Abstract
The mechanism by which renal cell carcinoma (RCC) colonizes the lung microenvironment during metastasis remains largely unknown. To investigate this process, we grafted human RCC cells with varying lung metastatic potential in mice. Gene expression profiling of the mouse lung stromal compartment revealed a signature enriched for neutrophil-specific functions that was induced preferentially by poorly metastatic cells. Analysis of the gene expression signatures of tumor cell lines showed an inverse correlation between metastatic activity and the levels of a number of chemokines, including CXCL5 and IL8. Enforced depletion of CXCL5 and IL8 in these cell lines enabled us to establish a functional link between lung neutrophil infiltration, secretion of chemokines by cancer cells, and metastatic activity. We further show that human neutrophils display a higher cytotoxic activity against poorly metastatic cells compared to highly metastatic cells. Together, these results support a model in which neutrophils recruited to the lung by tumor-secreted chemokines build an antimetastatic barrier with loss of neutrophil chemokines in tumor cells acting as a critical rate-limiting step during lung metastatic seeding.
INTRODUCTION
Metastatic RCC is one of the most lethal cancers, with a median survival of less than 12 months in the absence of effective therapy. In the United States, kidney cancer is the 7th most common malignancy among men and 12th among women. A third of the patients presents with metastatic disease with a median survival of 7–11 months and a 5-year survival of 0–10%. (1, 2). There has been no significant improvement in the mortality rate of RCC reflecting the relative ineffectiveness of currently available therapies for metastatic disease.
The majority of carcinomas are composed of neoplastic epithelial cells immersed in a dynamic framework of fibroblasts, adipocytes, and hematopoietic and endothelial cells. Studies from several laboratories over the years support the hypothesis that tumor microenvironment influences tumor initiation and progression (3, 4). Moreover, characterization of molecular signatures of tumor stroma has yielded information of prognostic value (5, 6). Hematopoietic-derived cells are highly represented in tumors. Although it was initially believed that immune cells are involved in antitumor immunity, subsequent clinical and experimental evidence showed that in many cases they enhance tumor progression to malignancy (7). A variety of studies have highlighted the paradoxical role of the immune system during tumor progression. Whereas acute activation of immune cells in response to tumor might result in the eradication of malignant cells, the latter can also escape immune responses and polarize the immune components toward a tumor promoting phenotype (7). Infiltration of tumors by hematopoietic cells involves a variety of cell types, including macrophages (TAMs) (8), dendritic cells (DCs) (9), myeloid-derived suppressor cell (MDSCs) (10), neutrophils (11), mast cells (12), T cells, and B cells (7). A wealth of studies has demonstrated that interactions between tumor and stroma are mediated by a large number of cytokines, chemokines, and growth factors (3).
The establishment of metastasis depends on the ability of cancer cells to subvert tissue homeostasis and reproduce a secondary tumor in a distant site through the deregulation of genetic networks in both stromal and tumor compartments. Metastasis is a multistep process in which each step by itself may be inefficient. At each stage of metastatic progression, local microenvironment plays a key role and the interaction between tumor and stromal cells determines the fate of the metastatic cell. The concept that the local tissue microenvironment or metastatic “niche” is important was first proposed more than a century ago by Stephen Paget in his ‘seed and soil’ hypothesis to explain the non-random pattern of metastasis (13). According to this hypothesis, the unique characteristics of the local microenvironment dictate the affinity and compatibility of the tumor cell for a specific metastatic site. Recently, the work of several laboratories has provided experimental support for this model. Studies involving the selection of tumor variants with specific tissue tropisms and transcriptome analysis of such variants have shed some light on the cellular and molecular mechanisms of tumor-stromal interactions at the metastatic site (14, 15). In recent years, evidence has emerged to show that growth factors secreted by the primary tumor prime certain tissues for tumor cell engraftment (16). In response to tumor-secreted products, several hematopoietic cell types are recruited to the “pre-metastatic niches”, creating a microenvironment that potentiates tumor growth.
Perturbations of cell-cell interactions are a key feature of tumor progression and metastatic spread. However, systematic changes in tumor microenvironment during metastatic progression are largely unexplored. In this study, we have used a chimeric mouse model composed of human tumor cells and mouse stromal cells to uncover the molecular and cellular interactions associated with RCC metastasis to the lung. By using species-specific profiling we were able to uncover changes specifically associated with the stromal lung compartment during metastasis. Analysis of the gene expression signatures preferentially regulated by poorly metastatic cells revealed a gene expression pattern typical of neutrophils. Finally, by associating the modulated stromal functions with the gene expression changes in tumor cells during metastatic conversion we were able to characterize the paracrine networks that underlie cellular changes during lung colonization of RCC.
