Abstract
Genome-scale studies hold great promise for revealing novel plant biology. Because of the complexity of these techniques, numerous considerations need to be made before embarking on a study. Here we focus on the Arabidopsis model system because of the wealth of available genome-scale data. Many approaches are available that provide genome-scale information regarding the state of a given organism (e.g. genomics, epigenomics, transcriptomics, proteomics, metabolomics interactomics, ionomics, phenomics, etc.). Integration of all of these types of data will be necessary for a comprehensive description of Arabidopsis. In this review we propose that ‘triangulation’ among transcriptomics, proteomics and metabolomics is a meaningful approach for beginning this integrative analysis and uncovering a systems level perspective of Arabidopsis biology.
Introduction
The completion of the Arabidopsis genome sequence facilitated extraordinary progress toward understanding plant biology. In particular, complete genomic sequence data drove the development of genome-wide transcriptional approaches, such as microarrays. Genome-scale studies (hereafter -omics) include, but are not limited to, analysis of RNAs, proteins, and metabolites. Significant progress has been made annotating and determining the function of many Arabidopsis genes. However, a plant is not just the sum of its genes, but a complex system where gene product interactions result in emergent properties. Therefore, with the ultimate intention of studying the biology of the whole organism, it is important to frame the next long-term goals for plant scientists.
We propose that a key long-term goal is the integration of different genome-scale approaches. The first steps in this direction have already occurred although the tools for the integration, visualization, and modeling of -omics data are still at a relatively early stage [(for reviews, see 1,2)]. In this review, we focus on gene expression, protein and metabolite profiling data, briefly introducing each of the individual approaches, and then highlighting recent efforts to integrate these -omics (Figure 1).
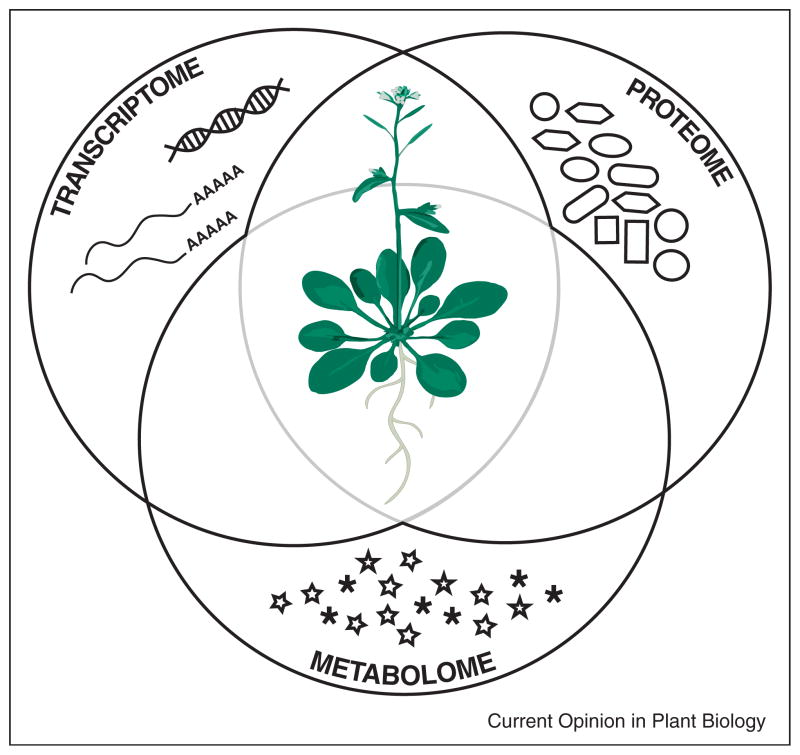
Figure 1.
The biology of the whole organism: integration of different -omics. A simplified schematic representation of -omics. Transcriptomics, proteomics and metabolomics are measured comprehensively by genome-scale methods. The integration of these -omics (as shown by the intersecting Venn-diagram) provides insight into systems-level understanding of Arabidopsis.
