Abstract
Congenital heart disease (CHD) occurs in ∼1% of newborns. CHD arises from many distinct etiologies, ranging from genetic or genomic variation to exposure to teratogens, which elicit diverse cell and molecular responses during cardiac development. To systematically explore the relationships between CHD risk factors and responses, we compiled and integrated comprehensive datasets from studies of CHD in humans and model organisms. We examined two alternative models of potential functional relationships between genes in these datasets: direct convergence, in which CHD risk factors significantly and directly impact the same genes and molecules and functional convergence, in which risk factors significantly impact different molecules that participate in a discrete heart development network. We observed no evidence for direct convergence. In contrast, we show that CHD risk factors functionally converge in protein networks driving the development of specific anatomical structures (e.g., outflow tract, ventricular septum, and atrial septum) that are malformed by CHD. This integrative analysis of CHD risk factors and responses suggests a complex pattern of functional interactions between genomic variation and environmental exposures that modulate critical biological systems during heart development.
Keywords: genetics, transcriptional profiles, systems biology, developmental biology
Congenital heart disease (CHD) is a prevalent birth defect that can occur from genetic variations, environmental exposures, and other factors. Genetic risk factors for CHD include Mendelian mutations, copy number variants (CNVs), translocations, and single nucleotide polymorphisms (SNPs) (1–3). The cardiac responses to some genetic risks have been identified through transcriptional analyses of model organisms engineered to carry human CHD mutations (4). There are many environmental risk factors for CHD, including pesticides and therapeutic agents (5), and some of these are known to target particular genes. Although the cardiac molecular responses to environmental teratogens remain largely unexplored, transcriptional profiles of zebrafish exposed to retinoic acid have been characterized (6). As CHD risk factors and responses rarely incriminate the same genes nor directly impact a limited set of biological pathways, whether these converge on discrete molecular programs involved in heart development is unknown.
We used systems biology analyses to integrate the heterogeneous risk factors and response involved in CHD. Given the extraordinary conservation of genes and processes involved in heart development across species (7–10), it is possible to integrate data from humans and model organisms in a straight forward manner. Moreover, our analyses capitalize on a series of previously published and validated functional molecular networks, which direct the development of distinct anatomical structures of the human heart (11). These networks provided a developmental scaffold onto which risk and responder datasets of CHD were assessed. The hypothesis underlying our work is that these integrative analyses might elucidate unsuspected functional relationships and provide unique insights into the molecular basis of heart defects.
To address this hypothesis, we assembled gene sets of CHD risk factors (risk genes hereafter), and transcriptional responses to CHD risk factors (responder genes hereafter). Risk genes included previously reported Mendelian mutations, SNPs, CNVs, translocations, and genes that are the direct targets of teratogens, such as retinoic acid receptors (environmental target genes hereafter). Responder genes included those with altered cardiac expression in model organisms with human monogenic CHD mutations (Mendelian responders hereafter) or following exposure to teratogens (environmental responders hereafter). We interrogated the risk and responder datasets for “direct convergence” in which the same genes were directly impacted and for “functional convergence” in which genes encoding different proteins within a discrete network of heart development (11) were impacted (Fig. 1). Unlike functional convergence, direct convergence can be analyzed by overlapping gene sets without prior knowledge of developmental programs.
