Abstract
The Podoviridae phage C1 was one of the earliest isolated bacteriophages and the first virus documented to be active against streptococci. The icosahedral and asymmetric reconstructions of the virus were calculated using cryo-electron microscopy. The capsid protein has an HK97 fold arranged into a T = 4 icosahedral lattice. The C1 tail is terminated with a φ29-like knob, surrounded by a skirt of twelve long appendages with novel morphology. Several C1 structural proteins have been identified, including a candidate for an appendage. The crystal structure of the knob has an N-terminal domain with a fold observed previously in tube forming proteins of Siphoviridae and Myoviridae phages. The structure of C1 suggests the mechanisms by which the virus digests the cell wall and ejects its genome. Although there is little sequence similarity to other phages, conservation of the structural proteins demonstrates a common origin of the head and tail, but more recent evolution of the appendages.
Keywords: bacteriophage C1 tail, tail knob, X-ray crystallography
Bacteriophage C1 infects group C streptococci and belongs to the Podoviridae family of phages. It was one of the earliest isolated bacteriophages and the first documented bacteriophage to be active against streptococci (1). Phage C1 is highly specific to group C streptococci, which are animal pathogens. As a result of its specificity, C1 was used for classification of bacterial strains, thus leading to the development of analytical phage typing (2). Although C1 only infects group C streptococci, the bacteriophage lysin, released during the last stage of infection, can rapidly kill groups A, C, and E streptococci as well as Streptococcus uberis and Streptococcus equi. Thus, in contrast to many bacteriophages that recognize the same epitope by their tail fibers and by the lysins (3–5), the bacteriophage C1 tail spike has evolved to become highly specific to only one group of streptococci. Although bacteriophage C1 lysin protein has been extensively studied (6–9), the structure of the phage has remained unknown.
Bacteriophage C1 has a small 16,687 bp genome, which encodes for twenty open reading frames (ORFs). Functions of only nine of these have yet been assigned (7). Bacteriophage C1 has been classified as a member of the 44AHJD-like genus, none of which have been investigated structurally. Analysis of the C1 genome showed very limited similarity to other phages. Four bacteriophage C1 proteins (the polymerase, the holin, the connector, and the major tail protein) have distant homology to structural proteins of phages from the φ29 family (7). Φ29-like phages have a terminal protein associated with the genome and utilize a protein-primed mechanism of replication (10, 11). The presence of a terminal protein has been also shown for bacteriophage C1, although the identity of this protein has not yet been determined (7). In contrast to characterized φ29-like and 44AHJD-like phages, which have short appendages at the head-to-tail junction (10), bacteriophage C1 has a skirt of unusually long appendages around the tail.
In the current work, the structure of bacteriophage C1 virus has been studied by cryo-electron microscopy (cryo-EM) and image reconstruction. Additionally, the structure of the C1 major tail protein, forming a tail knob, was determined by X-ray crystallography. The current study is the first structural investigation of a bacteriophage that infects streptococci. The reconstruction showed that phage C1 has an isometric capsid with a diameter of 500 Å. The capsid protein is organized into a T = 4 icosahedral lattice and has a HK97 fold. This fold of the capsid protein has been observed in all tailed phages (12–18). The short C1 tail is surrounded by a skirt of 12 long appendages, each terminating with a flexible globular domain, unlike those of tailed phages studied previously. The cylindrical portion of the C1 tail is terminated by a knob, which is a hexamer of the major tail protein, gene product (gp)12. The crystal structure of C1 knob protein, gp12, was determined to 3 Å resolution by X-ray crystallography. The small N-terminal domain of the C1 tail protein has a protein fold commonly found in neck and tail tube proteins of Sipho- and Myoviridae phages (19), but not previously found in Podoviridae phage proteins. Apart from the 120 residue N-terminal domain, the gp12 structure has a novel fold, which might also occur in knob proteins of φ29-like phages.
Results and Discussion
Protein Identification.
Several C1 structural proteins have been previously identified, including the major capsid, the major tail, the connector and the collar proteins (7). Only capsid and connector proteins were visible on SDS-PAGE of the purified virus. As a result of a modified viral purification, leading to higher virus concentration and increased sample purity, more proteins were resolved on an SDS gel. Based on N-terminal sequencing and mass-spectroscopy, the purified virus sample contains the major capsid protein, the major tail protein, the connector, the collar protein, and five proteins of unknown function: ORF5, ORF13, ORF17, ORF18, and ORF19. The latter three proteins are very small (the largest is 64 residues) and their location in the virus remains undetermined. A BLAST search (20) showed that ORF5 could have peptidase activity, suggesting that it could be associated with the tail. The protein ORF13 is possibly a bacteriophage appendage. Although ORF13 does not have any similarity to known phage proteins based on amino acid sequence analysis, using the HHpred and the PAIRCOIL structural prediction software (21–23) ORF13 was identified to have a coiled-coil domain and a C-terminal eight-stranded α/β barrel (TIM barrel) domain. Currently, there is no example of a tail spike structure with a TIM barrel domain in the Protein Data Bank (PDB). However, a possible tail spike protein, gp29 of Siphoviridae phage ISP (SPO1-like phage), is also predicted to have a TIM barrel domain by HHpred. Although the probability score for the C1 ORF13 structural prediction is not high, such a score was sufficient to predict domain structures for other proteins (21, 23–25). It had been previously suggested that ORF6 could be an appendage, based on sequence comparison (7). However, ORF6 was not present among the viral structural proteins analyzed from the gel. Furthermore, ORF6 is the only protein in the C1 sequence that has Walker A and B motifs, suggesting it could be a packaging ATPase.