RESULTS
Metastatic activity of RCC cells to the lung inversely correlates with neutrophil infiltration
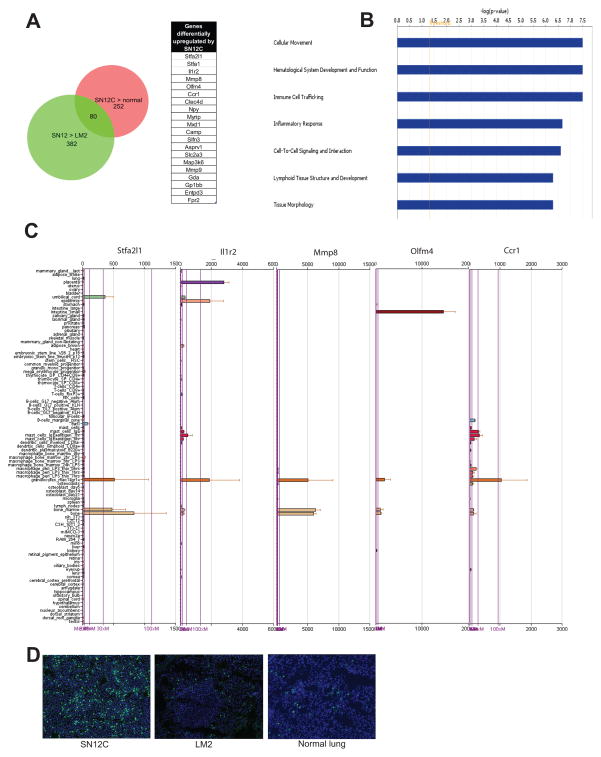
To investigate the mechanisms of lung colonization by tumor cells during RCC tumorigenesis, we used a previously described RCC tumor xenograft model (17, 18). The SN12C cell line is poorly metastatic and the derived variant LM2 cell line shows a profound enhancement of metastatic activity to the lung. To characterize the host changes associated with metastatic progression, we isolated RNA from lungs of mice injected orthotopically with either SN12C or LM2 cells and hybridized to mouse oligonucleotide arrays. To analyze the gene expression changes induced in the mouse tissues we exploited species differences between the interacting components of the system. To control for potential interspecies cross hybridization we subjected the human tumor cell line RNAs to the mouse array to identify probes that hybridized to the human sequences. The probes on the mouse array that hybridized with strong signals to human tumor RNA were eliminated from further analysis. Expression profiles of normal mouse lung comprised the controls. Using this approach, we identified 252 unique genes that were highly expressed in the lung stromal compartment of mice injected with SN12C cells in comparison to lungs of normal mice. Class comparison between differentially expressed genes in lung stroma of mice injected with either SN12C or LM2, followed by cross referencing with the 252 unique mouse genes obtained from the above analysis, identified 80 genes that were preferentially induced by SN12C cells in the lung stroma (Fig. 1A). This set of genes was then subjected to Ingenuity Pathway Analysis (www.ingenuity.com) to broadly characterize the cellular and molecular processes altered during tumor progression in the lung. This analysis revealed the deregulation of several functional processes related to inflammatory response, with high enrichment for functions associated with neutrophil activation (Fig. 1B, Table S1). To investigate this further, we used the Novartis gene expression atlas (19) that allows the determination of tissue-specific pattern of mRNA expression, and found that the vast majority of stroma-upregulated genes were almost exclusively expressed by granulocytes or tissues rich in granulocytes (Fig. 1C). The microarray results were validated by immunohistochemical staining using neutrophil-specific anti-Ly-6G antibody, which showed that mice injected with SN12C cells displayed a significantly higher number of neutrophils in the lung than mice injected with LM2 cells (Fig. 1D). To investigate whether tumor cells also induced recruitment of neutrophils in a paracrine fashion upon seeding the lung, we injected SN12C and LM2 cell lines into the lung through the tail vein and monitored neutrophil infiltration after 3 days. As expected, we observed that only lungs of mice injected with SN12C cells displayed a robust infiltration of neutrophils (Fig. S1). Globally, these results suggest that non-metastatic RCC cells promote recruitment of neutrophils to the lung through an endocrine and paracrine mechanism, and metastatic activity inversely correlates with the ability of tumor cells to recruit and activate these immune cells.
Figure 1. Poorly metastatic cells induce lung infiltration of neutrophils.
Groups of 5 mice were injected in the kidney subcapsule with either SN12C or LM2 cells. After 2 months, mice were sacrificed and lungs were dissected and gene the expression profile was analyzed. (A) Venn analysis showing the gene expression changes induced by SN12C in the lung stromal compartment. The list shows the genes that were significantly upregulated by SN12C in the lung stroma in comparison with normal mouse lung and lung stroma of mice injected with LM2 cells. (B) Ingenuity pathway analysis showing the top 7 canonical pathways based on significance. (C) Tissue expression pattern of the indicated genes (Top 5 in Fig. 1A) in mouse tissues using the Novartis gene expression atlas revealed expression in granulocytes or tissues rich in granulocytes (Bone marrow, Placenta). (D) Immunofluorescence staining of Gr1 cells in representative lung sections of mice injected with the indicated tumor cells.
