |
| +L.IYNEALK.- | 425.92 | 2 | 849.46 | 0.37 | |
| a #Calmodulin 1 | +M.acetyl-ADQLTEEQIAE.F | 644.92 | 2 | 1287.58 | 0.24 |
| a #Calmodulin 2 | +E.FVQMMTAK.- | 478.40 | 2 | 954.47 | 0.32 |
| CD2 molecule | +T.DVELKL.Y | 716.42 | 1 | 715.41 | 0.00 |
| a #Elongation factor 1-beta | &*M. GFGDLKTPAGLQVLNDYL. A | 961.05 | 2 | 1920.00 | 0.09 |
| +M.GFGDLKTPAGLQVLND.Y | 822.93 | 2 | 1643.85 | -0.01 | |
| Enolase | +L.YTAKGLFR.A | 955.53 | 1 | 954.53 | -0.01 |
| Galectin-1 | *E.VASDAKSFVLNL.G | 632.43 | 2 | 1262.69 | 0.16 |
| *V.RGEVASDAKSFVLNL.G | 803.53 | 2 | 1604.85 | 0.19 | |
| Glyceraldehyde-3-phosphate dehydrogenase | +L.MAYMASKE.- | 465.91 | 2 | 929.40 | 0.41 |
| +G.YSNRVVDL.M | 965.50 | 1 | 964.50 | -0.01 | |
| +G.KVDIVAINDPFIDL.N | 786.43 | 2 | 1570.86 | -0.02 | |
| +D.PANIKWGDAGAEY.V | 696.53 | 2 | 1390.65 | 0.39 | |
| +L.ISWYDNEYGYSNRVVDL.M | 1047.19 | 2 | 2091.95 | 0.41 | |
| aHeat shock 27 protein | +E.ARAQIGGPESEQSGAK.- | 793.60 | 2 | 1584.79 | 0.40 |
| +F.EARAQIGGPESEQSGAK.- | 857.92 | 2 | 1713.83 | 0.00 | |
| b +M. acetyl-TERRVPFSLLR.S | 472.61 | 3 | 1414.80 | 0.00 | |
| a #Macrophage migration inhibitory factor | &*L.AQATGKPAQYIAVHVVPDQLMTF.S | 829.15 | 3 | 2484.28 | 0.14 |
| &*L.AQATGKPAQYIAVHVVPDQL.M | 1053.60 | 2 | 2105.13 | 0.06 | |
| &*V.PRASVPEGFLSELTQQL.A | 936.55 | 2 | 1870.98 | 0.11 | |
| a #Peptidylprolyl isomerase A | +F.DITADGEPLGR.V | 572.48 | 2 | 1142.56 | 0.39 |
| b &*M.VNPTVFFDITADDEPLGRVSF.E | 1170.07 | 2 | 2338.15 | -0.02 | |
| b &*F.EDENFILKHTGPGILSM.A | 951.05 | 2 | 1899.94 | 0.15 | |
| +D.ITADGEPLGR.V | 514.77 | 2 | 1027.53 | 0.00 | |
| +F.FDITADGEPLGR.V | 646.08 | 2 | 1289.63 | 0.52 | |
| +F.FDITADGEPLGRVC.F | 747.10 | 2 | 1491.70 | 0.48 | |
| +F.DITADGEPLGRVCF.E | 746.67 | 2 | 1491.70 | -0.38 | |
| +L.FADKVPKTAENFR.A | 761.90 | 2 | 1521.79 | -0.01 | |
| a #Peroxiredoxin-5 | b *C.SLAPNILSQL.- | 528.35 | 2 | 1054.60 | 0.08 |
| #Phosphatidylethanolamine-binding protein | &+M.acetyl-AADISQWAGPL.S | 585.65 | 2 | 1169.57 | -0.29 |
| +M.acetyl-AADISQWAGPLS.L | 629.52 | 2 | 1256.60 | 0.42 | |
| #Pyruvate kinase isozyme M1 | +T.NTMRVVPVP.- | 1012.54 | 1 | 1011.55 | -0.02 |
| Sec31L1 protein | +L.KVVLSQASKLGV.- | 614.88 | 2 | 1227.76 | -0.01 |
| a #Thymosin beta-4 | +M. acetyl-SDKPDMAEIEKFD.K | 783.69 | 2 | 1565.69 | -0.33 |
| Thyroid hormone receptor-binding protein | +R.oxMPoxMPVNTPLGSNSRKoxMVY.Q | 1035.50 | 2 | 2068.97 | 0.01 |
| Transgelin (smooth muscle protein 22-alpha) | +R.GASQAGMTGYGRP.R | 627.06 | 2 | 1251.57 | 0.54 |
| +L.FEGKDMAAVQRTVMALGSL.A | 1012.61 | 2 | 2023.02 | 0.18 | |
| Tubulin polyglutamylase TTLL5 | *R.PLSASDAEoxMKNLVASAR.E | 888.50 | 2 | 1774.89 | 0.10 |
| Vimentin | +D.GQVINETSQHHDDLE.- | 861.58 | 2 | 1720.77 | 0.38 |
| +K.TVETRDGQVINETSQHHDD.L | 1091.16 | 2 | 2179.97 | 0.33 | |
| +K.TVETRDGQVINETSQHHDDLE.- | 1212.06 | 2 | 2422.10 | 0.01 | |
| +L.RETNLESLPLVD.T | 693.37 | 2 | 1384.72 | 0.01 | |
| #Voltage-dependent anion channel protein 2 | &+N.AGGHKLGLALELEA.- | 689.88 | 2 | 1377.76 | -0.02 |
Peptides identified in the mouse astrocytic cell lines;
peptides identified in the rat primary cultured astrocytes;
protein precursors also identified in the previous peptidomics analysis on mouse brain tissue;33
peptides also identified in the previous peptidomics analysis on mouse brain tissue;33
protein precursors also identified in human cell lines;32