Structure of the C1 Capsid.
Cryo-EM reconstruction showed that the C1 capsid has a diameter of 500 Å and a T = 4 icosahedral arrangement, making it one of the smallest of tailed phages. The majority of structurally studied tailed phages have a larger capsid with a T = 7 or higher icosahedral arrangement (P22, T7, HK97, ε15, K1-E, N4) (12, 16, 26–29). Bacteriophage φ29 forms a prolate head with T = 3 and Q = 5 triangulation numbers (14, 30). A mutant of φ29 is capable of producing T = 3 isometric heads, with a small portion of the virus making larger heads with T = 4 and possibly T = 7 symmetry (31). Another known example of a T = 4 capsid among tailed phages is the head of bacteriophage P4, a genetically unrelated satellite virus of P2 of the P2/P4 bacteriophage system. Both P2 and P4 utilize the same capsid protein, but bacteriophage P2 assembles into T = 7 capsids, whereas P4 assembles into T = 4 capsids due to the presence of a specific scaffolding protein (32, 33).
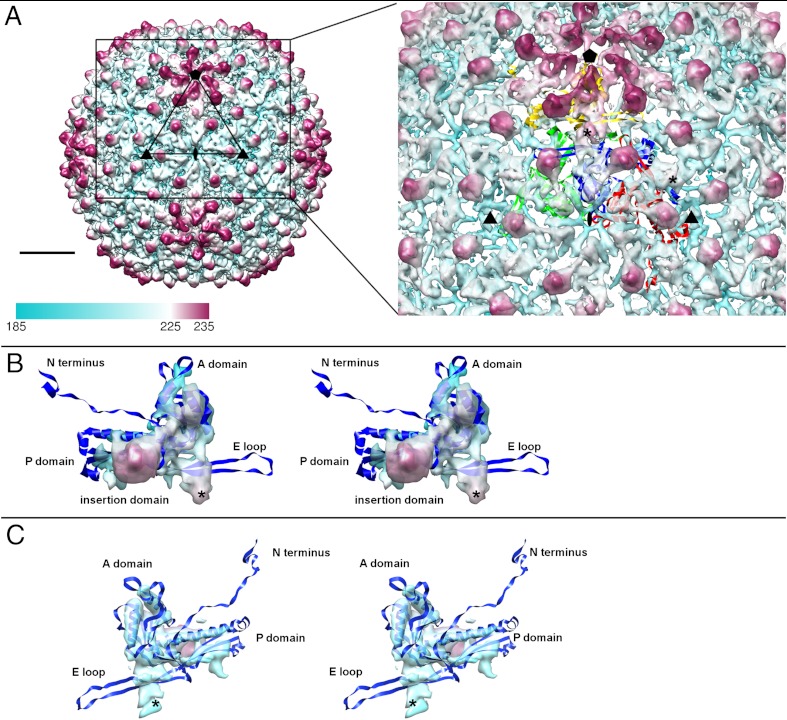
Bacteriophage C1 icosahedral reconstruction was calculated to 8.3 Å resolution (Fig. 1). The C1 major capsid protein, gp16, is homologous to capsid proteins from the 44AHJD genus and does not have any detectable similarity to capsid proteins from other phage groups. However, the distance between the capsomers in the cryo-EM reconstruction was calculated to be approximately 140 Å. This is the distance between the centers of capsomers, observed in HK97 and all other tailed dsDNA phages studied to date (12–14, 16–18, 27, 28, 34). The HK97 subunit consists of an axial A domain, a peripheral P domain and two extensions, an N-terminal arm and an E-loop (27).
Fig. 1.
Icosahedral reconstruction of the C1 capsid. (A) The surface of the capsid colored radially from cyan to magenta. The fit of HK97 subunits into four C1 capsid protein densities that form an asymmetric unit is shown on the right. The “*” symbol shows the position of two quasi-2-fold axes in the asymmetric unit. A scale bar corresponds to 100 Å. (B) A stereo view of the capsid protein density segmented from the map. (C) A stereo view of the capsid protein density segmented from the map. The orientations shown in B and C are related by 180° rotation.