Omics technology development: transcriptomics, proteomics and metabolomics
Transcriptomics
The completion of the Arabidopsis genome sequence has facilitated whole genome transcriptomic (gene expression) studies. The development of microarray technology enabled the simultaneous examination of thousands of genes; thus providing a comprehensive view of gene activity. Microarray gene expression data now cover organs, tissues, cell-types and developmental events, as well as responses to a variety of environmental perturbations [3–5,6••,7,8]. For profiling transcripts in model organisms with well-annotated genomes such as Arabidopsis, microarrays have played an invaluable role in our understanding of plant gene expression. Despite the power of microarrays, they are limited to providing relative abundance information about identified genes and gene models. Therefore, deep sequencing of transcripts (RNA-seq) provides an alternative to microarray technology [9,10]. Additionally, because RNA-seq does not depend on genome annotation, RNA-seq has emerged as the method of choice for transcriptional profiling in nonmodel organisms. RNA-seq approaches aim to detect diverse RNA molecules, including mRNA, noncoding RNA and small RNAs [11,12]. The unparalleled ability of RNA-seq to provide sequence information at single basepair-resolution enables the identification of novel genes, alternative-splicing, single nucleotide polymorphisms, and transcript abundance upon DNA methylation-state modification [13,14•,15]. Recently RNA-seq in specific cell types and developmental regions of the Arabidopsis root detected over 60 novel miRNAs [16••]. As a new technology, there are unique challenges that come with analyzing RNA-seq data including developing methods, algorithms and pipelines (e.g. library preparation procedures, RNA quantification, isoform detection and quantification, etc.). Despite these challenges, and because of the improved throughput and lower cost, RNA-seq has already shed light on the complexity and regulation of the plant transcriptome.
Proteomics
The Arabidopsis genome sequence enabled the prediction of genes and the proteins they encode. Comprehensive proteomic analysis seeks to determine the localization, quantity and post-translational modifications of all proteins in an organism. This information is complementary to transcriptomic analyses as it provides the functional readout of gene expression profiles. However, proteomic analysis has been more challenging than transcriptomic analysis for a number of reasons described below.
As generally practiced, proteomics first detects peptides and then assigns them to a gene model [(see 17)]. When dealing with complex samples, protein representation can be biased, with an overrepresentation of large or abundant proteins compared to small proteins [18••]. Additionally, information about protein accumulation and posttranslational modifications are required to fully understand a plant as a whole. One could envision that the in vivo protein concentrations could be measured by targeting specific cell-types and tissues. The Arabidopsis root offers an ideal system for proteomic analysis at cellular resolution comparable to what has been achieved for transcriptional analysis. Technological advances should improve detection and identification issues, enabling complete proteome analysis in the future.
There are multiple approaches for proteomic profiling [(for a review, see 20)]. The traditional approaches are gel-based such as SDS-PAGE, which is useful for protein ‘fingerprinting’ of complex extracts for protein quantities and post-translational modifications [19,21]. Advances in mass spectrometry (MS) measurements have enabled protein quantification from complex samples. Shotgun proteomics, which combines liquid chromatography and MS, has emerged as a promising method for comprehensive proteomics [22,23]. MS-based proteomic studies in plants have focused on identification of proteins in various organelles (e.g. plastids and mitochondria) in an effort to reduce protein complexity [20,24–27]. Quantitative proteomics encompassing whole plants requires analysis of complex samples containing many unique protein species. A recent proteome survey in Arabidopsis identified almost 50% of predicted protein-coding genes by tandem MS [18••]. This group found over 50 novel or alternative gene models highlighting the utility of this approach to identify known and novel genes through high throughput proteomic methods.
A protein-interaction map, using a yeast-2-hybrid system, was generated for Arabidopsis providing interaction information for ~8000 protein coding genes [28]. This map has already been used to gain insight into the response of plants to pathogen attack [29•]. These studies demonstrate the utility of these large-scale projects. As detection techniques improve, more proteins and protein complexes will be profiled enhancing our knowledge of protein localization, abundance and interactions.
