Abstract
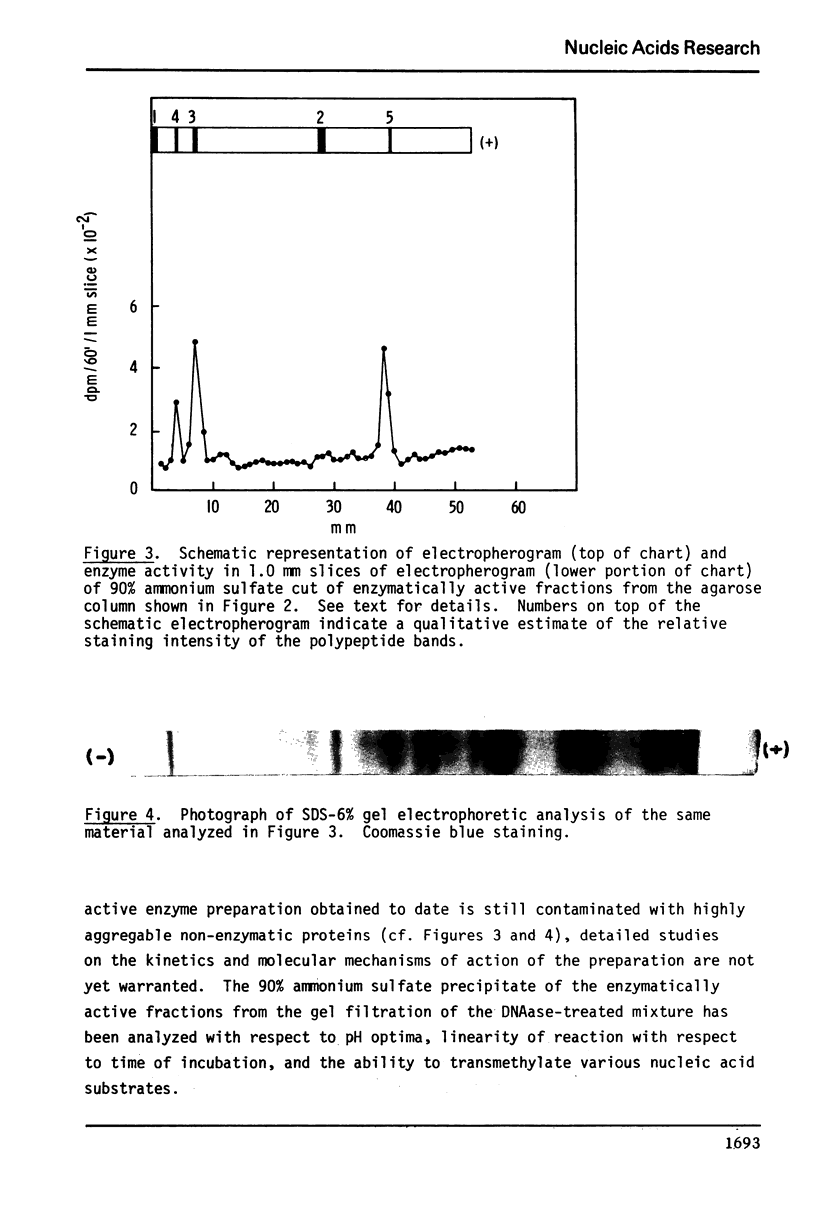
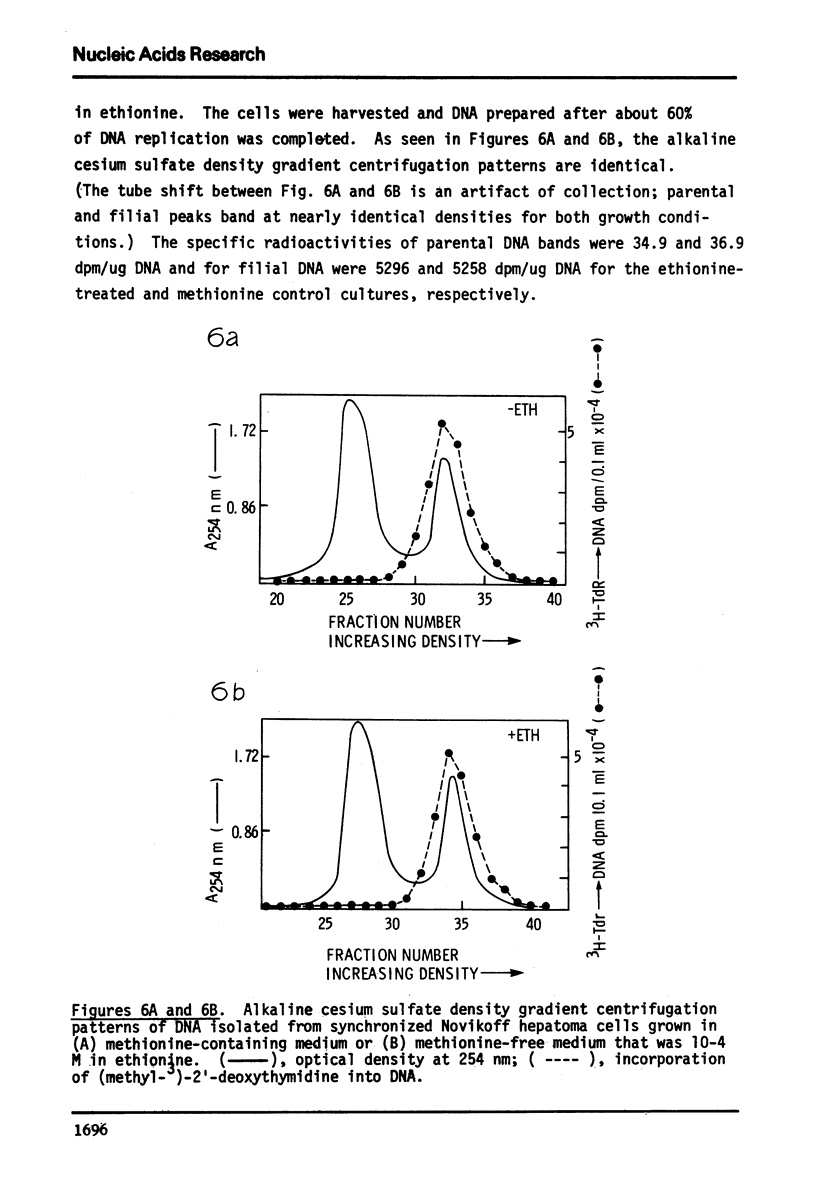
Partial purification of DNA methylase from Novikoff rat hepatoma cells is described. Contamination with other proteins persists although the enzyme preparation has a high specific activity and is purified 980-fold over homogenate activity. Evidence suggests, but does not prove, that there may be more than one species of DNA methylase in these cells. The enzyme has two broad pH optima at pH 7.0 and 7.5 and most readily methylates heterologous denatured DNAs although complex reaction kinetics indicate that native DNAs may eventually be methylated to an equal or greater level. The preparation of undermethylated DNA from Novikoff cells is also described. Undermethylated homologous DNA is an 85-fold greater acceptor of methyl groups than fully methylated Novikoff cell DNA. In contrast to other DNA substrates, the enzyme preparation methylates native undermethylated homologous DNA at a 3.5-fold greater than denatured undermethylated homologous DNA.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W., Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- Burdon R. H., Douglas J. T. The influence of subcellular fractions on the enzymic methylation of DNA in ascites cell nuclei. Nucleic Acids Res. 1974 Jan;1(1):97–103. doi: 10.1093/nar/1.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon R. H., Martin B. T., Lal B. M. Synthesis of low molecular weight ribonucleic acid in tumour cells. J Mol Biol. 1967 Sep 14;28(2):357–371. doi: 10.1016/s0022-2836(67)80015-9. [DOI] [PubMed] [Google Scholar]