RCC cells lose neutrophil chemokines during metastatic conversion
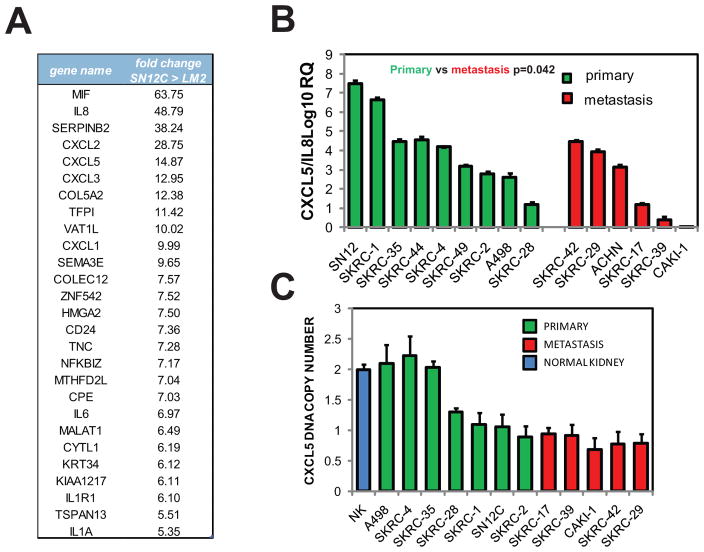
Gene expression analysis of SN12C and LM2 cells by microarray uncovered a chemokine expression signature that was highly enriched in the SN12C cell line. Several neutrophil chemokines, including CXCL1, CXCL2, CXCL3, CXCL5, and IL8 were downmodulated in the highly metastatic LM2 cells in comparison with the poorly metastatic SN12C cells, suggesting a link between chemokine expression and metastatic activity (Fig. 2A). We selected CXCL5 and IL8 for further analysis because they were among the top ranked downmodulated genes in the highly metastatic LM2 cells. To confirm and extend those observations we examined a panel of established RCC cell lines with distinct metastatic behavior (20). Q-PCR analysis indicated that cell lines derived from primary RCC tumors displayed significantly higher levels of combined CXCL5 and IL8 expression compared to cell lines derived from metastatic lesions (Fig 2B). We also analyzed the expression of CXCL5 and IL8 individually in the same panel of cell lines and found a reduced expression in the group of metastatic cell lines relative to the group of primary cell lines (Fig S2). To enrich tumor cells Tan et al. performed short-term cell culture of fresh RCC tumor biopsies and subjected them to gene expression analysis (21). Analysis of the differentially expressed genes in this dataset revealed that the expression of several CXCL chemokines was downmodulated in the metastatic cells (Fig. S3). To explore the possible genomic basis for the observed chemokine cluster downregulation, we subjected the DNA isolated from the established RCC cell lines investigated above to Taqman copy number assay. This assay revealed a substantial loss of DNA at the CXCL locus in all the metastasis-derived cell lines, while a significant percentage of cell lines that originated from primary RCC tumors (3/7) retained 2 copies (Fig. 2C). Since no homozygous deletion of the CXCL loci was detected in any of the analyses, we considered the possibility that the retained CXCL alleles may be silenced by methylation of their promoters. However, quantitation of CpG methylation using EpiTYPER (Sequenom) mass-spectrometry analysis of bisulfite-converted DNA at these loci did not identify negative for methylation changes (data not shown).
Figure 2. CXCL chemokines expression decreased during RCC metastatic progression.
(A) Genes showing 5-fold or greater alteration in expression in the SN12C cell line compared to the LM2 cell line. (B) Expression of IL8 + CXCL5 in a panel of RCC cell lines isolated from primary or metastatic tumors determined by Q-PCR. Error bars represent STDEV. P-values were obtained using Student’s t-test of comparing primary cells versus metastatic cells. (C) Comparison of CXCL5 DNA copy number in primary and metastatic cell lines using TaqMan copy number assay
These results together suggest that the metastatic microenvironment imposes a selection pressure on tumor cells toward a less immunogenic phenotype by downregulation of several neutrophil chemokines, as a result of a deletion of the physical loci of the encoding genes.
CXCL5 and IL8 regulate metastatic activity of RCC cells
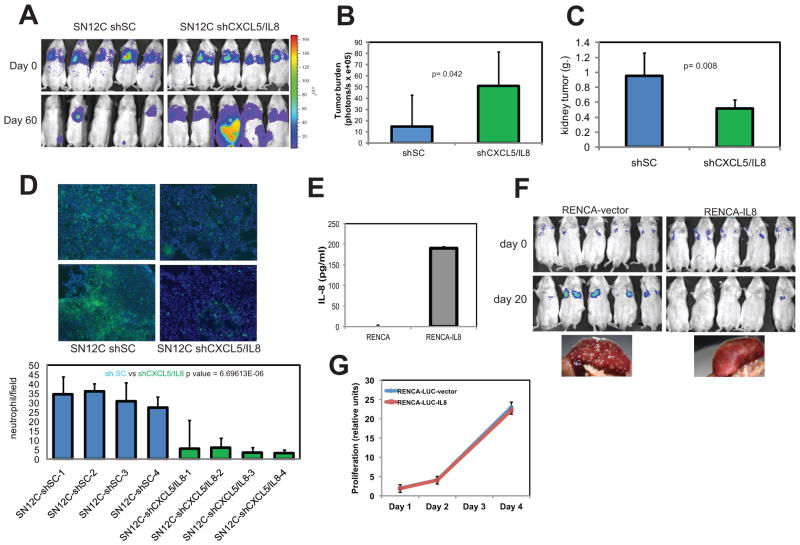
To demonstrate that neutrophil chemokines directly control tumor progression, we engineered the poorly metastatic SN12C cells to express reduced levels of CXCL5 and/or IL8 through shRNA knockdown (Fig. S4). We selected the above 2 chemokines because they were among the top ranked downmodulated genes in the highly metastatic LM2 cells. The delivery of SN12C cells with decreased levels of CXCL5 and IL8 (SN12C-shCXCL5/IL8) to the lungs of NOD/SCID mice consistently induced higher metastatic burden than SN12C control cells (SN12C-shSC) (Fig. 3A, B). Metastases originating from CXCL5 or IL8-depleted SN12C cells (SN12C-shCXCL5/IL8) developed slowly and were only visible after 3 months of implantation (Fig. S5). Thus, concurrent depletion of 2 chemokines had a synergistic effect on SN12C metastatic activity suggesting that depletion of additional chemokines may further