Metabolomics
The objective of metabolomics is to identify and quantify all metabolites in plants. Metabolomics is challenging in part because of the vast range of compounds found in plants; a single accession of Arabidopsis contains more than 5000 metabolites [30]. Because a major effort is needed for unequivocal identification of metabolites and no single approach can detect all compounds, combinations of different and complementary extraction and detection techniques are necessary to increase the coverage of a metabolome [31–33]. Keeping in mind that we would like to detect the complete repertoire of metabolites in a cell and understand how different metabolic pathways are coordinated across the entire organism, state-of-the-art metabolomic techniques are necessary. Metabolic profiles of different organs, tissues and even cell-types will provide greater insight into plant complexity. Arabidopsis, and especially the Arabidopsis root, provides an excellent system to start such metabolomic studies.
To cover the vast range of the metabolome, one of the most promising approaches is the combination of gas chromatography (GC — for primary metabolites), liquid chromatography (LC — for secondary metabolites) or capillary electrophoresis (CE — for ionic metabolites) with MS or metabolic fingerprinting using nuclear magnetic resonance (NMR) [34–38]. Metabolic fingerprinting techniques using NMR or Fourier transform-infrared spectroscopy (FT-IR) can also be used to investigate the dynamics of a metabolic network. If the primary goal is to have a better efficiency in sample separation, two-dimensional GC can then be combined with fast acquisition of rate mass spectroscopy (GC × GC–TOF-MS) [39].
The combination of these techniques has generated a vast amount of metabolomics data, which must be properly annotated and tracked to yield fruitful results. Although integration of data from multiple research groups has been difficult, efforts to construct common data repositories [40–42] and data analysis software are ongoing [1,43].
Data integration from molecular information: the gene–protein–metabolite relationship
Transcriptomics–proteomics
As gene expression profiling and proteomic methods improve, data can be combined to achieve a better understanding of Arabidopsis as a system. One important aspect of future investigation will involve the identification of variably spliced transcripts and the discrete proteins they encode. Before next generation sequencing analysis, estimates of alternatively spliced genes based on EST analysis ranged from 22 to 30% [44–47]. More recent information using RNA-seq predicts that 42% of Arabidopsis genes with introns are alternatively spliced [14•]. Questions remain regarding the functional significance of these isoforms (e.g. what percentage are expressed as proteins?). These questions will soon be addressable by examining the full proteome and comparing it to next-generation gene expression data.
Several groups have used gene expression and proteomic data together to gain information unattainable from either approach on its own. Integrative transcriptomic and proteomic studies have been performed for organelles, cell-types and organs in Arabidopsis, including chloroplasts [24,48], pollen [49], guard cells [50], and trichomes [51]. The most comprehensive proteomic study in Arabidopsis to date integrated the proteome findings with gene expression data to reveal potential biomarkers for roots, flowers, leaves, seeds, siliques and cell culture [18••]. Colocalization of transcripts and proteins reduces the likelihood that either occurred by contamination or chance. Comparing gene expression levels and protein abundance is challenging due to differing mRNA and protein stability [52]. Nevertheless, a few studies including some in plants have shown a small yet significant positive correlation between mRNA and protein abundances [18••,24]. Studies of individual cell-types are now possible which will provide more refined information regarding the colocalization of transcripts and proteins. Proteomic data can also be used to inform genome annotation and characterize post-translational modification as has been demonstrated in a number of recent studies [18••,53,54]. These studies revealed both missing and improperly annotated genes, highlighting the advantage of using proteomics for gene annotation.
Transcriptomics–metabolomics
Identification of specific compounds from experimental data (i.e. MS and NMR spectra) is a noteworthy, challenging task. Correlating mass peaks with transcripts could be a powerful strategy for identifying metabolites in complex extracts. Gene-to-metabolite associations have now been characterized for stress responses, plant defense and hormone-induced responses [55,56,57•,58,59]. Early integration of transcriptomics and metabolomics studies looked at the global and dynamic response during sulfur and nitrogen depletion at the system-level [60–63]. Detailed analyses resulting from this type of integration have identified several genes including those involved in glucosinolate biosynthesis, anthocyanin biosynthesis, chain elongation enzymes and glucosinolate transport [64–66]. Recently, large-scale dynamic transcriptomic and metabolomic studies have been undertaken to gain a comprehensive understanding of how biological systems respond to other stresses such asm elevated CO2 and salinity [67]. Additionally, integration of transcriptomic and metabolomic data from multiple related species and/or genotypes has been useful for identifying genes and processes underlying complex traits [68].