peptides also identified in human cell lines.32
Because molecules released in a physiologically dependent manner are more likely to be involved in cell-cell signaling,21, 23, 28, 51 multiple chemical stimuli reported to induce a Ca2+ increase in astrocytes were applied to the astrocyte cell lines for the peptide secretion studies. Among the released peptides, four were mass-matched to the identified astrocyte peptide list (Table 2) and 59 to peptides previously identified in mouse brain tissue.33 The four peptides were further compared to the peptides identified in previous peptidomics investigations on mouse brain tissue and protein precursors in astrocytes. Finally, the complex peptide profiles induced by different chemical stimuli were summarized (Table S1, Supporting Information), suggesting that astrocytes are actively involved in peptide secretion. Twenty-six peptides observed in multiple stimulation experiments but not in controls are more likely to be the subset released in a Ca2+-dependent manner.
Table 2.
Putatively identified peptides secreted by mouse astrocytic cell lines in response to different chemical stimuli. X denotes observed peptides.
| Proteins | Peptide Sequence | (M+H)+theor. | Bradykinin | KCl | Ionomycin |
|---|---|---|---|---|---|
| Peroxiredoxin-5 | C.SLAPNILSQL.- | 1055.61 | X | ||
| Galectin-1 | V.RGEVASDAKSFVLNL.G | 1605.86 | X | X | |
| Macrophage migration inhibitory factor | L.AQATGKPAQYIAVHVVPDQL.M | 2106.13 | X | X | |
| Elongation factor 1-beta | M.GFGDLKTPAGLQVLNDYL.A | 1921.01 | X |
Peptides in Astrocytes
Peptide extracts from mouse astrocytic cell cultures and rat primary cultured astrocytes were analyzed by both LC–ESI-IT MS/MS and CE–ESI-Qq-TOF MS/MS. A number of compounds in the peptide mass range were detected, among which 57 peptides were unambiguously identified by database search (Table 1). A representative peptide identified by ESI-IT MS/MS is shown in Figure 2. Although we cannot rule out the possibility that some peptides may have resulted from protease-catalyzed protein degradation during the extraction procedure, most are not likely to be protein degradation products because the sample preparation protocols we used here are common to peptidomics analyses.23, 46 During the sample preparation, protease activity was heat-inactivated prior to sample processing and large proteins were removed before LC–MS analysis.
Figure 2.
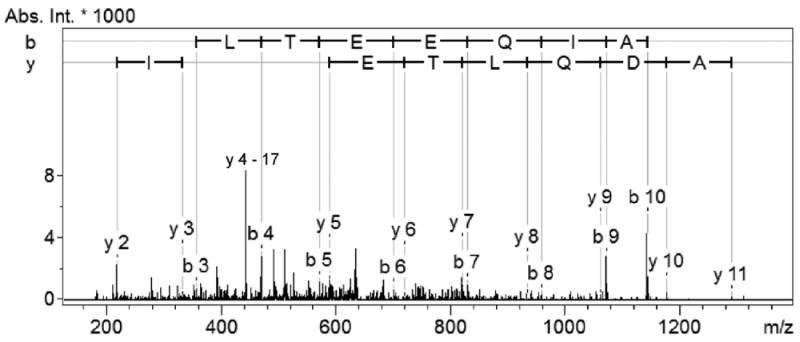
ESI-IT MS/MS spectrum of the N-terminal peptide of calmodulin-1 in mouse astrocytic cell lines. The identified peptide (m/z 644.92, z = 2+) was unambiguously assigned based on the matched sequence (acetyl-ADQLTEEQIAE) in the mouse protein database.
Previous studies have reported the observation of several prohormone transcripts52-55 and the presence of neuropeptide processing enzymes such as carboxypeptidase E56 and peptidylglycine alpha-amidating monooxygenase,57, 58 indicating that astrocytes have the potential to process prohormones in order to produce classically processed neuropeptides. However, only a limited number of neuropeptides have been reported.23, 59 In addition, carboxypeptidase E is secreted from astrocytes,56 which raises the possibility that neuropeptide processing in astrocytes is distinct from neurons, or that peptide processing could be affected by the astrocyte state and/or environment.