boost this activity. To study the effect of loss of CXCL5 and IL8 on primary tumor formation, SN12C-shSC and SN12C-shCXCL5/IL8 cell lines were injected in the kidney sub-capsule of 5 mice. At sacrifice, mice injected with SN12C cells lacking CXCL5 and IL8 showed a significant decreased in growth rate when compared with wild type SN12C cells (Fig. 3C). These findings indicate that the role of tumor-secreted chemokines depends on the anatomical site where the tumor arises. To establish a link between chemokine depletion and neutrophil activity, we injected SN12C or SN12C-shCXCL5/IL8 cells in the kidney sub-capsule and analyzed the degree of neutrophil infiltration in the lung after 2 months. Lungs of mice transplanted with SN12C cells showed significantly higher number of neutrophils than those of mice injected with SN12C-shCXCL5/IL8 cells (Fig. 3D). To exclude the potential confounding effects of the skewed immune reactions in immunocompromised mice, we validated the above results in the RENCA kidney cancer syngeneic transplantation model using immunocompetent mice (22). The pattern of growth of this tumor accurately mimics that of adult human RCC, particularly with regard to spontaneous metastasis to lung and liver. We therefore examined the consequence of introducing tumor cells engineered to secrete IL8 (RENCA-IL8) (Fig. 3E), in Balb/cCr mice. When RENCA-vector cells were injected in mice through the tail vein, they colonized the lung quite efficiently. However, when RENCA-IL8 cells were delivered to the lungs of mice via tail vein, they were strongly rejected (Fig. 3F). As expected, RENCA-IL8 grew at approximately the same rate as the wild type RENCA when grown in vitro or injected orthotopically (Fig. 4G, S5C), suggesting that the IL8 effect is metastasis-specific. In summary, these results suggest that CXCL5 and IL8 expression decrease the competence of RCC cells for lung metastasis.
Fig. 3. CXCL5 and IL8 inhibit lung metastasis.
SN12C-LUC cells were infected with lentivirus encoding a control hairpin, or with shRNAs targeting CXCL5/IL8 (A) 1×106 cells of control or combined knockdown samples were injected I.V. in groups of NOD/SCID mice. After 3 months luciferase activity of whole animals was measured using an IVS system. (B) Lung colonization was measured by bioluminescence, quantified and normalized. Error bars represent STDEV. P-values were obtained using Student’s t-test. (C) The indicated cell populations of SN12C-LUC-derived cells were injected in the kidney subcapsule. After 2 months mice were sacrificed and kidney tumor weight was assessed. Error bars represent STDEV. P-values were obtained using Student’s t-test. (D) Immunofluorescence staining of Gr1 positive cells in representative lung sections of mice injected with the indicated tumor cells. The graph indicates the number of neutrophils per field of mouse lung and the right panels show representative pictures. (E) RENCA cells were transduced with lentiviruses carrying the empty vector or IL8. IL-8 expression of RENCA cells transduced with a vector encoding IL-8. (F) BALB/c mice were injected I.V. with RECA-vector or RENCA-IL8 cell lines and monitored for metastatic growth at the indicated times by bioluminescence imaging using an IVS system. (G) RENCA-vector and RENCA-IL8 were plated in complete medium under sparse conditions and counted at the indicated times.
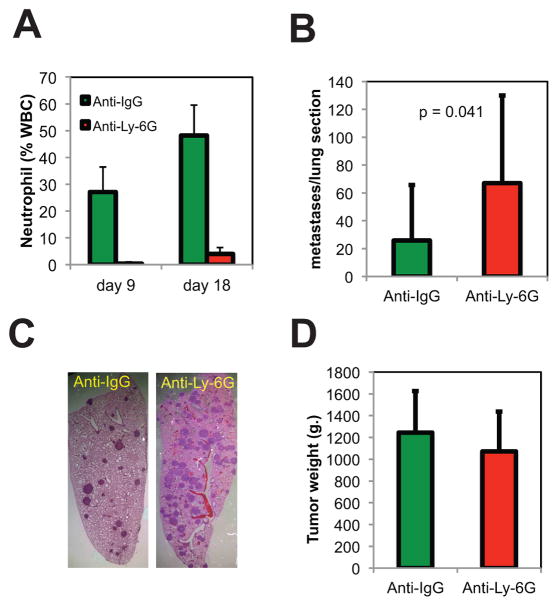
Fig. 4. Neutrophils inhibit lung metastasis in a syngeneic transplantation model of kidney cancer.
BALB/c mice were injected in the renal subcapsule with the RENCA cell line (N=20). After 2 days mice were randomized into two groups, receiving either anti-IgG antibody (n=10) or negative control anti-Ly-6G (n=10) (50 μg/mouse) administered by intraperitoneal injection (IP) daily during 16 days. (A) Percentage of neutrophils in blood of animals treated with the indicated antibodies. (B) Average number of tumor colonies per tumor section. Error bars represent STDEV. P-values were obtained using Student’s t-test. (C) Photomicrograph of H & E-stained, 3 μm formalin-fixed lung tissue. (D) Primary tumor growth in the indicated groups of mice.