Integrated transcriptomic and metabolomic analyses have been successfully combined with reductionist approaches to investigate regulatory mechanisms involved in gene expression and metabolites. Specifically, the altered expression of the transcription factors that regulate anthocyanin biosynthesis [64] allowed the identification of genes involved in later steps of this metabolic process [69,70]. Additionally, ectopic expression of a transcription factor that regulates the cold response also showed metabolomic changes [56]. Moreover, mutations in the abscisic acid (ABA) biosynthesis pathway together with the integrated analysis of the transcriptome and metabolome demonstrated that ABA can reconfigure metabolite levels as a response to dehydration stress [59].
The hypothesis that a correlation exists between gene expression and metabolite accumulation patterns has proved valid when trying to identify the function of genes [71]. Therefore, to facilitate the integration of gene expression data in the context of a functional/metabolic pathway, software packages such as MapMan, PageMan and MetGenMAp were developed [72–74]. These programs have been successfully used to identify genes and metabolic pathways involved in response to nitrogen deficiency, diurnal cycles, and more recently, seed dormancy and germination [63,75–77]. This ‘guilt-by-association’ approach was used to predict the function of genes coregulated under given conditions and identify genes involved in metabolite biosynthesis and transcriptional regulators of many different pathways [78,79]. Taken together, these results suggest that co-occurrence of transcripts and metabolites is a powerful approach for deciphering gene function.
Conclusion and future prospectives
As the studies that are highlighted here demonstrate, the integration of multiple genomic-scale studies can reveal novel biology. A comprehensive systems-level understanding of Arabidopsis will require -omics methods to be integrated and combined [80,81]. In the near future, each of these -omics approaches will be used in an integrative fashion to inform and validate the findings of other genome-scale projects. Proteomics has been used to predict metabolic activity in the roots and shoots of Arabidopsis demonstrating the interconnectivity of these -omics efforts [82]. These studies explore the relationship between genomic information and the products directly and indirectly encoded by the genome, which will lead to novel testable hypotheses regarding the connection between genotype and phenotype.
Acknowledgments
We are grateful to the members of the Benfey lab for helpful comments. Work in our lab on this area is funded by grants from the NIH (R01 GM-43778 and P50-GM081883) and the NSF (IOS-1021619). LML is funded by a fellowship from the Jane Coffin Childs Fund for Medical Research.
Footnotes
Conflict of interests
The authors declare that they have no competing interests.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1•.Brady SM, Provart NJ. Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell. 2009;21:1034–1051. doi: 10.1105/tpc.109.066050. In this review, the authors highlighted well-developed Web-based tools that have integrated data from several sources. They focused on transcriptomic data as an example of all of the large-scale data types available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruffel S, Krouk G, Coruzzi GM. A systems view of responses to nutritional cues in Arabidopsis: toward a paradigm shift for predictive network modeling. Plant Physiol. 2010;152:445–452. doi: 10.1104/pp.109.148502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 4.Brady SM, Orlando DA, Lee J-Y, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 5.Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 6••.Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell. 2011;21:770–782. doi: 10.1016/j.devcel.2011.09.009. A meta-analysis was conducted on transcriptional data from whole roots under 14 stress conditions and from 4 stresses at a cell type and developmental zone-specific resolution. The authors found that for a given stress the most transcriptionally responsive cell type could often be associated with a stress-induced phenotypic change. Additionally, from the whole root analysis, no universal transcriptional stress response was identified at the level of specific cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 8.Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Weber AP, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS. SNP discovery via 454 transcriptome sequencing. Plant J. 2007;51:910–918. doi: 10.1111/j.1365-313X.2007.