In typical neuropeptidomic measurements, both prohormone-derived peptides and protein products are detected. Here the astrocyte peptides were derived from proteins. Although some of the identified peptides were produced by cleavages of proteins at basic sites (e.g., lysine or arginine), most were N- or C-terminal peptides from proteins. This observation has been reported before.33, 60 Because general protein degradation is expected to produce a broader spectrum of protein fragments, these peptides may be selectively generated. Common post-translational modifications such as N-terminal acetylation have also been identified on some of the peptides; these modifications may stabilize the peptides compared to their protein degradation counterparts.61 In general, the identified astrocyte peptides have similar features to non-classical neuropeptides derived from proteins, with the term non-classical indicating that they are not processed from prohormones through the classical secretory pathway; such peptides have been discussed in the literature with regard to neuropeptidomic investigations of biological tissue.33, 60 One feature is that a common cleaved amino acid residue is alanine, and the cleavages frequently occur at hydrophobic residues such as leucine. If proteins are selectively processed in astrocytes, one would expect to find the corresponding processing enzymes in astrocytes. The calpains, a class of calcium-activated cytosolic enzymes, may be candidate enzymes because they are known to cleave proteins, often at hydrophobic sites, and have been reported in astrocytes.62, 63 However, a recent study on human cell lines identifying similar peptides showed that calpains are not involved with peptide production.32 Further studies are needed to identify the enzymes involved in the formation of the peptidome of astrocytes.
Most previous neuropeptidome studies have used brain tissue homogenates that included both glia and neurons; thus, the possibility that astrocyte peptides have contributed to prior neuropeptidomics characterization studies should not be neglected. Thirteen out of 24 protein precursors and eight out of 57 peptides in this work (as shown in Table 1) were also found in the previous peptidomics analysis of mouse brain tissue.33 In addition, a recent study revealed considerable overlap in the peptidomes of three human cell lines (HEK293, MCF7, and SH-SY5Y).31 Ten of 24 protein precursors and four of 57 peptides detected in this work were also found in human cell lines (Table 1). On the one hand, it’s interesting to observe the same protein precursors and even the same processed peptides detected with 100% sequence conservation across different species. For example, the N-terminal peptide of heat shock 27 protein with acetylation was only identified in the MCF7 cells, but not in the HEK293 or SH-SY5Y cells.31 The same peptide was identified in the rat primary cultured astrocytes here. Similarly, the C-terminal peptide of peroxiredoxin-5 reported in HEK293 and MCF7 was identified in the mouse astrocytic cell lines in this study. On the other hand, some peptides from common protein precursors have not been reported in previous peptidomic studies. This lack of prior detection may be from differences in sample preparation or the measurement process; alternatively, perhaps these peptides were not detected in the peptidomes of brain tissue homogenates that included astrocytes, other glia, and neurons because they were lost by including the peptides from non-astrocyte cells. Our results on specific astrocyte cell populations provide evidence for the presence of peptides in astrocytes. Further studies are needed to determine the pathways and potential physiological functions of these peptides derived from proteins.
Stimulation-dependent Peptide Release from Astrocytes
The identification of astrocyte peptides has resulted in the creation of a dataset of potential signaling molecules. The study of stimulation-induced peptide release from astrocytes is of particular interest because peptides released in a physiologically dependent manner are more likely to be involved in cell-cell signaling. A variety of chemical stimuli known to trigger a Ca2+ increase in astrocytes were applied to the astrocyte cell line cultures: ionomycin,22, 36, 37 KCl,38-40 serotonin,41-43 and bradykinin.21, 23, 44 By comparing the peptide content of astrocyte-conditioned media with and without chemical stimulation, we were able to study the stimulation-dependent release of peptides.
Because secreted peptides are in low abundance, neither a labeling or label-free quantitation protocol enabled satisfactory quantitative analyses. Therefore, this secretion study did not include quantitative measures of peptide level changes. Instead, we focused on the peptides that were released and detected in the post-stimulated astrocyte-conditioned medium, but not in the associated controls. Although the control and experimental samples were prepared and analyzed in parallel, given the possible variations in biological samples and sample preparation/detection, we made a full list of peptides from all the control samples. In this list, ~40% of the peptides were observed more than once among the controls. The peptide contents from each experimental sample were compared with this combined control peptide list, and the peak observed in any control was subtracted from the experimental peptide contents. Accordingly, a total of 26 peptides were observed in more than one experimental sample. These peptides are of great interest for astrocyte peptide secretion studies due to the high possibility of their release in response to chemical stimuli in astrocytes (Table S1, Supporting Information). Although more experiments are needed to confirm that some of the peptides observed only once in the experimental samples were released in a stimulation-dependent manner, we included all of these peptides to aid follow-up studies.