- Drahovsky D., Morris N. R. The mechanism of action of rat liver DNA methylase. 3. Nucleotide requirements for binding and methylation. Biochim Biophys Acta. 1972 Aug 25;277(2):245–250. doi: 10.1016/0005-2787(72)90404-2. [DOI] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. I. Interaction with double-stranded methyl-acceptor DNA. J Mol Biol. 1971 May 14;57(3):475–489. doi: 10.1016/0022-2836(71)90104-5. [DOI] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. II. Interaction with single-stranded methyl-acceptor DNA. J Mol Biol. 1971 Oct 28;61(2):343–356. doi: 10.1016/0022-2836(71)90384-6. [DOI] [PubMed] [Google Scholar]
- Hilliard J. K., Sneider T. W. Repair methylation of parental DNA in synchronized cultures of Novikoff hepatoma cells. Nucleic Acids Res. 1975 Jun;2(6):809–819. doi: 10.1093/nar/2.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjertén S., Jerstedt S., Tiselius A. Electrophoretic "particle sieving" in polyacrylamide gels as applied to ribosomes. Anal Biochem. 1965 May;11(2):211–218. doi: 10.1016/0003-2697(65)90007-2. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Kalousek F., Morris N. R. Deoxyribonucleic acid methylase activity in pea seedlings. Science. 1969 May 9;164(3880):721–722. doi: 10.1126/science.164.3880.721. [DOI] [PubMed] [Google Scholar]
- Kalousek F., Morris N. R. The purification and properties of deoxyribonucleic acid methylase from rat spleen. J Biol Chem. 1969 Mar 10;244(5):1157–1163. [PubMed] [Google Scholar]
- Morris N. R., Pih K. D. The preparation of soluble DNA methylase from normal and regenerating rat liver. Cancer Res. 1971 Apr;31(4):433–440. [PubMed] [Google Scholar]