Neutrophils attenuate the formation of lung metastasis in a syngeneic mouse model of kidney cancer
To directly demonstrate the anti-metastatic action of neutrophiles, RENCA cells were orthotopically inoculated in mice and treated either with the specific neutrophil depleting antibody anti-Ly-6G (1A8) or control antibody anti-IgG. Anti-Ly-6G antibody efficiently depleted circulating neutrophils even after 16 days of treatment (Fig. 4A). Histological examination of the lungs of neutrophil-depleted tumor bearing mice revealed an increased rate of metastatic colonization when compared with control mice (Fig. 4B, C). Importantly, neutrophil depletion did not impact the growth of tumor cells at the primary site (Fig. 4D). Similar results were obtained when neutrophils were depleted using an anti-GR-1 antibody (clone RB6-8C) (Fig. S6). All these results together indicate that neutrophils recruited to the lung attenuate tumor growth.
Poorly metastatic cells activate human neutrophils in vitro
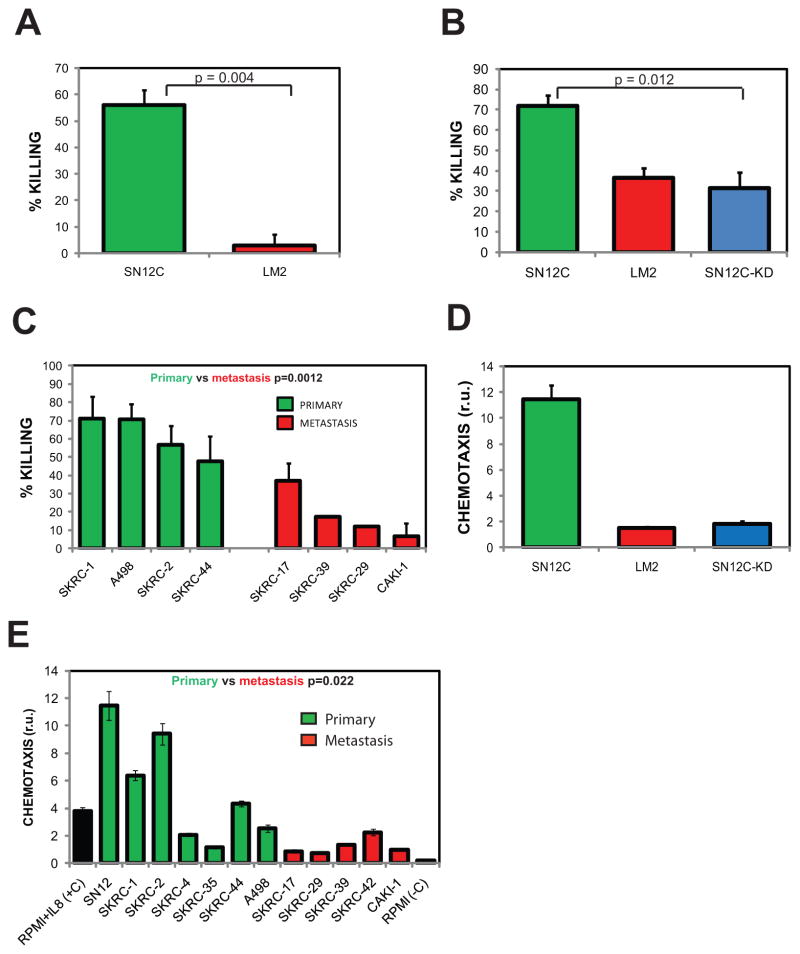
To demonstrate that neutrophils preferentially kill poorly metastatic cells, normal blood neutrophils were co-cultured with SN12C or LM2 cells, and cytotoxicity toward tumor cells was assessed after 24 hours. At ratios of 20 neutrophils to each tumor cell, we found that SN12C cells were killed more efficiently than LM2 cells (50% vs 3%) (Fig. 5A). To investigate the contribution of CXCL5 and IL8 to tumor cell killing, we co-cultured neutrophils with SN12C or SN12C-shCXCL5/IL8 tumor cells. As expected, SN12C cells were killed more efficiently than LM2 cells and depletion of CXCL5/IL8 by shRNA in SN12C cells allowed cells to escape the cytotoxic effects of neutrophils (Fig. 5B). To generalize the results above we examined the neutrophil-mediated cytotoxicity toward a panel of established RCC cell lines isolated from various anatomical sites and found that cell lines isolated from the kidney tumor were more efficiently killed than cell lines isolated from various metastatic sites (Fig. 5C). We next evaluated the effect of chemokine-activated neutrophils on highly metastatic LM2 cells. As predicted, IL8 enhanced the cytotoxicity of neutrophils toward LM2 cells (Fig. S7A). As mentioned above, neutrophils do not inhibit the growth of SN12C in the primary site. One potential explanation could be the secretion of immunomodulatory cytokines such as TGF-β by the tumor cells. However, TGF-β expression did not correlate with the metastatic activity of tumor cells (Fig. S7B–D). Moreover, doping the killing assays with TGF-β did not block neutrophil cytotoxicity towards SN12C cells (Fig. S7A). It has been well documented that neutrophils migrate towards sites of infection or inflammation by chemotaxis, responding to chemotactic gradients of a large number of chemokines including IL8 and CXCL5. To determine if chemokines secreted by RCC tumor cells were able to modulate neutrophil chemotactic activity, serum-free and cell-free-conditioned medium from SN12C, LM2, and SN12C-shCXCL5/IL8 cell populations was collected and assayed in a Boyden chamber using human neutrophils. As showed in Fig. 5D, SN12C-conditioned medium induced significantly higher chemotactic activity than LM2-conditioned medium. In addition, conditioned medium from SN12C cells deficient in CXCL5 and IL8 induced neutrophil chemotaxis at a much lower rate. To validate and extend the previous results, we examined a panel of 12 established RCC cell lines, 5 isolated from metastatic lesions and 7 from primary kidney lesions. We found that conditioned medium from cell lines isolated from the primary sites generally induced higher chemotactic activity than conditioned medium from cultures of metastatic cell lines (Fig. 5E). We also monitored neutrophil activation by measuring the release of H2O2 of neutrophils co-cultured with tumor cells and found a correlation between metastatic activity and H2O2 (Fig. S8). Collectively, the above results indicate that blood neutrophils are mobilized and activated by tumor-secreted chemokines, acting as a critical metastatic barrier that metastatic cells overcome through the loss of expression of those chemokines.