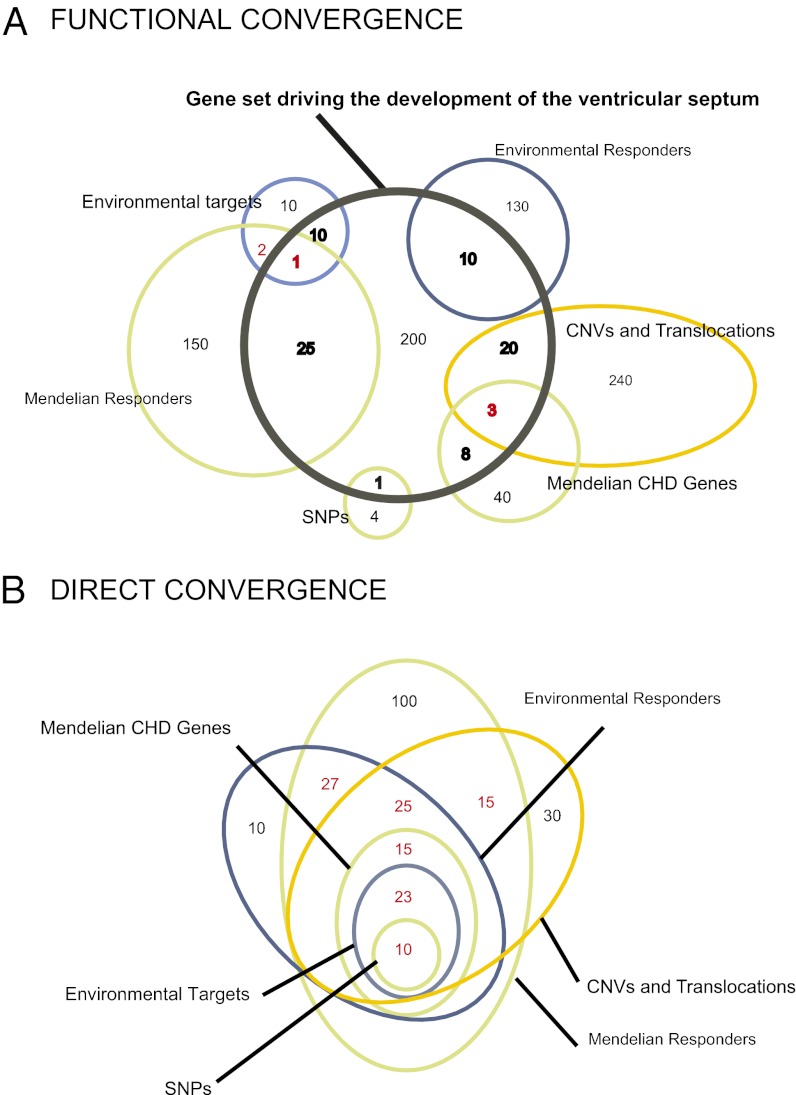
Fig. 1.
Functional convergence versus direct convergence of CHD datasets. Risk and responder datasets are indicated by green, gold, and blue circles or ellipses, respectively. Numbers represent theoretical gene counts and are different for datasets with the same name in A and B. (A) Illustrates six risk and responder datasets with different genes that are all functionally converging on a predefined developmental program of the ventricular septum (black circle, containing a total of 278 genes). The example illustrates how different risk and responder datasets can contain many genes involved in a common polygenic developmental program (bolded black and red numbers), while only sparsely overlapping with each other (red numbers). (B) Theoretical example of direct convergence, where the risk and responder datasets can be directly overlapped to reveal a common subset of genes (red numbers).
Whereas we observe little evidence for direct convergence, our analyses revealed significant functional convergence of the risk and responder datasets in the context of well-defined, but highly polygenic gene sets that participate in biological networks at specific stages of heart development. Significant functional convergence of the risk and responder datasets occurred in gene sets that form networks driving the development of the ventricular septum, atrial septum, outflow tract, and other cardiac structures often malformed in CHDs. We suggest that this strategy can be exploited to understand biologic relationships among molecules identified through studies of CHD genetics, epigenetics, teratogens, and the molecular responses that these incite.
Results
Genetic and Environmental CHD Risk and Responder Datasets.
We assembled and analyzed both previously published and experimental data generated for this study on CHD from human patients and model organisms. These datasets comprise genes directly affected by genetic or environmental perturbations (denoted as CHD risk datasets, Table 1), and genes with significantly altered expression due to genetic or environmental perturbations (denoted as CHD responder datasets, Table 2). All risk and responder gene sets are available (www.cbs.dtu.dk/suppl/dgf/).
Table 1.