Compared to ESI-IT and ESI-Qq-TOF MS, LC–MALDI-TOF MS has better detection limits and yields only singly charged ions, which results in less complex spectra.25, 27, 30 Therefore, we employed LC–MALDI-TOF MS to investigate the peptide (m/z 1000–3000) differences in the astrocyte-conditioned media with and without chemical stimulation. The resulting stimulation-dependent secreted peptides were then mass-matched to the peptides identified in astrocytes in this work and those in the previous peptidomics analysis of mouse brain tissue.33 Since mouse astrocytic cell lines were used to target the secreted peptides, and both mouse astrocytic cell lines and rat primary cultured astrocytes were employed to create an astrocytic peptide database, the sequences of mass-matching rat peptides were confirmed to also be present in the mouse database.
The chemical stimuli used in this study have been reported to trigger a Ca2+ increase in astrocytes.21-23, 36-44 Ionomycin is known as an ionophore that forms pores in the membrane, allowing extracellular Ca2+ to enter into cells.64, 65 Elevated levels of extracellular K+ are known to depolarize astrocytes and increase the permeability of gap junctions.40 We observed that both 5 μM ionomycin and 55 mM KCl caused detachment of some cells in the mouse astrocytic cell line cultures. One peak (m/z at about 1605.90) was observed in both the ionomycin and the K+ post-stimulation media, and this peak has been putatively identified as a peptide derived from galectin-1 (discussed below).
Bradykinin is a selective stimulus as it induces an increase of internal Ca2+ in astrocytes but not in neurons.44 In this work, the astrocyte cell line cultures were exposed to different concentrations of bradykinin. No differences in the peptide mass range were observed in the post-stimulated medium with 100 nM bradykinin stimulation compared to the control sample, which is consistent with previous data.23 Here, 1 μM bradykinin was selected for further analysis with serum-free medium (instead of 100 μM or 1 mM) because this concentration proved optimal for inducing astrocyte secretion and caused minimal interference with the MS detection. Although more experiments are needed to better target the specific peptides released by bradykinin stimulation, our study provided initial results on this. For example, a peak (m/z at about 1076.76) putatively identified as an internal peptide derived from phosphatidylethanolamine-binding protein was observed in all five bradykinin-stimulation experiments (Table S1, Supporting information). Another three peptides derived from the same protein were respectively observed in three out of five bradykinin stimulation experiments, one out of five bradykinin-stimulation experiments, and a KCl-stimulation experiment, lending credence to this protein being involved in astrocyte release. In addition, the N-terminal peptide whose sequence is conserved between rat and mouse from this protein was identified in astrocytes. This combined evidence suggested that phosphatidylethanolamine-binding protein could be a precursor of the “non-classical neuropeptide” as discussed above. Similar to bradykinin, serotonin, a neurotransmitter and secretagogue in the CNS,66 can be taken up by astrocytes43 and causes an increase of intracellular Ca2+ concentration.41, 42, 67 Among the seven peptides observed in both bradykinin-stimulation and serotonin-stimulation, one was putatively identified as a peptide derived from procholecystokinin.
Four mouse peptides were identified in the profile of astrocyte secretion, as shown in Table 2. The protein precursors of these four peptides are not in the list of proteins identified in previous proteomics studies of astrocyte-conditioned medium,18-20 indicating that these peptides are likely to be stimulation-dependent. Two of the four observed peptides were reported in a previous neuropeptidomics study on brain tissue extracts.33 These peptides are the N-terminal peptide of elongation factor 1 beta and the peptide cleaved at leucine of the macrophage migration inhibitory factor. Although elongation factor 2 has been reported to be in and secreted from astrocytes,9, 10, 18-20 no similar information is available for elongation factor 1. The presence of these two peptides in brain tissue, isolated astrocytes and astrocyte stimulation-dependent secretion, and the absence of their intact proteins in the previous astrocyte protein secretion studies, suggest that the two peptides are processed in astrocytes and released in a Ca2+-dependent manner. One of the other two peptides is the C-terminal peptide of peroxiredoxin-5. This specific peptide was not observed in previous neuropeptidomics studies, but different peptides derived from peroxiredoxin-5 have been reported in brain tissue extracts.33 These may have resulted from peptide processing differences related to cell environments; our work is based on isolated astrocytes, while the previous neuropeptidomics investigation33 was done with brain tissue in which both glia and neurons were present. In addition, although peroxiredoxin-5 and -6 were identified in astrocyte proteomics studies,9, 10 only peroxiredoxin-1, -2, -3, and -4 were reported in astrocyte secretion studies.18, 20 Here, we observed that a peptide derived from peroxiredoxin-5 was secreted from astrocytes upon stimulation. Finally, we observed an intriguing peptide derived from galectin-1, a multi-functional glycoprotein previously reported to be present in astrocytes but not in neurons.10, 68 The peptide is produced by the cleavages of the N-terminal of arginine and C-terminal of leucine when astrocytes are stimulated. While galactin-3 has been reported in astrocyte protein secretion,18 no evidence supporting the secretion of galactin-1 was available prior to this study.