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Filichkin S, Priest H, Givan S, Shen R, Bryant D, Fox S, Wong W, Mockler T. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. This study is the first to use RNA-seq to assess the degree to which alternative splicing occurs in Arabidopsis. Their data reveal extensive alternative splicing and estimate that as many as 42% of Arabidopsis genes with introns are alternatively spliced. Additionally they document several cases of stress-induced alternative splicing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Breakfield NW, Corcoran DL, Petricka JJ, Shen J, Sae-Seaw J, Rubio-Somoza I, Weigel D, Ohler U, Benfey PN. High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res. 2012;22:163–176. doi: 10.1101/gr.123547.111. Breakfield and coauthors present small RNA datasets comprising over 200 million aligned Illumina sequence reads covering all major cell types and distinct developmental zones of the Arabidopsis root. Using a new computational pipeline, PIPmiR the authors identified 66 new high-confidence miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck M, Claassen M, Aebersold R. Comprehensive proteomics. Curr Opin Biotechnol. 2011;22:3–8. doi: 10.1016/j.copbio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 18••.Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. This study was the first comprehensive proteomic study performed in Arabidopsis. They demonstrate the utility of proteomic studies in Arabidopsis and show the organ specificity of proteome profiles and proteotypic peptides. Their analysis is strengthened by their integrative approach comparing their results to genome-wide gene expression studies. [DOI] [PubMed] [Google Scholar]
- 19.Taylor NL, Heazlewood JL, Millar AH. The Arabidopsis thaliana 2-D gel mitochondrial proteome: refining the value of reference maps for assessing protein abundance, contaminants and post-translational modifications. Proteomics. 2011;11:1720–1733. doi: 10.1002/pmic.201000620. [DOI] [PubMed] [Google Scholar]
- 20.Jorrín-Novo JV, Maldonado AM, Echevarría-Zomeño S, Valledor L, Castillejo MA, Curto M, Valero J, Sghaier B, Donoso G, Redondo I. Plant proteomics update (2007–2008): second-generation proteomic techniques, an appropriate experimental design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage and expand biological knowledge. J Proteomics. 2009;72:285–314. doi: 10.1016/j.jprot.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 21.Supek F, Peharec P, Krsnik-Rasol M, Smuc T. Enhanced analytical power of SDS-PAGE using machine learning algorithms. Proteomics. 2008;8:28–31. doi: 10.1002/pmic.200700555. [DOI] [PubMed] [Google Scholar]
- 22.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 23.Ahrens CH, Brunner E, Qeli E, Basler K, Aebersold R. Generating and navigating proteome maps using mass spectrometry. Nat Rev Mol Cell Biol. 2010;11:789–801. doi: 10.1038/nrm2973. [DOI] [PubMed] [Google Scholar]
- 24.Baginsky S, Kleffmann T, von Zychlinski A, Gruissem W. Analysis of shotgun proteomics and RNA profiling data from Arabidopsis thaliana chloroplasts. J Proteome Res. 2005;4:637–640. doi: 10.1021/pr049764u. [DOI] [PubMed] [Google Scholar]
- 25.Oeljeklaus S, Meyer HE, Warscheid B. Advancements in plant proteomics using quantitative mass spectrometry. J Proteomics. 2009;72:545–554. doi: 10.1016/j.jprot.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Palma JM, Corpas FJ, del Río LA. Proteome of plant peroxisomes: new perspectives on the role of these organelles in cell biology. Proteomics. 2009;9:2301–2312. doi: 10.1002/pmic.200700732. [DOI] [PubMed] [Google Scholar]
- 27.Wienkoop S, Baginsky S, Weckwerth W. Arabidopsis thaliana as a model organism for plant proteome research. J Proteomics. 2010;73:2239–2248. doi: 10.1016/j.jprot.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Consortium AIM. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Mukhtar MS, Carvunis A-R, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. This study uses the most complete interactome data set available in Arabidopsis to create a plant pathogen interactome network. This approach reveals that pathogen proteins act indirectly with plant defense pathway proteins. They also show that evolutionarily diverse pathogens act on similar pathways to infect their host. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, Saito K, Ohta D, Shibata D, Kanaya S. KNApSAcK: a comprehensive species-metabolite relationship database. Plant Metabol. 2006;57:165–181. [Google Scholar]