Fig. 5. Poorly metastatic RCC cell lines activate the cytotoxic and migratory functions of human neutrophils.
Neutrophil were isolated from human buffy coats by density centrifugation. (A) (B) Indicated tumor cells and human neutrophils were co-cultured in the conditioned medium of the indicated tumor cell, at 20:1 neutrophil: tumor cell ratio. After 24 hours, the percentage of tumor cells killed by neutrophils was calculated. Conditioned medium was obtained by culturing the cell lines at confluence for 2 days and collecting the culture medium. (C) (D) Neutrophil chemotactic activity modulated by the conditioned medium of the indicated cells lines was performed as indicated in the methods section. (E) The indicated tumor cells were co-cultured with neutrophils at 20:1 neutrophil : tumor cell ratio. After 24 hrs, the percent tumor cells killed was calculated. Error bars represent STDEV. P-values were obtained using Student’s t-test.
DISCUSSION
In this study we have characterized some of the cellular and molecular interactions between RCC cells and the lung stromal microenvironment. We found that lung colonization by tumor cells is associated with a massive infiltration of neutrophils and the extent of this infiltration correlated with metastatic activity. Finally, analysis of the genes deregulated during metastatic conversion in tumor cells uncovered a paracrine network that controls the recruitment of neutrophils to the lung.
Tumorigenesis involves sequential changes in the tumor cells and in an entire variety of surrounding stromal cell types. In order to obtain a comprehensive view of those changes we analyzed the expression signatures of lung metastases and stromal cells exploiting the species differences between the tumor and the host by using species-specific microarrays. Genome-wide analysis of the tumor microenvironment during cancer progression has been limited to date. Pioneering studies by Allinen and colleagues identified a set of 417 genes expressed specifically in the most abundant cell types in breast cancer (5). More recently, Finak and colleagues obtained expression signatures from both epithelial and stromal compartments from the same tumor biopsy via laser capture microdissection (23). The same type of analysis performed in prostate cancer identified 544 unique genes higher in expression and 606 genes lower in expression in the reactive stroma, when compared with normal stroma (24). Similar studies dissecting the molecular pathways deregulated in the metastatic stroma are lacking.
By comparing the gene expression changes induced in the lung by SN12C or LM2 tumors we uncovered a molecular fingerprint typically associated with neutrophils preferentially induced by SN12C cells. Historically, neutrophils have been considered as agents of host defense against cancer. However, most of the recent clinical studies reported a tight correlation between tumor-associated neutrophils and poor clinical outcome, including decreased survival (25–27). According to these results, recruitment of neutrophils could be driven by the tumor, and does not necessarily represent a means of host defense. In opposition to this view, some studies demonstrated that neutrophils show tumoricidal activity against tumors engineered to overexpress some cytokines or chemokines (28, 29). Neutrophils infiltrate early melanomas to inhibit growth (30). Recently, studies by Fridlender and colleagues supported a model where neutrophils can be alternatively activated. They proposed that neutrophils have 2 different polarization states: the N1 (highly cytotoxic) and the N2 (tumor growth promoting). Interestingly, their data also suggest that TGF-β within the tumor microenvironment induces a population of neutrophils with a pro-tumor phenotype. However, treatment of mice with a TGF-β inhibitor caused massive infiltration of cytotoxic neutrophils and blunted tumor growth. (11). Our results are consistent with a scenario where neutrophil activation has an anti-metastatic activity at least in the lung while it has no effect on the growth of the primary tumor, most likely due to the immunosuppressive microenvironment in the bulky tumor. In contrast to our results, Kowanetz et al. recently suggested that metastatic breast cancer tumors overexpressing granulocyte-colony stimulating factor (G-CSF) mobilize Ly6G+Ly6C+ granulocytes and facilitate their subsequent homing at distant organs even before the arrival of tumor cells, promoting lung metastasis (31). This discrepancy between the two results could be explained in part by the different tumor models under study. In addition, the differing nature of the paracrine networks involved in the two tumor models could also lead to the recruitment of granulocytes with contrasting properties that would modulate metastatic growth in disparate ways. During the preparation of this manuscript Granot et al. reported, that tumor-entrained neutrophils attenuate lung metastasis in a model of breast cancer (32). Chemokine networks are central to the movement of cells in both homeostatic and pathological processes. Most cancers express an extensive array of chemokines and chemokine receptors (33). Chemokines have pleiotropic effects; therefore, it is logical to presume that in cancer, chemokines will also have wide-ranging functions. Chemokines have been shown to play pro-tumorigenic roles, including growth promotion, angiogenesis, manipulation of the local immune environment, cancer cell invasion, and metastasis. On the other hand, chemokines also have been shown to display anti-cancer actions. However, studies showing the anti-cancer effects of chemokines are very limited. For example CXCL2 has been show to have anti-cancer properties in pancreatic cancer and exert this effect through the recruitment of cytotoxic macrophages (34). CXCL9 induced by interferon-gamma secreted by tumor cells is critical for T-cell–mediated suppression of cutaneous tumors (35). In RCC, tumors expressing CXCL4, CXCL5, CXCL9, and CXCL10 did not recur after surgery while intratumoral expression of CXCL10, CXCL9 and CCL4 correlated positively with CD8+ T cells (36).