Fifty-nine peptides secreted from the mouse astrocytic cell lines were mass-matched to peptides previously identified in mouse brain tissue (Table S1, Supporting Information). Some of these peptides are derived from the proteins (e.g., phosphatidylethanolamine-binding protein and peptidyl-prolyl isomerase A) from which different peptide segments have been identified in mouse astrocytes (Table 1). Interestingly, some peptides are derived from neuropeptide precursors such as proenkephlin, prosomatostatin, and proSAAS. Although MS/MS data on the corresponding peptides is needed to confirm the peptide identities, the existence of the transcripts of proenkephlin and prosomatostatin53-55 in astrocyte cultures and proSAAS in the astrocyte secretion proteome18 supports these putative identifications.
The potential functions of the astrocyte peptides secreted in a Ca2+-dependent manner are not necessarily related to the known functions of their protein precursors, and are the subject of future studies.
CONCLUSIONS
We have identified 57 astrocyte peptides, four of which were likely released from astrocytes in response to chemical stimuli. In the secretion study, 26 peptides were observed in more than one stimulation sample, and 59 were mass-matched to peptides previously identified in mouse brain tissue. Our results demonstrate that peptides derived from proteins may play an important role in astrocyte-mediated, cell-to-cell communication, and that the secretion of peptides may be induced by Ca2+-related mechanisms. While it has been challenging to identify the observed secreted peptides due to their low concentrations in a complex medium, future work will employ larger-scale sample enrichment methods and more sensitive characterization techniques. In this work, we focused on isolated astrocytes. Applying the techniques presented in this study to context-specific secretion will provide information regarding the intricacies of glia-to-neuron communication.
Supplementary Material
Acknowledgments
We thank Matthew D. Whim for providing a detailed sample preparation protocol for the primary astrocyte cultures, and Peter Nemes and Suresh P. Annangudi for their technical assistance. The project described was supported by the following award numbers and agencies: P30 DA018310 from the National Institute on Drug Abuse; T32 HD007333 from the National Institute of Child Health and Development; 1R01 HL092571 from the National Heart, Lung and Blood Institute; and CHE-05-26692 and CHE-11-11705 from the National Science Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the award agencies.
Footnotes
The MS/MS spectra for peptide identifications are available in Figure S1. The observed peptides in the astrocyte secretion study are listed in Table S1. These materials are free of charge via the Internet at http://pubs.acs.org.
References
- 1.Strand FL. Neuropeptides: general characteristics and neuropharmaceutical potential in treating CNS disorders. Prog Drug Res. 2003;61:1–37. doi: 10.1007/978-3-0348-8049-7_1. [DOI] [PubMed] [Google Scholar]
- 2.Miller G. Neuroscience: the dark side of glia. Science. 2005;308(5723):778–781. doi: 10.1126/science.308.5723.778. [DOI] [PubMed] [Google Scholar]
- 3.Volterra A, Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat Rev Neurosci. 2005;6:626–640. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- 4.Haydon PG, Carmignoto G. Astrocyte control of synaptic transmission and neurovascular coupling. Physiol Rev. 2006;86(3):1009–1031. doi: 10.1152/physrev.00049.2005. [DOI] [PubMed] [Google Scholar]
- 5.Charles AC, Merrill JE, Dirksen ER, Sandersont MJ. Intercellular signaling in glial cells: Calcium waves and oscillations in response to mechanical stimulation and glutamate. Neuron. 1991;6(6):983–992. doi: 10.1016/0896-6273(91)90238-u. [DOI] [PubMed] [Google Scholar]
- 6.Hassinger TD, Guthrie PB, Atkinson PB, Bennett MVL, Kater SB. An extracellular signaling component in propagation of astrocytic calcium waves. Proc Natl Acad Sci U S A. 1996;93(23):13268–13273. doi: 10.1073/pnas.93.23.13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halassaa MM, Fellina T, Haydon PG. The tripartite synapse: roles for gliotransmission in health and disease. Trends Mol Med. 2007;13(2):54–63. doi: 10.1016/j.molmed.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Perea G, Navarrete M, Araque A. Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 2009;32(8):421–431. doi: 10.1016/j.tins.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Yang J-W, Suder P, Silberring J, Lubec G. Proteome analysis of mouse primary astrocytes. Neurochem Int. 2005;47(3):159–172. doi: 10.1016/j.neuint.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Yang J-W, Rodrigo R, Felipo V, Lubec G. Proteome analysis of primary neurons and astrocytes from rat cerebellum. J Proteome Res. 2005;4(3):768–788. doi: 10.1021/pr049774v. [DOI] [PubMed] [Google Scholar]