- 31.De Vos RC, Moco S, Lommen A, Keurentjes JJ, Bino RJ, Hall RD. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protoc. 2007;2:778–791. doi: 10.1038/nprot.2007.95. [DOI] [PubMed] [Google Scholar]
- 32.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 33.Ward JL, Baker JM, Beale MH. Recent applications of NMR spectroscopy in plant metabolomics. FEBS J. 2007;274:1126–1131. doi: 10.1111/j.1742-4658.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- 34.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunn WB. Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Phys Biol. 2008;5:011001. doi: 10.1088/1478-3975/5/1/011001. [DOI] [PubMed] [Google Scholar]
- 36.Eisenreich W, Bacher A. Advances of high-resolution NMR techniques in the structural and metabolic analysis of plant biochemistry. Phytochemistry. 2007;68:2799–2815. doi: 10.1016/j.phytochem.2007.09.028. [DOI] [PubMed] [Google Scholar]
- 37.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee do Y, Lu Y, Moon S, Nikolau B. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 38.Scherling C, Roscher C, Giavalisco P, Schulze ED, Weckwerth W. Metabolomics unravel contrasting effects of biodiversity on the performance of individual plant species. PLoS One. 2010;5:e12569. doi: 10.1371/journal.pone.0012569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kempa S, Hummel J, Schwemmer T, Pietzke M, Strehmel N, Wienkoop S, Kopka J, Weckwerth W. An automated GC 3 GC– TOF-MS protocol for batch-wise extraction and alignment of mass isotopomer matrixes from differential 13C-labelling experiments: a case study for photoautotrophic–mixotrophic grown Chlamydomonas reinhardtii cells. J Basic Microbiol. 2009;49:82–91. doi: 10.1002/jobm.200800337. [DOI] [PubMed] [Google Scholar]
- 40.Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, Hirai MY, Sakurai T, Kikuchi J, Saito K. PRIMe: a Web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 2008;8:339–345. [PubMed] [Google Scholar]
- 41.Chikayama E, Sekiyama Y, Okamoto M, Nakanishi Y, Tsuboi Y, Akiyama K, Saito K, Shinozaki K, Kikuchi J. Statistical indices for simultaneous large-scale metabolite detections for a single NMR spectrum. Anal Chem. 2010;82:1653–1658. doi: 10.1021/ac9022023. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Savage LJ, Larson MD, Wilkerson CG, Last RL. Chloroplast 2010: a database for large-scale phenotypic screening of Arabidopsis mutants. Plant Physiol. 2011;155:1589–1600. doi: 10.1104/pp.110.170118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tohge T, Fernie AR. Web-based resources for mass-spectrometry-based metabolomics: a user’s guide. Phytochemistry. 2009;70:450–456. doi: 10.1016/j.phytochem.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang B-B, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen F, Wang S, Chaw S, Huang Y, Chuang T. Plant gene and alternatively spliced variant annotator. A plant genome annotation pipeline for rice gene and alternatively spliced variant identification with cross-species expressed sequence tag conservation from seven plant species. Plant Physiol. 2007;143:1086–1095. doi: 10.1104/pp.106.092460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbazuk W, Fu Y, Mcginnis KM. Genome-wide analyses of alternative splicing in plants. opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 48.Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjölander K, Gruissem W, Baginsky S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14:354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 49.Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S. Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics. 2005;5:4864–4884. doi: 10.1002/pmic.200402011. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Z, Zhang W, Stanley BA, Assmann SM. Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell. 2008;20:3210–3226. doi: 10.1105/tpc.108.063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kryvych S, Kleessen S, Ebert B, Kersten B, Fisahn J. Proteomics — the key to understanding systems biology of Arabidopsis trichomes. Phytochemistry. 2011;72:1061–1070. doi: 10.1016/j.phytochem.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 52.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baginsky S, Gruissem W. Arabidopsis thaliana proteomics: from proteome to genome. J Exp Bot. 2006;57:1485–1491. doi: 10.1093/jxb/erj130. [DOI] [PubMed] [Google Scholar]