The role of neutrophil chemokines such as IL8 and CXCL5 on tumor biology is controversial. This controversy can probably be explained in part by their multiple biological functions. Several of those functions appear to involve pro-tumorigenic activity. IL8 has been shown to induce tumor growth, migration, angiogenesis, and metastasis in some tumor models (37–39). In contrast, Lee et al. reported that IL8 has anti-tumorigenic effect on human ovarian cancer cells (40). Moreover, they also demonstrated that inhibition of tumor progression in this system is associated with a massive infiltration of neutrophils. Several lines of evidence further show that CXCL5 is an important factor in tumor biology. It has been reported to enhance endothelial cell proliferation and invasion in vitro (41), to promote tumor angiogenesis in vivo in non-small cell lung carcinoma, and to modulate tumor growth and metastasis (42). CXCL5 is overexpressed in gastric, prostate, endometrial, squamous cell, pancreatic, and colon cancers (43, 44). In opposition to the previous reports and in agreement with our results, Speetjens et al. have shown that CXCL5 blocks tumor progression in colorectal cancer (45).
Genetic studies characterizing the molecular mechanisms that disrupt gene expression during RCC progression have implicated a number of chromosomal loci in the biology of kidney cancer (46). However, relatively little is known about the genetic aberrations associated with the metastatic event. To our knowledge, our study showing that the CXCL1-8 locus is genetically inactivated during RCC metastatic progression, is the first to document the precise nature of genetic change that turn certain genes into suppressors of metastasis in this tumor type. Although we have not conducted an in depth analysis of the mechanisms by which the remaining CXCL1-8 alleles are inactivated we anticipate that other genetic or epigenetic events might be involved. Notably, some studies have shown that loss of 4q cytoband (harboring CXCL1-8 locus) is a common event in RCC progression and metastasis (47, 48).
In conclusion, our results show that neutrophil infiltration in the lung establishes an immunological barrier that impairs the growth of tumor cells. Our results also suggest that metastatic cells escape surveillance by those immune cells by losing the expression of a number of chemokines responsible for the chemo-attraction of neutrophils. This new paradigm could be exploited in the design or new therapeutic strategies for the treatment of metastatic RCC.
METHODS
Cell lines
The SN12C cell line was established in culture from a primary RCC from a 43 year-old male and was described previously (49). LM2 cells were isolated from lung metastasis of SN12C cells in NOD/SCID mice and were also described previously (17). The human RCC cell lines ACHN, A498 and KRCY were obtained from Dr. Harry Drabkin (University of Colorado Health Science Center, Denver, CO) and cultured under standard conditions. The SK-RC RCC cell lines were obtained from the Ludwig Institute for Cancer Research at the Memorial Sloan-Kettering Cancer Center (MSKCC) (20) and maintained in Eagles minimal essential medium (MEM) supplemented with 10% fetal calf serum (20).
RNA isolation, labeling and microarray hybridization
RNA was isolated from tumor tissues and cell lines using the RNeasy minikit (Qiagen). Labeling and hybridization of the samples to mouse 430 2.0 Affymetrix gene expression array were performed by the MSKCC Genomics Core Facility using standard methodology. For Human gene expression profiling HG-133 Plus 2.0 array was used (Affymetrix). Background subtraction, normalization, and log2 transformation of the array data were performed using the Robust Multi-Array Average (29) method within the Bioconductor for R software package. Significantly differentially expressed genes were identified using the Significance Analysis of Microarrays algorithm (50). Microarray data have been deposited in NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=dlqdhkwgeqwyyna&acc=GSE29688)
Immunofluorescence
For immunofluorescence studies, mouse lungs were perfused with a 1:1 mixture of PBS/OCT postexcision before embedding in OCT (Tissue-Tek). To detect neutrophils we used anti-Ly-6G antibody (eBioscience). 10-μm-thick frozen sections were fixed in acetone, preincubated with 1X PNB-blocking buffer, and incubated with the primary antibody of interest overnight at 4°C.
Analyisis of mRNA expression
Total RNA (1 μg) was reverse transcribed and cDNA was used for quantitative PCR. Indicated Taqman gene expression assays (Applied Biosystems) and the Taqman universal PCR master mix (Applied Biosystems) were used to quantify expression. Quantitative expression data were acquired and analyzed using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems).
DNA isolation and Taqman copy number assays
DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). The relative change of CXCL5 copy number between normal and tumor pairs or different cell lines was determined by real-time polymerase chain reaction (RT-PCR) assay. The TaqMan assay (Applied Biosystems) used for CXCL5 was Hs02708942_cn. The target and reference genes (RNase P) were amplified using the ABI PRISM 7500 Sequence Detection System (Applied Biosystems). PCR was performed according the manufacturer’s recommendations. For all PCR assays, copy number was established using the copycaller software (Applied Biosystems).