- SRINIVASAN P. R., BOREK E. ENZYMATIC ALTERATION OF NUCLEIC ACID STRUCTURE. Science. 1964 Aug 7;145(3632):548–553. doi: 10.1126/science.145.3632.548. [DOI] [PubMed] [Google Scholar]
- Scarano E. The control of gene function in cell differentiation and in embryogenesis. Adv Cytopharmacol. 1971 May;1:13–24. [PubMed] [Google Scholar]
- Sheid B., Srinivasan P. R., Borek E. Deoxyribonucleic acid methylase of mammalian tissues. Biochemistry. 1968 Jan;7(1):280–285. doi: 10.1021/bi00841a034. [DOI] [PubMed] [Google Scholar]
- Sneider T. W. Methylation of mammalian deoxyribonucleic acid. II. The distribution of 5-methylcytosine in pyrimidine deoxyribonucleotide clusters in Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1971 Aug 10;246(15):4774–4783. [PubMed] [Google Scholar]
- Sneider T. W., Potter V. R. Methylation of mammalian DNA: studies on Novikoff hepatoma cells in tissue culture. J Mol Biol. 1969 Jun 14;42(2):271–284. doi: 10.1016/0022-2836(69)90043-6. [DOI] [PubMed] [Google Scholar]
- Sneider T. Methylation of mammalian deoxyribonucleic acid. 3. Terminal versus internal location of 5-methylcytosine in oligodeoxyribonucleotides from Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1972 May 10;247(9):2872–2875. [PubMed] [Google Scholar]
- Tosi L., Granieri A., Scarano E. Enzymatic DNA modifications in isolated nuclei from developing sea urchin embryos. Exp Cell Res. 1972 May;72(1):257–264. doi: 10.1016/0014-4827(72)90588-5. [DOI] [PubMed] [Google Scholar]
- Wildenauer D., Gross H. J. Methyldeficient mammalian 4s RNA: evidence for L-ethionine-induced inhibition of N6-dimethyladenosine synthesis in rat liver tRNA. Nucleic Acids Res. 1974 Feb;1(2):279–288. doi: 10.1093/nar/1.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]