Human risk datasets
| Type of risk factor | Description | Source for dataset |
| CNVs and translocations: Cohort A | Four hundred three rare structural variants (spanning 709 genes) identified in 136 trios with isolated CHD. | Ref. 3 and this study. Details, chromosomal coordinates, and gene identifiers are provided in SI Appendix and Dataset S1. |
| CNVs and translocations: Cohort B | One thousand five hundred forty-four rare structural variants (spanning 657 genes) identified in 526 probands with isolated or syndromic CHD. | This study. Details, chromosomal coordinates, and gene identifiers are provided in SI Appendix and Dataset S2. |
| Mendelian CHD genes | Forty-six genes with mutations that cause Mendelian forms of CHD. | Curated from the literature. Details and gene list are provided in SI Appendix and Dataset S3. |
| SNP-associated genes | Six genes containing SNPs that are associated with CHD. | Curated from the literature. Details and gene list are provided in SI Appendix and Dataset S3. |
| Environmental target genes | Seven genes (e.g., retinoic acid receptor) that are direct targets of environmental CHD teratogens (e.g., retinoic acid). | Curated from the literature. Details and gene list are provided in SI Appendix and Dataset S3. |
Table 2.
Genetic and environmental responder datasets
| Type of responder | Description | Source for dataset |
| Mendelian responder genes | Six hundred seventy-three differentially expressed genes in the hearts of mice with CHD mutations in Gata4 and Nkx2-5. | This study. Details, and gene list are provided in SI Appendix and Dataset S6. |
| Environmental responder gene | Two hundred forty differentially expressed genes in the hearts of zebrafish after embryonic exposure to teratogenic doses of retinoic acid. | Curated from the literature. Gene list is provided in ref 5. |
Risk datasets (Table 1 and Methods) include genes from the literature that have been demonstrated to contain sequence variants (Mendelian mutations or SNPs) that cause or increase the risk for CHD and genes that lie within rare structural variants in the genome (CNVs or translocations) in two CHD cohorts studied here. Rare (<1% population frequency) structural variants identified in cohort A, 136 cases with isolated, nonsyndromic CHD and in cohort B, 526 cases with both isolated and syndromic CHD were identified (SI Appendix and Datasets S1 and S2). Risk datasets also include genes that are direct targets of previously defined environmental factors.
Responder datasets (Table 2 and Methods) are the human orthologs of genes that exhibit significantly altered downstream expression in experimental models of CHD. These datasets (SI Appendix and Dataset S3) are derived from transcriptional analyses of heart tissues from mice with haploinsufficiency of the cardiac transcription factors Gata4 (12, 13) or Nkx2-5 (14), generated here. Comparable human mutations in these genes result in CHD. Additional responder genes are those with significantly altered expression in zebrafish hearts following environmental exposure to teratogenic levels of retinoic acid (6), annotated from the literature.
Functional Convergence of CHD Risk and Responder Datasets in Heart Developmental Networks.
We previously reported (11) a series of experimentally validated protein networks (available at www.cbs.dtu.dk/suppl/dgf) that drive the development of 19 discrete anatomical cardiac structures (e.g., septa, valves, and outflow tract). Disruption of genes encoding the proteins in these networks, when introduced into mouse models, can cause heart malformations that recapitulate human CHD. Onto these networks we mapped the human risk and responder datasets. The statistical propensity of functional convergence was assessed by measuring the number of genes from the risk and responder datasets (Tables 1 and 2) represented in each of the heart developmental networks (as exemplified in Fig. 1A) and by comparing to a random expectation.
We illustrate the general tendency of functional convergence between CHD risk factors using networks involved in outflow tract development (Fig. 2A), ventricular septal development (Fig. 2B), and atrial septal development (Fig. 2C). Malformations of these structures occur commonly in CHD patients. For example, ventricular septal defects occur in ∼40% of all CHD cases (15). Despite the considerable heterogeneity of factors annotated in Tables 1 and 2 we observed significant functional convergence between the seven datasets of risk and responder genes in each of these three networks (P < 1.0e-4 for each network, using a permutation test). For clarity, the risk and responder datasets have been grouped in Fig. 2. The specific risk or responder dataset that a given protein belongs to is shown in Fig. 3.