In the asymmetric unit of the T = 4 C1 capsid, one subunit comes from a pentameric capsomer and three from an adjacent hexameric capsomer (Fig. 1A). The HK97 capsid protein structure was fitted into the C1 capsid shell using the Emfit program (35). The electron density of the capsid protein monomer contains three rod-like densities that are 50, 25, and 15 Å long. These densities correspond to the long helix α3 and two short helixes α5 and α6 of the HK97 capsid protein structure (numbered according to the HK97 protein convention) (27). Most differences between the HK97 and C1 capsid proteins occur in the regions corresponding to the long E loop and the N-terminal arm of HK97 fold (Fig. 1 B and C). In the HK97 structure, these regions have the largest structural deviation among the seven subunits that form an asymmetric unit of the HK97 capsid (27). Also, these portions of the HK97 fold were shown to have the most differences among different phages (12, 14, 18, 36). The number of residues in gp16 of C1 is comparable to that of the HK97 capsid protein (392 a.a. versus 385 a.a.). However, the N-terminal 103 residues of the HK97 capsid protein form a delta-domain that is cleaved upon capsid assembly (37). In contrast to HK97, no cleavage of the C1 capsid protein was detected by N-terminal sequencing of the protein band from the virus preparation. The additional approximately 100 residues probably form an insertion domain, which corresponds to the protruding density observed in the cryo-EM reconstruction. Analysis of the C1 capsid protein molecular envelope showed the presence of two densities that were uninterpreted by fitting of the HK97 structure: a protrusion close to the location of the HK97 capsid protein E loop and an additional density close to the quasi-2-fold axes (Fig. 1). Other phages (for example, P22, φ29, K1-E, and T4) were also shown to contain protruding insertion domains in the capsid protein structure (12–14, 16). The structure and location of such domains is often different between different phages, although the rest of the structure has a HK97 fold. In some phages, such as T4 or φ29, an insertion domain was suggested to stabilize the capsid by bridging either neighboring subunits within a capsomer or capsid proteins from neighboring capsomers (13, 14). The position of the C1 insertion domain density is such that it could be assigned to either of two neighboring capsid proteins. It is located either over the peripheral P domain of the same molecule or is associated with the neighboring capsid protein in place of the E-loop of the HK97 fold. At the current resolution, the continuity of the electron density favors the first possibility (Fig. 1 B and C). The other additional density, located at the C1 capsid quasi-2-fold, bridges between neighboring hexameric capsomers and between neighboring hexameric and pentameric capsomers, thus aiding capsid stability. Similar densities at the capsomer interfaces had been observed in phages BPP-1, P-SSP7, and P22 (17, 18, 36). In Bordetella phage BPP-1 these densities were attributed to a cement protein. In P-SSP7 the capsid protein has an extension in the E-loop, creating an additional density at interfaces between capsomers. In P22 a density made by a loop in the insertion domain provides strong interactions connecting neighboring capsomers. As no additional band of comparable intensity to the capsid protein band was present in the SDS gel of the purified C1 virus, it is unlikely that the twofold density in phage C1 is an extra protein. Therefore, the twofold density probably corresponds to a loop in the capsid protein, similar to that in phage P-SSP7.
Asymmetric Reconstruction.
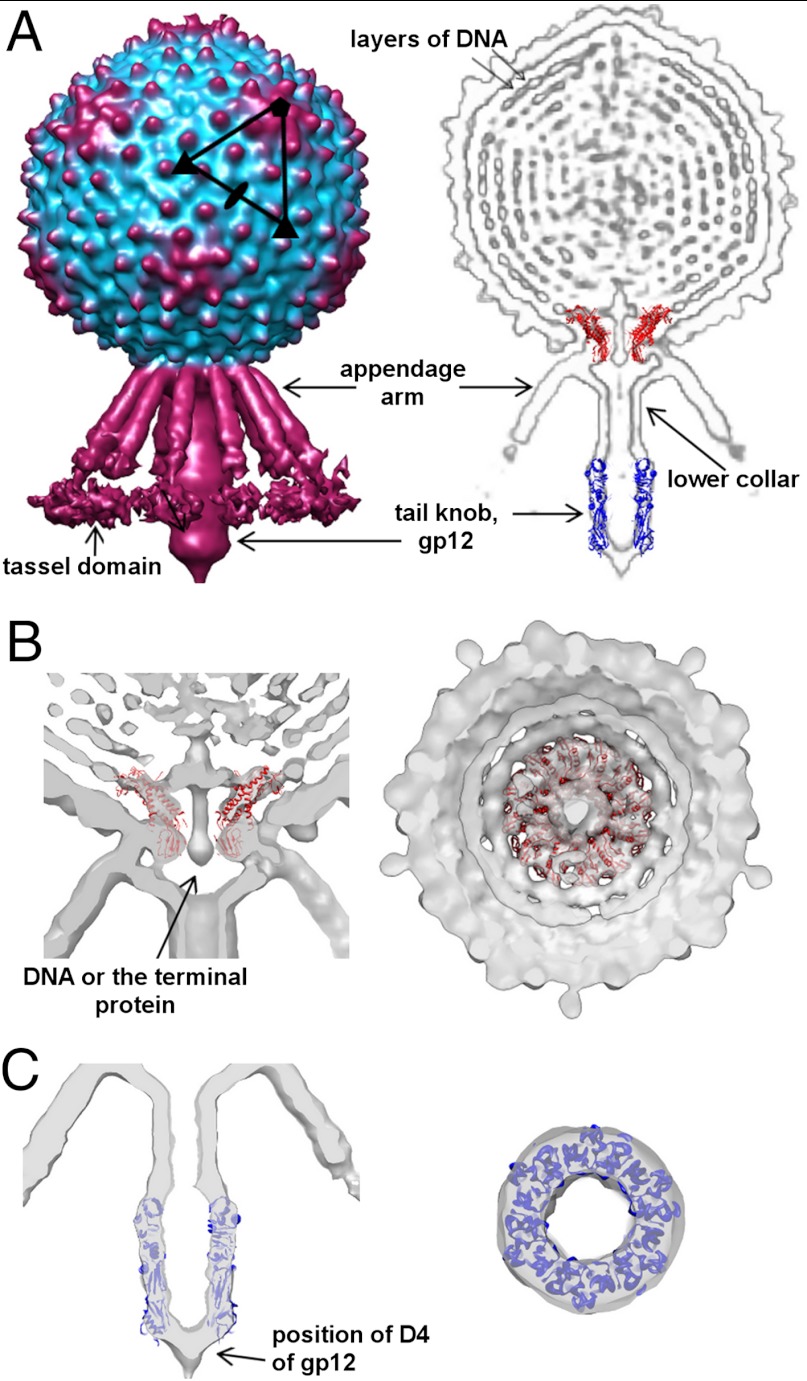
By assuming no symmetry in the reconstruction it was observed that there is a fixed orientation between the sixfold tail and the fivefold symmetric head in many tailed phages (14, 18, 28, 29, 38–40). A reconstruction without assuming any symmetry of the C1 virion showed that the tail consists of an about 250 Å long narrow cylinder, which terminates with a 150 Å long knob, surrounded by a skirt of twelve 180 Å long appendages (Fig. 2). Because no symmetry was imposed in the reconstruction, relative arrangement and oligomeric states of the different components could be observed. The connector, positioned at the interface of the head-tail symmetry mismatch, has twelve lobes of density, displaying approximate 12-fold symmetry. Here, as well as in other bacteriophage reconstructions, the twelve connector subunits are in different environments (28, 39). In the case of P-SSP7 it was shown that the connector does not follow strict 12-fold symmetry (18). Plasticity of the connector helps accommodate the symmetry mismatch and could play a