- 11.Verkhratsky A, Steinhäuser C. Ion channels in glial cells. Brain Res Rev. 2000;32:380–412. doi: 10.1016/s0165-0173(99)00093-4. [DOI] [PubMed] [Google Scholar]
- 12.Kimelberg HK, MacVicar BA, Sontheimer H. Anion channels in astrocytes: biophysics, pharmacology, and function. Glia. 2006;54(7):747–757. doi: 10.1002/glia.20423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibeshih Belachew VG. Synaptic and extrasynaptic neurotransmitter receptors in glial precursors’ quest for identity. Glia. 2004;48(3):185–196. doi: 10.1002/glia.20077. [DOI] [PubMed] [Google Scholar]
- 14.Bezzi P, Gundersen V, Galbete JL, Seifert G, Steinhäuser C, Pilati E, Volterra A. Astrocytes contain a vesicular compartment that is competent for regulated exocytosis of glutamate. Nat Neurosci. 2004;7:613–620. doi: 10.1038/nn1246. [DOI] [PubMed] [Google Scholar]
- 15.Coco S, Calegari F, Pravettoni E, Pozzi D, Taverna E, Rosa P, Matteoli M, Verderio C. Storage and release of ATP from astrocytes in culture. J Biol Chem. 2003;278(2):1354–1362. doi: 10.1074/jbc.M209454200. [DOI] [PubMed] [Google Scholar]
- 16.Martineau M, Baux G, Mothet J-P. D-Serine signalling in the brain: friend and foe. Trends Neurosci. 2006;29(8):481–491. doi: 10.1016/j.tins.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Jourdain P, Bergersen LH, Bhaukaurally K, Bezzi P, Santello M, Domercq M, Matute C, Tonello F, Gundersen V, Volterra A. Glutamate exocytosis from astrocytes controls synaptic strength. Nat Neurosci. 2007;10(3):331–339. doi: 10.1038/nn1849. [DOI] [PubMed] [Google Scholar]
- 18.Dowell JA, Johnson JA, Li L. Identification of astrocyte secreted proteins with a combination of shotgun proteomics and bioinformatics. J Proteome Res. 2009;8(8):4135–4143. doi: 10.1021/pr900248y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greco TM, Seeholzer SH, Mak A, Spruce L, Ischiropoulos H. Quantitative mass spectrometry-based proteomics reveals the dynamic range of primary mouse astrocyte protein secretion. J Proteome Res. 2010;9(5):2764–2774. doi: 10.1021/pr100134n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafon-Cazal M, Adjali O, Galéotti N, Poncet J, Jouin P, Homburger V, Bockaert J, Marin P. Proteomic analysis of astrocytic secretion in the mouse. J Biol Chem. 2003;278(27):24438–24448. doi: 10.1074/jbc.M211980200. [DOI] [PubMed] [Google Scholar]
- 21.Calegari F, Coco S, Taverna E, Bassetti M, Verderio C, Corradi N, Matteoli M, Rosa P. A regulated secretory pathway in cultured hippocampal astrocytes. J Biol Chem. 1999;274(32):22539–22547. doi: 10.1074/jbc.274.32.22539. [DOI] [PubMed] [Google Scholar]
- 22.Kržan M, Stenovec M, Kreft M, Pangršič T, Grilc S, Haydon PG, Zorec R. Calcium-dependent exocytosis of atrial natriuretic peptide from astrocytes. J Neurosci. 2003;23(5):1580–1583. doi: 10.1523/JNEUROSCI.23-05-01580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramamoorthy P, Whim MD. Trafficking and fusion of neuropeptide Y-containing dense-core granules in astrocytes. J Neurosci. 2008;28(51):13815–13827. doi: 10.1523/JNEUROSCI.5361-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fricker LD, Lim J, Pan H, Che F-Y. Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom Rev. 2006;25(2):327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- 25.Wei H, Nolkrantz K, Parkin MC, Chisolm CN, O’Callaghan JP, Kennedy RT. Identification and quantification of neuropeptides in brain tissue by capillary liquid chromatography coupled off-Line to MALDI-TOF and MALDI-TOF/TOF-MS. Anal Chem. 2006;78(13):4342–4351. doi: 10.1021/ac052196x. [DOI] [PubMed] [Google Scholar]