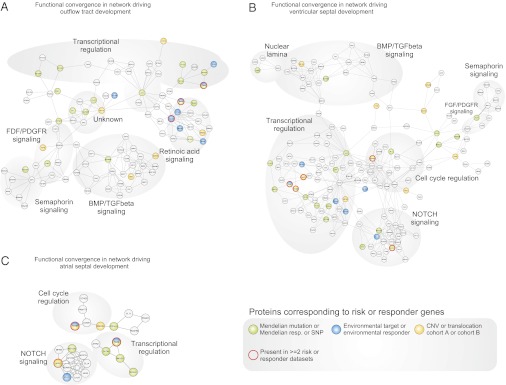
Fig. 2.
Functional convergence of CHD risk and responder datasets in cardiac developmental networks. (A–C) Functional convergence was analyzed in predefined developmental programs (11), deduced from protein–protein interaction networks driving the development of the outflow tract (A), ventricular septum (B), and atrial septum (C). Circles represent gene-encoded proteins (gene names viewable with the Adobe zoom tool or at www.cbs.dtu.dk/suppl/dgf/). Lines represent protein–protein interactions. Colors indicate datasets of Mendelian risk genes, SNPs, or Mendelian responder genes (green), CNVs and translocations from cohort A or B (gold), or environmental target or responder genes (blue). Red circles indicate proteins belonging to more than one dataset.
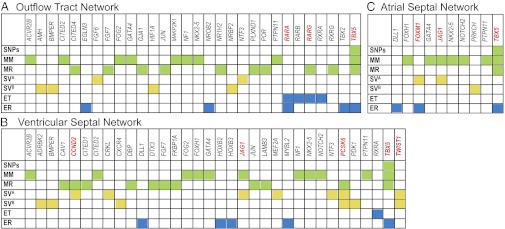
Fig. 3.
Few genes are shared between CHD risk and responder datasets. Each panel shows all genes from any risk or responder dataset that overlaps with the gene set represented in the outflow tract (A), ventricular septum (B), and atrial septum (C) developmental network. Datasets are color coded as detailed in Fig. 2 and abbreviated as: SNPs, single nucleotide polymorphisms; MM, Mendelian mutations; MR, Mendelian responders; SVA and SVB, structural variants in cohorts A and B respectively; ET, environmental targets; ER, environmental responders. Despite the significant functional convergence between the risk and responder datasets in each of the networks (Fig. 2), only a few genes (highlighted in red) are identified in multiple risk datasets.
Analyses of risk and responder gene sets within the context of the outflow tract development network (Fig. 2A) revealed significant functional convergence. Within this network we observed 1 protein encoded by a gene with CHD-associated SNPs, 9 proteins encoded by genes with Mendelian CHD mutations, 10 proteins encoded by Mendelian responder genes, 2 proteins encoded by genes disrupted by CNVs in cohort A, 4 proteins encoded by genes disrupted by CNVs in cohort B, 4 proteins encoded by genes that are targets of environmental teratogens, and 5 proteins encoded by environmental responder genes (Fig. 3A). This degree of functional convergence of the risk and responder datasets was compared with a random expectation using permutation tests (see SI Appendix for an example). Permutation tests were performed to assess how many genes in Tables 1 and 2 would be expected to functionally converge in the gene set represented in the outflow tract network by chance, given the size and composition of the individual risk and responder datasets, while taking into consideration the composition and properties of the gene set represented in the outflow tract network.
Similar analyses were conducted for 16 other developmental networks involved in distinct heart structures. Analyses of this larger group of networks (SI Appendix, Datasets S4 and S5, and SI Appendix, Figs. S1–S16) confirmed our initial findings in three networks (Fig. 2 A–C). Thus, in total we observed significant (P < 0.05 after adjustment for multiple hypotheses testing) functional convergence between the risk and responder datasets in 17 of 19 networks. Moreover, these results did not reflect shared genes among different networks being tested (details in SI Appendix).
Lack of Direct Convergence of the Risk and Responder Datasets.