role during DNA packaging. The structure of the φ29 connector fits well into the C1 phage connector density (Fig. 2B). Many bacteriophage connector proteins are bigger than the φ29 connector, ranging from 36 kDa in φ29 (41) to 83 kDa in P22 (42, 43). However, the C1 connector protein is comparable in size to that of φ29 (317 a.a. vs. 309 a.a.). In the connector channel there is a continuous density, which could be attributed to either DNA or the terminal protein (Fig. 2B). In the interior of the capsid there are several layers of highly ordered DNA in which individual strands of DNA are separated by about 25 Å (Fig. 2A). The rings of DNA are hexagonally packed and arranged as a spool wound around the central axis. The spool is very well ordered, but there is no internal body inside the C1 capsid unlike in many other phages, including ε15, T7, N4, K1-E, or P-SSP7 (16, 18, 26, 28, 29). In comparison, only two to three layers of ordered DNA are visible in the reconstruction of the prolate φ29 head (39). The narrow end of the connector protrudes from the C1 capsid and binds to the lower collar and possibly interacts with the appendages.
Fig. 2.
Asymmetric reconstruction of the C1 virion. (A) Surface representation of the C1 virion (Left) and cross-section through the bacteriophage, showing different components of the virus (Right). The structure of the φ29 connector and the C1 major tail protein was fitted into the electron density. (B) Two orthogonal cross-sections through the cryo-EM density map, showing the fit of the φ29 connector. (C) Two orthogonal cross-sections through the cryo-EM density showing the fit of gp12 into the knob.
The cylindrical tail structure of bacteriophage C1 consists of a bulge region, to which the appendages are connected, a narrow upper tube and a wider terminal knob. Based on the sequence and positional similarities with the tail of φ29, the upper portion corresponds to the lower collar protein, gp14, and the knob is made of the major tail protein, gp12, discussed below. In the case of C1, the knob density is cone-shaped with a narrower terminal end, whereas in the case of φ29 it is bell-shaped with a wider end (39). Bacteriophage φ29 has an extra protein, gp13, that is present at the very tip of the knob, which possesses peptidase activity (44). The electron density, corresponding to gp13, is visible only in the emptied φ29 particles after DNA ejection. In the case of phage C1, a protein encoded by ORF5, present in the virus sample, could have some peptidase activity based on the sequence analysis using a BLAST search. Although ORF5 is smaller than gp13 of φ29 (207 residues versus 365) and there is no detectable sequence similarity between these two proteins; ORF5 could be associated with the tip of the C1 tail in a way similar to gp13 of φ29. The cylindrical part of the C1 tail is surrounded by 12 appendages, which have a novel morphology. The appendages of φ29 form a very tight collar around the head-to-tail junction and do not extend all the way to the tail knob as in C1 (30, 39). In φ29, the appendages have short arms or ribs and terminate with a tail spike that has a ubiquitous β-helical fold (45, 46). As discussed above, the structure of the C1 appendage protein is probably different from that of φ29, with a TIM barrel terminal domain. The twelve C1 appendages radiate from the head-tail junction between the lower collar and the connector at about 45° to the cylindrical part of the tail (Fig. 2A). Based on the electron density, each appendage can be segmented into three domains: a proximal domain that binds to the virus, a long arm domain, and a terminal flexible tassel domain. The proximal domain of the appendage protein interacts with the connector and capsid, as well as with the lower collar protein. The appendage arm is about 140 Å long, twice as long as the arm or rib of a φ29 appendage. From the dimensions of the density, it is reasonable to suggest that the appendage arm has a coiled-coil structure. As a typical coiled-coil has a length of 150 Å per 100 residues, the coiled-coil domain of the appendage protein has to be about 95 residues, which is in agreement with the structural prediction of a coiled-coil structure in ORF13. The density of the terminal tassel domain is weak relative to the rest of the tail density, suggesting it is flexible. As in other Podoviridae phages, the terminal domain of the appendage probably has an enzymatic activity and is involved in binding and/or digesting the cell wall during infection. This domain probably corresponds to the TIM barrel fold, predicted at the C-terminal part of ORF13. The flexibility of the tassel domain might be beneficial for its enzymatic function.
Crystal Structure of the Major Tail or Knob Protein, gp12.