- 26.Svensson M, Sköld K, Svenningsson P, Andren PE. Peptidomics-based discovery of novel neuropeptides. J Proteome Res. 2003;2(2):213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 27.Bora A, Annangudi SP, Millet LJ, Rubakhin SS, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Neuropeptidomics of the supraoptic rat nucleus. J Proteome Res. 2008;7(11):4992–5003. doi: 10.1021/pr800394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatcher NG, Atkins N, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Mass spectrometry-based discovery of circadian peptides. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Profiling metabolites and peptides in single cells. Nat Methods. 2011;8(4s):S20–S29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubakhin SS, Sweedler JV. Quantitative measurements of cell–cell signaling peptides with single-cell MALDI MS. Anal Chem. 2008;80(18):7128–7136. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millet LJ, Bora A, Sweedler JV, Gillette MU. Direct cellular peptidomics of supraoptic magnocellular and hippocampal neurons in low-density cocultures. ACS Chem Neurosci. 2009;1(1):36–48. doi: 10.1021/cn9000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD. Peptidomic Analysis of Human Cell Lines. J Proteome Res. 2011;10(4):1583–1592. doi: 10.1021/pr100952f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fricker LD. Analysis of mouse brain peptides using mass spectrometry-based peptidomics: implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol Biosys. 2010;6(8):1355–1365. doi: 10.1039/c003317k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dircksen H, Neupert S, Predel R, Verleyen P, Huybrechts J, Strauss J, Hauser F, Stafflinger E, Schneider M, Pauwels K, Schoofs L, Grimmelikhuijzen CJP. Genomics, transcriptomics, and peptidomics of Daphnia pulex neuropeptides and protein hormones. J Proteome Res. 2011;10(10):4478–4504. doi: 10.1021/pr200284e. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Sweedler JV. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu Rev Anal Chem. 2008;1(1):451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 36.Venance L, Stella N, Glowinski J, Giaume C. Mechanism involved in initiation and propagation of receptor-induced intercellular calcium signaling in cultured rat astrocytes. J Neurosci. 1997;17(6):1981–1992. doi: 10.1523/JNEUROSCI.17-06-01981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeftinija SD, Jeftinija KV, Stefanovic G, Liu F. Neuroligand-evoked calcium-dependent release of excitatory amino acids from cultured astrocytes. J Neurochem. 1996;66(2):676–684. doi: 10.1046/j.1471-4159.1996.66020676.x. [DOI] [PubMed] [Google Scholar]
- 38.Morgan ACA, Chang H-Y, Liu JSH, Hua LL, Lee SC. High extracellular potassium modulates nitric oxide synthase expression in human astrocytes. J Neurochem. 2000;74(5):1903–1912. doi: 10.1046/j.1471-4159.2000.0741903.x. [DOI] [PubMed] [Google Scholar]
- 39.Mongin AA, Cai Z, Kimelberg HK. Volume-dependent taurine release from cultured astrocytes requires permissive [Ca2+]i and calmodulin. Am J Physiol - Cell Physiol. 1999;277(4):C823–C832. doi: 10.1152/ajpcell.1999.277.4.C823. [DOI] [PubMed] [Google Scholar]
- 40.Kristian Enkvist MO, McCarthy KD. Astroglial gap junction communication is increased by treatment with either glutamate or high K+ concentration. J Neurochem. 1994;62(2):489–495. doi: 10.1046/j.1471-4159.1994.62020489.x. [DOI] [PubMed] [Google Scholar]
- 41.Haak LL, Heller HC, van den Pol AN. Metabotropic glutamate receptor activation modulates kainate and serotonin calcium response in astrocytes. J Neurosci. 1997;17(5):1825–1837. doi: 10.1523/JNEUROSCI.17-05-01825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jalonen TO, Margraf RR, Wielt DB, Charniga CJ, Linne ML, Kimelberg HK. Serotonin induces inward potassium and calcium currents in rat cortical astrocytes. Brain Res. 1997;758(1-2):69–82. doi: 10.1016/s0006-8993(97)00163-7. [DOI] [PubMed] [Google Scholar]
- 43.Anderson EJ, McFarland D, Kimelberg HK. Serotonin uptake by astrocytes in situ. Glia. 1992;6(2):154–158. doi: 10.1002/glia.440060210. [DOI] [PubMed] [Google Scholar]
- 44.Parpura V, Basarsky TA, Liu F, Jeftinij K, Jeftinija S, Haydon PG. Glutamate-mediated astrocyte–neuron signalling. Nature. 1994;369:744–747. doi: 10.1038/369744a0. [DOI] [PubMed] [Google Scholar]
- 45.Alliota F, Pessac B. Astrocytic cell clones derived from established cultures of 8-day postnatal mouse cerebella. Brain Res. 1984;306(1-2):283–291. doi: 10.1016/0006-8993(84)90377-9. [DOI] [PubMed] [Google Scholar]
- 46.Dowell JA, Heyden WV, Li L. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J Proteome Res. 2006;5(12):3368–3375. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- 47.Yin P, Hou X, Romanova EV, Sweedler JV. Neuropeptidomics: mass spectrometry-based qualitative and quantitative analysis. In: Merighi A, editor. Methods Mol Biol. Vol. 789. Springer Science+Business Media, LLC; 2011. pp. 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lapainis T, Rubakhin SS, Sweedler JV. Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Anal Chem. 2009;81(14):5858–5864. doi: 10.1021/ac900936g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knolhoff AM, Nemes P, Rubakhin SS, Sweedler JV. MS-based methodologies for single-cell metabolite detection and identification. In: Lutz N, Wevers R, Sweedler JV, editors. Metabolomics Methods, Methods in Molecular Biology Series. Cambridge University Press; 2012. in press. [Google Scholar]