Despite observing that some genes are represented in more than one of the risk and responder datasets, these genetic and environmental risk factors did not converge on a limited gene set or pathway. Fig. 3 illustrates the very few genes (boxed in red) present within three networks (Fig. 2) that were identified in more than one risk or responder dataset. Analyses of the larger set of networks (SI Appendix, Figs. S1–S16) confirmed this observation.
To statistically test for direct convergence between the risk and responder datasets, we made pairwise comparisons without mapping the datasets to developmental networks (Fig. 4). Among responder datasets, we observed a trend toward direct convergence, but this generally did not extend across all of the risk datasets and sharply contrasted to the functional convergence observed when the risk and responder datasets were integrated into the developmental networks. Notably, the genetic and environmental risk factors rarely converged on the same genes. Moreover, whereas some CHD risks impacted well-characterized developmental signaling pathways, these pathways were individually associated with only a few of the risk factors examined here. Thus, directly comparing the risk and responder datasets would suggest that these genes were not functionally related or involved in a common molecular program for heart development.
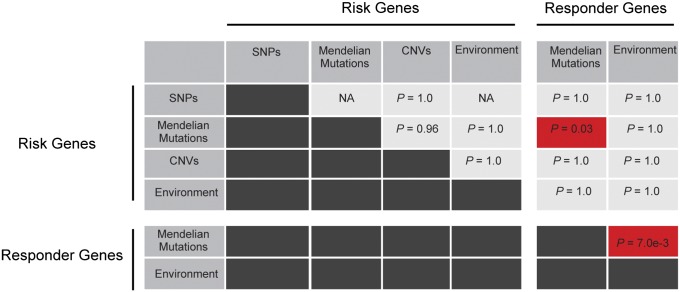
Fig. 4.
Pairwise comparisons of direct convergences of risk and responder datasets. Genes (Table 1) were compared using hypergeometric statistics to assess signficance of overlap between datasets of varying sizes and corrected for multiple testing. Shading denotes significant (red) or insignificant (grey) direct convergence. SNPs within Mendelian CHD genes and environmental target genes are not independent and were excluded (NA).
Enrichment of Heart Development Genes in Risk and Responder Datasets.
We considered whether the lack of direct convergence of the risk and responder datasets reflected “noise,” i.e., the inclusion of genes in the datasets that are not directly involved in heart development. To allow a normalized comparison between the risk and responder datasets despite considerable differences in gene counts (and experimental platforms underlying these data), we examined dataset-specific enrichments of heart development genes in each of the individual risk and responder datasets compared with null distributions. We observed that each of the individual datasets were significantly enriched for genes present in the 19 developmental networks underlying cardiac anatomical structures (Fig. 5A; P < 5.0e-4 to P = 0.019 for the individual datasets using a permutation test), indicating noise in each of these datasets cannot explain the observed lack of direct convergence.
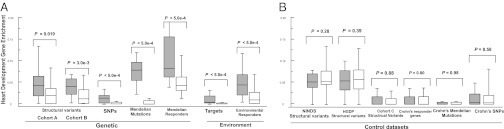
Fig. 5.
Risk and responder datasets are enriched for heart developmental genes. (A) Box-and whisker plots showing the fraction of genes shared between the individual risk and responder datasets (Tables 1 and 2) and each of the 19 cardiac developmental networks, compared with dataset-specific null distributions. Each dataset shows significant enrichment for heart developmental genes represented in one or more of the networks (P < 5.0e-4 to P = 0.016 for the individual datasets of risk or responder genes, respectively) after taking into account the size and composition of each dataset. Note that the enrichment of genes within structural variants identified in cohort A are replicated in an independent dataset from cohort B. (B) Analyses of control datasets of comparable complexity to the CHD risk and responder datasets showed no enrichment for genes within 19 heart developmental networks. Distributions are plotted as their median, first, and third quartile (box) and minimum and maximum (whiskers). Gray boxes, dataset-specific enrichment; white boxes, dataset-specific null distribution.