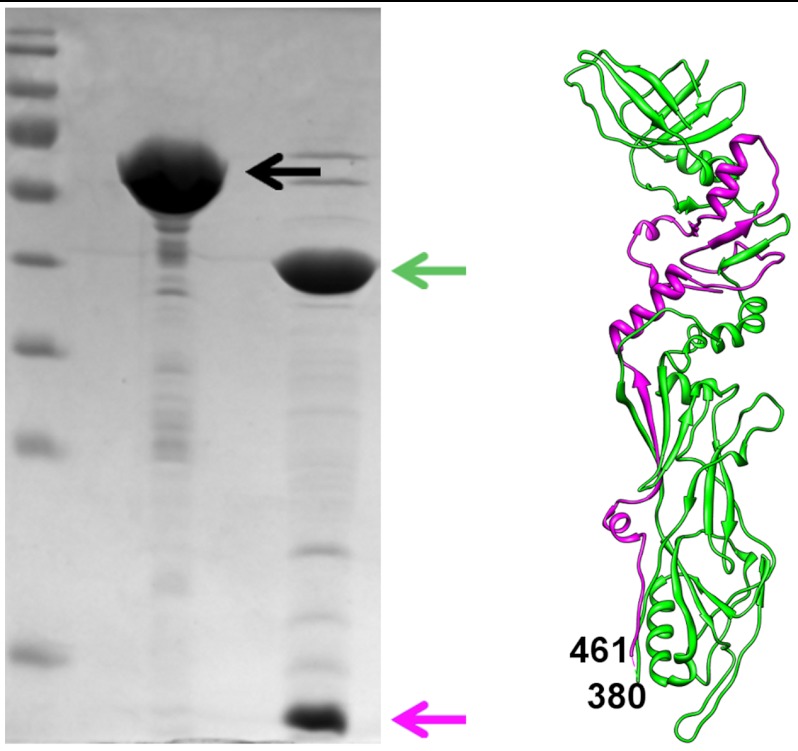
The full-length major tail protein, gp12, was expressed in E. coli and purified using a C-terminal hexa-histidine tag (Materials and Methods). The full-length protein contains 573 amino acid residues and was estimated to form a hexamer in solution using size-exclusion chromatography. Because the full-length protein did not produce crystals, trypsin digestion was used to obtain a stable fragment of gp12. It was found that trypsin cut gp12 into two fragments that could be separated by an SDS PAGE. N-terminal sequencing results showed that the higher molecular weight band corresponded to the N-terminal fragment of gp12 and the lower molecular weight band corresponded to the fragment starting with residue 461 (Fig. 3). Despite the presence of a hexa-histidine tag at the C-terminus of the protein, it was impossible to separate the two fragments by Ni-NTA affinity chromatography. Moreover, the protein remained a hexamer in solution, as determined by size-exclusion chromatography. Thus, the two fragments were structurally intertwined. The protein was crystallized in the P21212 space group with a hexamer of gp12 in the asymmetric unit and in C2 space group with two hexamers in the asymmetric unit. The structure of the trypsin resistant part of gp12 was determined by X-ray crystallography, using the Multiple Anomalous Dispersion method (Materials and Methods and Table S1). The structure showed that trypsin digestion removed a small 80-residue insertion domain (381–460) resulting in two polypeptide chains. The larger linear fragment corresponds to residues 1–381 and the smaller linear fragment corresponds to residues 461–573, which is consistent with the N-terminal sequencing results. Thus, the gp12 structure consists of two fragments, which are part of the same fold and can only be separated under denaturing conditions (Fig. 3).
Fig. 3.
Trypsin resistant fragment of gp12. The lanes on the SDS-gel show the marker, full length gp12 (black arrow) and gp12 after trypsin digestion (green and magenta arrows). Corresponding fragments are colored in green and magenta in the gp12 structure shown on the right.
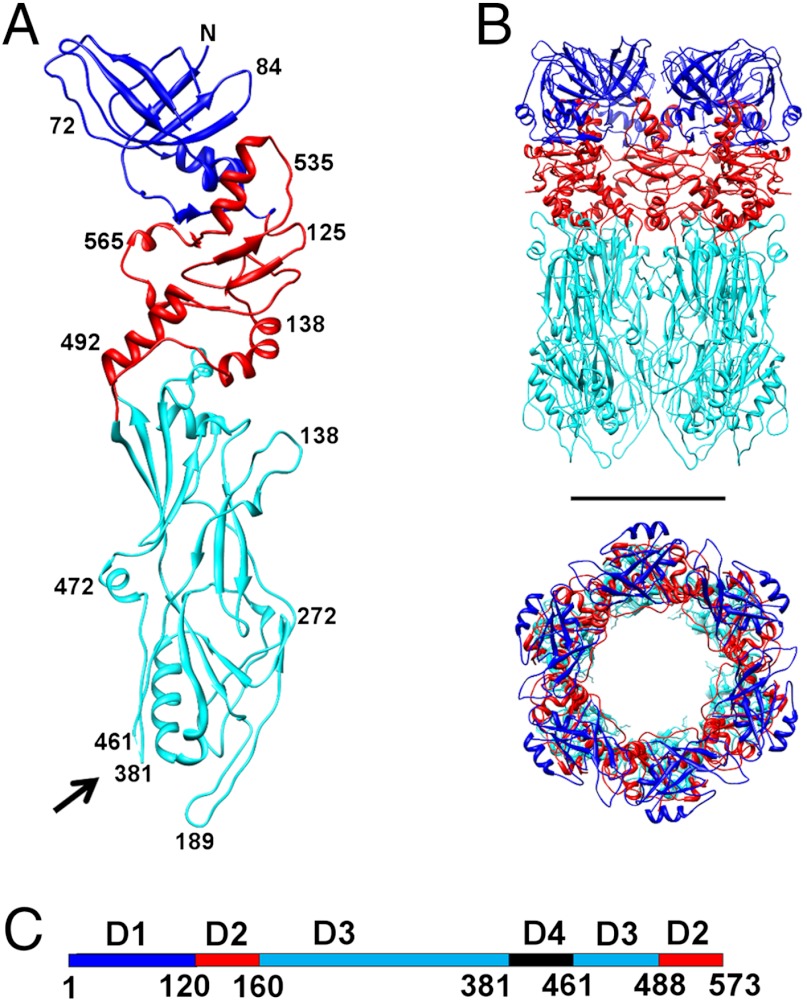
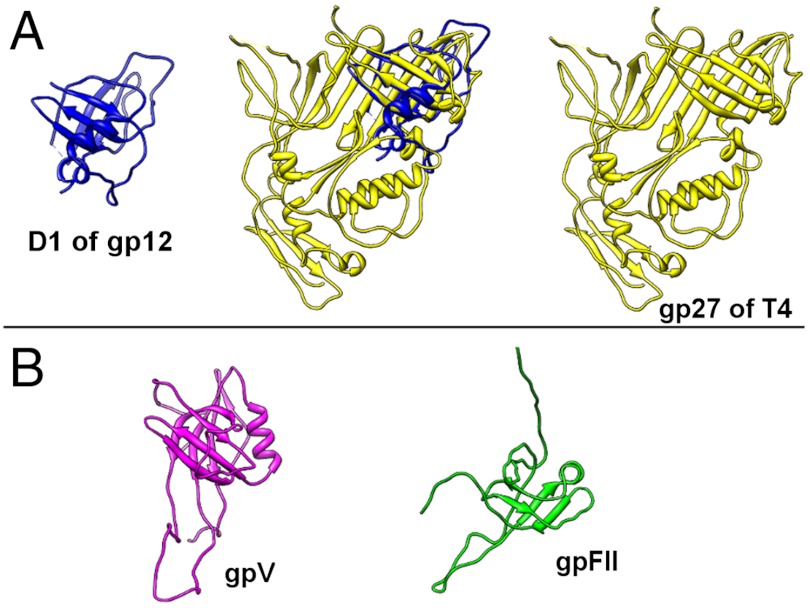
The hexamer of gp12 is a 140 Å-long hollow cylinder with an outer diameter of about 90 Å and an inner diameter of about 40 Å (Fig. 4B). The orientation and position of gp12 hexamer in the electron density of the tail knob was determined by the EMfit program (Fig. 2C, SI Materials and Methods, Table S2). The gp12 monomer is a long, narrow molecule that can be separated into three domains: an N-terminal domain (D1), a short middle