- 50.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. Metabolic differentiation of neuronal phenotypes by single-cell capillary electrophoresis–electrospray ionization-mass spectrometry. Anal Chem. 2011;83:6810–6817. doi: 10.1021/ac2015855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Squires LN, Talbot KN, Rubakhin SS, Sweedler JV. Serotonin catabolism in the central and enteric nervous systems of rats upon induction of serotonin syndrome. J Neurochem. 2007;103(1):174–180. doi: 10.1111/j.1471-4159.2007.04739.x. [DOI] [PubMed] [Google Scholar]
- 52.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vilijn MH, Vaysse PJ, Zukin RS, Kessler JA. Expression of preproenkephalin mRNA by cultured astrocytes and neurons. Proc Natl Acad Sci U S A. 1988;85(17):6551–6555. doi: 10.1073/pnas.85.17.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shinoda H, Marini AM, Cosi C, Schwartz JP. Brain region and gene specificity of neuropeptide gene expression in cultured astrocytes. Science. 1989;245(4916):415–417. doi: 10.1126/science.2569236. [DOI] [PubMed] [Google Scholar]
- 55.Shinoda H, Marini AM, Schwartz JP. Developmental expression of the proenkephalin and prosomatostatin genes in cultured cortical and cerebellar astrocytes. Dev Brain Res. 1992;67(2):205–210. doi: 10.1016/0165-3806(92)90220-q. [DOI] [PubMed] [Google Scholar]
- 56.Vilijn M-H, Das B, Kessler JA, Flicker LD. Cultured astrocytes and neurons synthesize and secrete carboxypeptidase E, a neuropeptide-processing enzyme. J Neurochem. 1989;53(5):1487–1493. doi: 10.1111/j.1471-4159.1989.tb08542.x. [DOI] [PubMed] [Google Scholar]
- 57.Rhodes CH, Xu RY, Angeletti RH. Peptidylglycine alpha-amidating monooxygenase (PAM) in Schwann cells and glia as well as neurons. J Histochem Cytochem. 1990;38(9):1301–11. doi: 10.1177/38.9.2387985. [DOI] [PubMed] [Google Scholar]
- 58.Klein RS, Fricker LD. Cultured astrocytes express mRNA for peptidylglycine-α-amidating monooxygenase, a neuropeptide processing enzyme. Brain Res. 1992;596(1–2):202–208. doi: 10.1016/0006-8993(92)91548-s. [DOI] [PubMed] [Google Scholar]
- 59.Ubink R, Calza L, Hökfelt T. ‘Neuro’-peptides in glia: Focus on NPY and galanin. Trends Neurosci. 2003;26(11):604–609. doi: 10.1016/j.tins.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 60.Gelman J, Fricker L. Hemopressin and other bioactive peptides from cytosolic proteins: are these non-classical neuropeptides? AAPS J. 2010;12(3):279–289. doi: 10.1208/s12248-010-9186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mo XY, Cascio P, Lemerise K, Goldberg AL, Rock K. Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J Immunol. 1999;163(11):5851–5859. [PubMed] [Google Scholar]
- 62.Vosler P, Brennan C, Chen J. Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol Neurobiol. 2008;38(1):78–100. doi: 10.1007/s12035-008-8036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shields DC, Tyor WR, Deibler GE, Hogan EL, Banik NL. Increased calpain expression in activated glial and inflammatory cells in experimental allergic encephalomyelitis. Proc Natl Acad Sci U S A. 1998;95(10):5768–5772. doi: 10.1073/pnas.95.10.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chatila T, Silverman L, Miller R, Geha R. Mechanisms of T cell activation by the calcium ionophore ionomycin. J Immunol. 1989;143(4):1283–1289. [PubMed] [Google Scholar]
- 65.Yoshida S, Plant S. Mechanism of release of Ca2+ from intracellular stores in response to ionomycin in oocytes of the frog Xenopus laevis. J Physiol (Lond) 1992;458(1):307–318. doi: 10.1113/jphysiol.1992.sp019419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wise TN, Fishbain DA, Holder-Perkins V. Painful physical symptoms in depression: a clinical challenge. Pain Med. 2007;8(s2):S75–S82. doi: 10.1111/j.1526-4637.2007.00352.x. [DOI] [PubMed] [Google Scholar]
- 67.Sandén N, Thorlin T, Blomstrand F, Persson PAI, Hansson E. 5-Hydroxytryptamine2B receptors stimulate Ca2+ increases in cultured astrocytes from three different brain regions. Neurochem Int. 2000;36(4-5):427–434. doi: 10.1016/s0197-0186(99)00134-5. [DOI] [PubMed] [Google Scholar]
- 68.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16(11):137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.