That significant enrichment of heart development genes was observed in the analyses of structural variants was particularly noteworthy, given the inclusion of many genes within each deleted or duplicated genomic segment. As some of these genes are unlikely to be involved in CHD, including them should limit the statistical power of these analyses. To validate the positive signal associated with the analysis of cohort A (Dataset S1), we studied a second independent dataset of rare CNVs derived from cohort B (Dataset S2). Despite the considerable diversity of heart malformations, and the likely inclusion of many genes not involved in CHD in these CNVs, data from cohort B replicated the findings for cohort A (Fig. 5A, P = 3.0e-3, using a permutation test).
To confirm that the differences in sizes between the datasets were not confounding this analysis, we applied another test based on probability distributions of the observed overlaps between networks and individual datasets (which normalizes for size differences between the datasets; SI Appendix). This test corroborated and strengthened the results in Fig. 5A: All risk and responder datasets were enriched for heart developmental genes (P < 5.0e-4 to P = 0.002 for the individual datasets using a permutation test; SI Appendix, Fig. S17).
As a control for both tests, we also performed comparable analyses for six control datasets unrelated to CHD. Analogous to the risk and responder datasets assembled for CHD, we gathered risk and responder datasets for Crohn disease (SI Appendix and ref. 16) and additional datasets of genes altered by rare structural variants (CNVs <1% population frequency) in three independent cohorts of healthy individuals (the National Institute of Neurological Disorders and Stroke, the Human Genome Diversity Project, and cohort C, detailed in SI Appendix). Analyses of each of these six control datasets showed no enrichment for genes involved in the heart developmental networks (SI Appendix, Fig. S18 and Fig. 5B) and confirmed that the observed enrichment (Fig. 5A) is specific to the CHD-related datasets of risk and responder genes.
Discussion
By testing the relationships between datasets of genetic and environmental risk factors in CHD, we uncovered functional convergence among thousands of CHD risk factors. Surprisingly, although genetic and environmental factors involved in CHD impact genes that participate across many different molecular pathways (Fig. 2 A–C), these seemingly unrelated risk factors affect pathways that participate in larger, but discrete, protein interaction networks that drive the development of specific cardiac structures. In contrast, we observed no direct convergence between the risk and responder datasets on a set of genes, a result that is unlikely to be caused by noise in the individual datasets. These results are analogous to observations for other disorders (both Mendelian and common, complex disorders) supporting the increasing evidence that similar polygenic molecular networks could underlie other diseases (17–21).
An alternative interpretation of our results might be that these reflect a tissue-specific signal, unrelated to CHD, because several of the datasets were generated from heart tissue. If this were so, we would expect a high degree of direct convergence between the risk and responder datasets, which was not observed. For example, the datasets of genes with Mendelian CHD mutations and the environmental responder genes were both significantly enriched for genes represented in the developmental networks, although they directly converged on only one shared gene. Moreover, the CNV datasets provided an unbiased control, because they included whole-genome data unrelated to tissue expression. Both the CNV dataset from cohorts A and B were statistically enriched for genes encoding proteins in the networks.
We did not quantify the pathological relevance of genes in the risk or responder data, but considered them as a uniform set. As such, a CNV was considered to convey CHD risk even if it was observed in only one CHD patient, and Mendelian CHD genes were not weighted on the basis of the numbers of mutations demonstrated to cause CHD. Similarly, responder genes were identified on the basis of statistically significant differential expression in experimental models; we neither factored the degree to which levels were altered nor considered whether changes were primary or secondary. We therefore expected that all datasets were biased toward noise rather than signal.
By scaffolding the risk datasets with biological insight into the 19 predefined heart developmental networks, our analyses benefited from increased statistical power. This analytical framework can be readily expanded to include larger CHD datasets derived from ongoing studies of the molecular responses to environmental exposures and genetic and epigenetic data emerging from the application of contemporary sequencing technologies. We suggest that this strategy may inform other complex and congenital disorders.