domain (D2), and a long domain (D3). The positions of the trypsin cleavage sites (residues 381 and 461) are close in space and indicate that the missing 80 residues (381–461) could form a fourth, or “tip,” domain (D4) at the end of the knob (Fig. 4 A and C). Thus, the tail protein structure has a modular architecture: D4 is an insertion in D3, which itself is an insertion in D2 (Fig. 4C). The DALI search (47) showed that only the fold of the N-terminal domain (D1) had been observed previously, whereas the rest of the gp12 structure has a novel fold. It was found that the 120 residue N-terminal D1 is similar to the fold observed in a very ubiquitous family of bacteriophage proteins (Fig. 5), which form closed ring structures found as a part of the tail tube, neck, or baseplate (19). This β-sandwich fold is observed, for example, in a bacteriophage T4 baseplate protein gp27, lambda tail tube protein gpV, lambda neck protein gpU, and gpFII (48–51). Most of these proteins are hexamers or pseudo-hexamers (i.e., trimers with a monomer that has domain duplication) like gp27 of T4. Similarly, the gp12 N-terminal domain forms a hexameric ring. However, in the case of gp12, the N-terminal domains do not form a continuous β-barrel through tight interactions within a hexamer, as observed in other tube proteins. Moreover, the N-terminal domains are the most disordered in the gp12 structure and have twice higher B-factors than the rest of the protein. When an electron density map was calculated without applying the sixfold non-crystallographic symmetry (NCS) averaging, the densities of the six NCS-related N-terminal domains were different. Only those N-terminal domains were ordered that formed crystal contacts with neighboring molecules. However, it is possible that, upon interaction with a lower collar protein which presumably forms the upper cylindrical part of the C1 tail, the gp12 N-terminal domains could become ordered and form a tighter hexameric ring.
Fig. 4.
Structure of the major tail protein, gp12. (A) A monomer of gp12 with domains D1, D2 and D3 colored in blue, red and cyan, respectively. Black arrow points to the location of D4 where the protein was cleaved by trypsin. (B) Side and top views of the gp12 hexamer. Scale bar is 50 Å long. (C) Location of the domains on the linear sequence of gp12, using the same color scheme as in A and B.
Fig. 5.
Structural similarity of the C1 major tail protein N-terminal domain (D1) to Siphoviridae and Myoviridae proteins. (A) Structural alignment of gp12 D1 to gp27 from bacteriophage T4. (B) Structures of gpV and gpFII from bacteriophage lambda that have the same fold as D1 of the C1 major tail protein.