- 54.Castellana N, Bafna V. Proteogenomics to discover the full coding content of genomes: a computational perspective. J Proteomics. 2010;73:2124–2135. doi: 10.1016/j.jprot.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albinsky D, Sawada Y, Kuwahara A, Nagano M, Hirai A, Saito K, Hirai MY. Widely targeted metabolomics and coexpression analysis as tools to identify genes involved in the side-chain elongation steps of aliphatic glucosinolate biosynthesis. Amino Acids. 2010;39:1067–1075. doi: 10.1007/s00726-010-0681-5. [DOI] [PubMed] [Google Scholar]
- 56.Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, et al. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 2009;150:1972–1980. doi: 10.1104/pp.109.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell. 2009;21:1659–1668. doi: 10.1105/tpc.109.066480. This study is an important example of the possibility to measure metabolite and gene expression with the goal of understanding the connection between transcriptome and metabolome at a cell type level using cell sorting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA. 2009;106:10348–10353. doi: 10.1073/pnas.0903478106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009;57:1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x. [DOI] [PubMed] [Google Scholar]
- 60.Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 2003;33:651–663. doi: 10.1046/j.1365-313x.2003.01658.x. [DOI] [PubMed] [Google Scholar]
- 61.Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 2005;138:304–318. doi: 10.1104/pp.104.053793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H, et al. Elucidation of gene-to-gene and metabolite-to-gene networks in arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem. 2005;280:25590–25595. doi: 10.1074/jbc.M502332200. [DOI] [PubMed] [Google Scholar]
- 65.Sawada Y, Kuwahara A, Nagano M, Narisawa T, Sakata A, Saito K, Hirai MY. Omics-based approaches to methionine side chain elongation in Arabidopsis: characterization of the genes encoding methylthioalkylmalate isomerase and methylthioalkylmalate dehydrogenase. Plant Cell Physiol. 2009;50:1181–1190. doi: 10.1093/pcp/pcp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sawada Y, Toyooka K, Kuwahara A, Sakata A, Nagano M, Saito K, Hirai MY. Arabidopsis bile acid:sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 2009;50:1579–1586. doi: 10.1093/pcp/pcp110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanani H, Dutta B, Klapa MI. Individual vs. combinatorial effect of elevated CO2 conditions and salinity stress on Arabidopsis thaliana liquid cultures: comparing the early molecular response using time-series transcriptomic and metabolomic analyses. BMC Syst Biol. 2010;4:177. doi: 10.1186/1752-0509-4-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan EKF, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 2011;9:e1001125. doi: 10.1371/journal.pbio.1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- 70.Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics — ‘majority report by precogs’. Trends Plant Sci. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 72.Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 73.Joung J-G, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei Z. Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol. 2009;151:1758–1768. doi: 10.1104/pp.109.145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 2006;7:R76. doi: 10.1186/gb-2006-7-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joosen RV, Kodde J, Willems LA, Ligterink W, van der Plas LH, Hilhorst HW. GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J. 2010;62:148–159. doi: 10.1111/j.1365-313X.2009.04116.x. [DOI] [PubMed] [Google Scholar]
- 78.Boavida LC, Borges F, Becker JD, Feijo JA. Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 2011;155:2066–2080. doi: 10.1104/pp.110.169813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei H, Persson S, Mehta T, Srinivasasainagendra V, Chen L, Page GP, Somerville C, Loraine A. Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol. 2006;142:762–774. doi: 10.1104/pp.106.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 81.Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, Conesa A, Tarraga J, Pascual-Montano A, Nogales-Cadenas R, Santoyo J, et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2008;38:W210–W213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mooney BP, Miernyk JA, Michael Greenlief C, Thelen JJ. Using quantitative proteomics of Arabidopsis roots and leaves to predict metabolic activity. Physiol Plant. 2006;128:237–250. [Google Scholar]