Our analyses incorporated risk and responder datasets that span more than 2,100 unique genes. Whereas each dataset was enriched for genes encoding proteins in the heart developmental networks, only 15 genes (ACTC1, CCND2, FOXM1, IGFBP5, JAG1, MYH6, NOTCH1, PCSK6, PLCB1, RAF1, RARG, RARA, TBX5, TMOD1, and TWIST1) were found in multiple risk or response datasets, as well as in one of the 19 heart developmental networks. Seven of these genes (CCND2, FOXM1, IGFBP5, PCSK6, PLCB1, TWIST1, and TMOD1) are not recognized as Mendelian causes of CHD; nor are these genes the direct targets of teratogens. Further study of these genes in cardiac development and CHD may be warranted (discussion in SI Appendix).
Another unexpected result from our analyses was that sequence and structural variants, both rare and common, converged on large polygenic gene sets required for heart development, but did not point to a shared subset of genes. Although more tests will be necessary to fully interpret these observations, it is tempting to speculate that functional convergence might help to explain the “missing heritability” paradox in complex disorders (22) including CHD. As the extent of individual genomic variation is more widely appreciated, we expect that both deleterious and mitigating influences on the highly polygenic molecular networks driving heart development will be discovered and that individual patients could have deleterious sequence or structural variation in several genes across a network.
Whereas we recognize limitations in these analyses (details in SI Appendix), recent studies support functional convergence of risk factors in heart and other developmental networks. For example, pairs of CNVs within a genome convey greater risk for severe developmental delay than individual CNVs (23), and de novo exon mutations in autism patients physically interact in protein interaction networks (19, 20). Moreover, emerging studies of the combined effect of environment and genetics in animal models also support functional convergence (discussion in SI Appendix).
Given these analyses and the complexity of CHD, we speculate that malformations arise from a multidimensional combinatorial space of perhaps tens or hundreds of sequence and structural variants that may perturb several different members of large developmental networks in a single patient. The individual genetic risk factors might be very rare, and the specific combination of risk factors in many patients unique, which could account for the considerable phenotypic heterogeneity observed in CHD patients with identical mutations in established CHD genes. Although speculative at this point, these hypotheses should be amenable to testing using systems-based approaches to integrate personal sequence variations in the context of heart developmental networks.
Improved understanding of the functional convergence of risk factors in highly polygenic disorders such as CHD may suggest new prevention strategies and diagnostic methods based on genetics. Despite critical advances in surgical and catheterization-based CHD therapies that have reduced childhood mortality to less than 5% in developed countries, life-long cardiovascular problems often remain after corrective procedures (24, 25). Moreover, the mortality of CHD is still very high in developing countries (26). Systems-based insights into cardiac developmental networks and CHD risk factors may lead to improved genetic diagnoses, treatment, and eventually prevention strategies and promote deeper understanding of how an individual’s genome and environmental interactions result in disease.
Methods
All studies conformed to the Helsinki Declaration of the World Medical Association. Human datasets were acquired using protocols approved by institutional human research committees. Animal studies were performed using protocols approved by the Animal Care and Use Committee of Harvard Medical School. All risk, responder, and control gene sets, the cardiac developmental networks, and the random gene sets used in the permutation analyses are available at www.cbs.dtu.dk/suppl/dgf/. Details for the risk and responder datasets are provided in SI Appendix.
Statistical Tests.
Functional convergence was assessed using permutation tests to measure the likelihood of observing similar or higher degrees of convergence on protein sets matched to the proteins in the specific cardiac development network being tested using data from the protein–protein interaction database InWeb (18, 27, 28). Direct convergence was assessed from the significance of pairwise overlaps between risk and responder datasets, calculated using a hypergeometric distribution. Enrichment of heart developmental genes in datasets was assessed by measuring the fraction of genes affected in each of the corresponding 19 protein networks and comparing the number of affected genes with a dataset-specific null distribution using a two-sample Kolmogorov–Smirnov test. (For more details, see SI Appendix).