The tail tube fold of the N-terminal D1 of gp12 had been previously observed only in the Myoviridae and Siphoviridae families of phages. To the best of our knowledge, this fold has not previously been found in a tail of a Podoviridae phage. Although the similarity between the C1 gp12 and the φ29 gp9 knob proteins is observed only in the C-terminal portion of the sequence, it is likely that φ29-like phages have the same knob protein fold as phage C1. The presence of common folds among diverse groups of bacteriophages that do not have any similarity at the level of DNA sequence indicates evolutionary ties between bacteriophage families and shows that they evolved from a common ancestor.
Infection Process.
Streptococcus C is a Gram-positive pathogen with a 250 Å-thick cell wall, which corresponds to about the length of the C1 tail. During infection the C1 phage probably has to digest the cell wall so that the tip of the tail can reach the cellular membrane and inject the viral DNA. By comparison of the group C host with the group A bacteria, it was deduced that bacteriophage C1 probably uses N-acetylglucoseamine side chains on a polyrhamnose backbone as part of its receptor (7). By analogy with other phages, like φ29 and P22, and due to the lack of any enzymatic activity suggested by the structure of gp12, it is reasonable to suggest that the appendages are involved in the processing of the cell wall and thus facilitate the approach of the tip of the virus tail to the cell membrane. Additionally, a TIM barrel fold, which most likely forms a tassel at the tip of the appendage protein, often has an enzymatic activity. Since there are twelve appendages per bacteriophage, the binding of one appendage to the receptor increases the probability of attachment by the other eleven. Thus, a large number of appendages around the tail increase the avidity of the receptor binding. This could facilitate the digestion of the bacterial cell wall, getting the tip of the tail closer to the membrane. If ORF5 is a peptidase, as suggested above, and is also associated with the tail, it could help digest the cross-linked peptidoglycan layer.
The fit of gp12 into the knob density indicates that the missing insertion domain (D4) is located at the very tip of the tail knob (Fig. 2 A and C). Thus, it probably forms the plug that seals the hollow cylindrical tail and prevents premature DNA ejection. Based on the secondary structure prediction, D4 is mostly α-helical. Unfortunately, it was not possible to find any homologs to D4 using a BLAST search or HHpred. Considering its location, D4 might be involved in interactions with the cell membrane during phage infection. The flexibility of D4 could be important for the opening of the tail cylinder, leading to the DNA release.
Most bacteriophages have a two-step infection process that involves initial receptor binding and/or cell wall digestion and a secondary interaction that triggers the release of the viral genome. It has been previously suggested for different tailed phages that a signal is propagated from the tail to the connector, changing its conformation and releasing the DNA (18, 39, 52–55). In C1 this trigger could propagate through the cylindrical tail part or could also come from the appendages, which interact directly with the connector. A signal from the appendages could initiate the DNA exit into the tail cylinder. Finally, the opening of the tail knob terminal tip could be triggered by its contact with the membrane or a membrane protein, leading to DNA ejection into the host cell.
Materials and Methods
The propagation and purification of C1 phage was based on the protocol published previously (7) (SI Materials and Methods). A combination of EMAN (56) and SPIDER (57) software packages were used for the image reconstructions (SI Materials and Methods and Fig. S1). Details of cloning, purification, crystallization and structure determination of gp12 are described in SI Materials and Methods. Data collection and refinement statistics are summarized in Table S1. The program EMfit (35) was used for the crystal structure fitting (SI Materials and Methods). The individual capsid protein density was segmented from the capsid map based on the connectivity using programs MAMA and AVE from the Uppsala Software Factory (58), as well as using the program SEGGER as part of the CHIMERA package (59, 60). The Figures were prepared using the program CHIMERA (59).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Anthony Battisti, Pavel Plevka, and Ye Xiang for stimulating discussions. We thank Petr Leiman and Mikhail Shneider for assistance with HHpred analysis of ORF13. We thank Sheryl Kelly for help in preparation of the manuscript. We thank the staff at the beamline 23 GM/CA-CAT of Advanced Photon Source, Argonne National Laboratory for help with the data collection. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences (Contract DE-AC02-06CH11357). Aspects of this work were supported by the National Science Foundation (MCB-1014547 awarded to M.G.R.) or the National Institutes of Health (AI081726 awarded to M.G.R and AI11822 to V.A.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.E.J. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates of the knob protein gp12 were deposited with the Protein Data Bank, www.pdb.org (PDB ID codes 4EO2 and 4EP0). Cryo-EM maps of the C1 capsid and the whole virus were deposited with the Electron Microscopy Data Bank, www.emdatabank.org (EMDB codes 5445 and 5446, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207730109/-/DCSupplemental.
References
- 1.Clark PF, Clark AS. A “bacteriophage” active against a hemolytic streptococcus. J Bacteriol. 1926;11:89. [Google Scholar]
- 2.Evans AC. Streptococcus bacteriophage: A study of four serological types. Public Health Rep. 1934;49:1386–1401. [Google Scholar]
- 3.Diaz E, Lopez R, Garcia JL. Chimeric phage-bacterial enzymes: A clue to the modular evolution of genes. Proc Natl Acad Sci USA. 1990;87:8125–8129. doi: 10.1073/pnas.87.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001;294:2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 5.Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418:884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 6.Köller T, et al. PlyC, a novel bacteriophage lysin for compartment-dependent proteomics of group A streptococci. Proteomics. 2008;8:140–148. doi: 10.1002/pmic.200700001. [DOI] [PubMed] [Google Scholar]
- 7.Nelson D, Schuch R, Zhu S, Tscherne DM, Fischetti VA. Genomic sequence of C1, the first streptococcal phage. J Bacteriol. 2003;185:3325–3332. doi: 10.1128/JB.185.11.3325-3332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: A multimeric bacteriophage lysin. Proc Natl Acad Sci USA. 2006;103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischetti VA. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vybiral D, et al. Complete nucleotide sequence and molecular characterization of two lytic Staphylococcus aureus phages: 44AHJD and P68. FEMS Microbiol Lett. 2003;219:275–283. doi: 10.1016/S0378-1097(03)00028-4. [DOI] [PubMed] [Google Scholar]
- 11.Meijer WJ, Horcajadas JA, Salas M. Phi29 family of phages. Microbiol Mol Biol Rev. 2001;65:261–287. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang W, et al. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- 13.Fokine A, et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morais MC, et al. Conservation of the capsid structure in tailed dsDNA bacteriophages: The pseudoatomic structure of φ29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leiman PG, et al. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J Mol Biol. 2007;371:836–849. doi: 10.1016/j.jmb.2007.05.083. [DOI] [PubMed] [Google Scholar]
- 17.Dai W, et al. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc Natl Acad Sci USA. 2010;107:4347–4352. doi: 10.1073/pnas.0915008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardarelli L, et al. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc Natl Acad Sci USA. 2010;107:14384–14389. doi: 10.1073/pnas.1005822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: Improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 23.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 24.Aksyuk AA, et al. Structural conservation of the Myoviridae phage tail sheath protein fold. Structure. 2011;19:1885–1894. doi: 10.1016/j.str.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG. Phage pierces the host cell membrane with the iron-loaded spike. Structure. 2012;20:326–339. doi: 10.1016/j.str.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Agirrezabala X, et al. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 28.Jiang W, et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi KH, et al. Insight into DNA and protein transport in double-stranded DNA viruses: The structure of bacteriophage N4. J Mol Biol. 2008;378:726–736. doi: 10.1016/j.jmb.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao Y, et al. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi KH, Morais MC, Anderson DL, Rossmann MG. Determinants of bacteriophage φ29 head morphology. Structure. 2006;14:1723–1727. doi: 10.1016/j.str.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Dokland T, Wang S, Lindqvist BH. The structure of P4 procapsids produced by coexpression of capsid and external scaffolding proteins. Virology. 2002;298:224–231. doi: 10.1006/viro.2002.1485. [DOI] [PubMed] [Google Scholar]
- 33.Marvik OJ, et al. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J Mol Biol. 1995;251:59–75. doi: 10.1006/jmbi.1995.0416. [DOI] [PubMed] [Google Scholar]
- 34.Fu X, Walter MH, Paredes A, Morais MC, Liu J. The mechanism of DNA ejection in the Bacillus anthracis spore-binding phage 8a revealed by cryo-electron tomography. Virology. 2011;421:141–148. doi: 10.1016/j.virol.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with X-ray crystallographic structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 36.Chen DH, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci USA. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duda RL, et al. Structural transitions during bacteriophage KH97 head assembly. J Mol Biol. 1995;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- 38.Morais MC, et al. Cryoelectron-microscopy image reconsruction of symmetry mismatches in bacteriophage φ29. J Struct Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang Y, et al. Structural changes of bacteriophage φ29 upon DNA packaging and release. EMBO J. 2006;25:5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang J, Sinkovits RS, Baker TS. Three-dimensional asymmetric reconstruction of tailed bacteriophage. Methods Enzymol. 2010;482:185–210. doi: 10.1016/S0076-6879(10)82008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson AA, et al. Structure of the bacteriophage φ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olia AS, Prevelige PE, Jr, Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang J, et al. Peering down the barrel of a bacteriophage portal: The genome packaging and release valve in P22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y, et al. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage φ29 tail. Proc Natl Acad Sci USA. 2008;105:9552–9557. doi: 10.1073/pnas.0803787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang Y, et al. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol Cell. 2009;34:375–386. doi: 10.1016/j.molcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinbacher S, et al. Crystal structure of P22 tailspike protein: Interdigitated subunits in a thermostable trimer. Science. 1994;265:383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- 47.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 48.Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 49.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage λ major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pell LG, et al. The X-ray crystal structure of the phage λ tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J Mol Biol. 2009;389:938–951. doi: 10.1016/j.jmb.2009.04.072. [DOI] [PubMed] [Google Scholar]
- 51.Maxwell KL, Yee AA, Arrowsmith CH, Gold M, Davidson AR. The solution structure of the bacteriophage λ head-tail joining protein, gpFII. J Mol Biol. 2002;318:1395–1404. doi: 10.1016/s0022-2836(02)00276-0. [DOI] [PubMed] [Google Scholar]
- 52.Leiman PG, et al. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. Available at http://www.virologyj.com/content/7/1/355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertin A, de Frutos M, Letellier L. Bacteriophage-host interactions leading to genome internalization. Curr Opin Microbiol. 2011;14:492–496. doi: 10.1016/j.mib.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Lhuillier S, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA. 2009;106:8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plisson C, et al. Structure of bacteriophage SPP1 tails reveals trigger for DNA ejection. EMBO J. 2007;26:3720–3728. doi: 10.1038/sj.emboj.7601786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 57.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 58.Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA. Around O. In: Rossmann MG, Arnold E, editors. International Tables for Crystallograhy, Vol F, Crystallography of Biological Macromolecules. Dordrecht/Boston/London: Kluwer Academic Publishers; 2001. pp. 353–356–366–367. [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Computat Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.