Supplementary Material
Acknowledgments
This work was supported by grants from the Danish Heart Foundation (L.A.L.), Forskningsrådet for Sundhed og Sygdom and the Eleanor and Miles Shore Fellowship (K.L.), the Pediatric Scientist Developmental Program (S.C.G.), the National Institutes of Health (NICHD HD055150-03 and 1P01HD068250-01 to K.L. and P.K.D. and NHLBI UO1 HL098166 and U01 HL098147 to A.E.R., W.T.P., H.W., J.M.G., C.E.S. and J.G.S.), and the Howard Hughes Medical Institute (C.E.S).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210730109/-/DCSupplemental.
References
- 1.Wessels MW, Willems PJ. Genetic factors in non-syndromic congenital heart malformations. Clin Genet. 2010;78:103–123. doi: 10.1111/j.1399-0004.2010.01435.x. [DOI] [PubMed] [Google Scholar]
- 2.Vannay A, et al. Single-nucleotide polymorphisms of VEGF gene are associated with risk of congenital valvuloseptal heart defects. Am Heart J. 2006;151:878–881. doi: 10.1016/j.ahj.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Greenway SC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JB, et al. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins KJ, et al. American Heart Association Council on Cardiovascular Disease in the Young Noninherited risk factors and congenital cardiovascular defects: Current knowledge: A scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Carney SA, Peterson RE, Heideman W. Comparative genomics identifies genes mediating cardiotoxicity in the embryonic zebrafish heart. Physiol Genomics. 2008;33:148–158. doi: 10.1152/physiolgenomics.00214.2007. [DOI] [PubMed] [Google Scholar]
- 7.Sperling SR. Systems biology approaches to heart development and congenital heart disease. Cardiovasc Res. 2011;91:269–278. doi: 10.1093/cvr/cvr126. [DOI] [PubMed] [Google Scholar]
- 8.Yang JH, Saucerman JJ. Computational models reduce complexity and accelerate insight into cardiac signaling networks. Circ Res. 2011;108:85–97. doi: 10.1161/CIRCRESAHA.110.223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava D. Making or breaking the heart: From lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Fishman MC, Olson EN. Parsing the heart: Genetic modules for organ assembly. Cell. 1997;91:153–156. doi: 10.1016/s0092-8674(00)80397-9. [DOI] [PubMed] [Google Scholar]
- 11.Lage K, et al. Dissecting spatio-temporal protein networks driving human heart development and related disorders. Mol Syst Biol. 2010;6:381. doi: 10.1038/msb.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopal SK, et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol. 2007;43:677–685. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M, et al. A mouse model of congenital heart disease: Cardiac arrhythmias and atrial septal defect caused by haploinsufficiency of the cardiac transcription factor Csx/Nkx2.5. Cold Spring Harb Symp Quant Biol. 2002;67:317–325. doi: 10.1101/sqb.2002.67.317. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 16.Wu F, et al. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: Insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 17.Cotsapas C, et al. FOCiS Network of Consortia Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossin EJ, et al. International Inflammatory Bowel Disease Genetics Constortium Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell MK, et al. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc Natl Acad Sci USA. 2012;109:2978–2983. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 23.Girirajan S, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verheugt CL, et al. Mortality in adult congenital heart disease. Eur Heart J. 2010;31:1220–1229. doi: 10.1093/eurheartj/ehq032. [DOI] [PubMed] [Google Scholar]
- 25.Warnes CA, et al. ACC/AHA 2008 Guidelines for the Management of Adults with Congenital Heart Disease: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (writing committee to develop guidelines on the management of adults with congenital heart disease) Circulation. 2008;118:e714–e833. doi: 10.1161/CIRCULATIONAHA.108.190690. [DOI] [PubMed] [Google Scholar]
- 26.Tchervenkov CI, et al. The improvement of care for paediatric and congenital cardiac disease across the World: A challenge for the World Society for Pediatric and Congenital Heart Surgery. Cardiol Young. 2008;18(Suppl 2):63–69. doi: 10.1017/S1047951108002801. [DOI] [PubMed] [Google Scholar]
- 27.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 28.Lage K, et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci USA. 2008;105:20870–20875. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.