Gene knockdown and overexpression in cell lines
For knockdown, the plasmid pLKO.1 encoding short hairpin RNA (shRNA) targeting CXCL5 and IL8 was obtained from Open Biosystems. Viral supernatants were generated by transfecting 293-FT cells with the shRNA constructs in combination with the packaging vectors pVSVG and pDR2. The efficiency of the knockdown and overexpression was confirmed by quantitative PCR TaqMan gene expression assays (Applied Biosystems). For bioluminescence tracking, cell lines were retrovirally infected with a plasmid encoding firefly luciferase (MSCV Luciferase PGK Hygro) developed in Scott Lowe’s laboratory (Cold Spring Harbor Laboratory, Cold spring Harbor, NY 11724) (Addgene plasmid 18782).
Supplementary Material
Acknowledgments
This study was supported by research grants from the National Cancer Institute (CA-121327), the V-Foundation, and a Syms kidney cancer award. We thank Marco Seandel and Eric Pamer for critical reading of the manuscript. The MSKCC Genomics Core Facility provided expert assistance in performing the gene expression profiling assays. Author R. Chaganti is a Board member and a paid consultant of Cancer Genetics, Inc., Rutherford, NJ.
References
- 1.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005 Dec 8;353(23):2477–90. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 2.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007 Jan 11;356(2):115–24. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 3.McAllister SS, Weinberg RA. Tumor-host interactions: a far-reaching relationship. J Clin Oncol. 2010 Sep 10;28(26):4022–8. doi: 10.1200/JCO.2010.28.4257. [DOI] [PubMed] [Google Scholar]
- 4.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009 Apr;9(4):239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004 Jul;6(1):17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004 Feb;2(2):E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006 Jan;6(1):24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 8.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010 Apr;22(2):231–7. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Talmadge JE, Donkor M, Scholar E. Inflammatory cell infiltration of tumors: Jekyll or Hyde. Cancer Metastasis Rev. 2007 Dec;26(3–4):373–400. doi: 10.1007/s10555-007-9072-0. [DOI] [PubMed] [Google Scholar]
- 10.Ostrand-Rosenberg S. Immune surveillance: a balance between protumor and antitumor immunity. Curr Opin Genet Dev. 2008 Feb;18(1):11–8. doi: 10.1016/j.gde.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 2009 Sep 8;16(3):183–94. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008 Aug;8(8):618–31. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 13.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989 Aug;8(2):98–101. [PubMed] [Google Scholar]
- 14.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003 Jun;3(6):453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007 May;8(5):341–52. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005 Dec 8;438(7069):820–7. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Lago MA, Thodima VJ, Guttapalli A, Chan TA, Heguy A, Molina AM, et al. Genomic Deregulation during Metastasis of Renal Cell Carcinoma Implements a Myofibroblast-Like Program of Gene Expression. Cancer Res. 2010 Oct 15; doi: 10.1158/0008-5472.CAN-10-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naito S, Walker SM, Fidler IJ. In vivo selection of human renal cell carcinoma cells with high metastatic potential in nude mice. Clin Exp Metastasis. 1989 Jul-Aug;7(4):381–9. doi: 10.1007/BF01753659. [DOI] [PubMed] [Google Scholar]
- 19.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebert T, Bander NH, Finstad CL, Ramsawak RD, Old LJ. Establishment and characterization of human renal cancer and normal kidney cell lines. Cancer Res. 1990 Sep 1;50(17):5531–6. [PubMed] [Google Scholar]
- 21.Tan X, Zhai Y, Chang W, Hou J, He S, Lin L, et al. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer. 2008 Sep 1;123(5):1080–8. doi: 10.1002/ijc.23637. [DOI] [PubMed] [Google Scholar]
- 22.Murphy GP, Hrushesky WJ. A murine renal cell carcinoma. J Natl Cancer Inst. 1973 Apr;50(4):1013–25. doi: 10.1093/jnci/50.4.1013. [DOI] [PubMed] [Google Scholar]
- 23.Finak G, Sadekova S, Pepin F, Hallett M, Meterissian S, Halwani F, et al. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast Cancer Res. 2006;8(5):R58. doi: 10.1186/bcr1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dakhova O, Ozen M, Creighton CJ, Li R, Ayala G, Rowley D, et al. Global gene expression analysis of reactive stroma in prostate cancer. Clin Cancer Res. 2009 Jun 15;15(12):3979–89. doi: 10.1158/1078-0432.CCR-08-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen HK, Donskov F, Marcussen N, Nordsmark M, Lundbeck F, von der Maase H. Presence of intratumoral neutrophils is an independent prognostic factor in localized renal cell carcinoma. J Clin Oncol. 2009 Oct 1;27(28):4709–17. doi: 10.1200/JCO.2008.18.9498. [DOI] [PubMed] [Google Scholar]
- 26.Bellocq A, Antoine M, Flahault A, Philippe C, Crestani B, Bernaudin JF, et al. Neutrophil alveolitis in bronchioloalveolar carcinoma: induction by tumor-derived interleukin-8 and relation to clinical outcome. Am J Pathol. 1998 Jan;152(1):83–92. [PMC free article] [PubMed] [Google Scholar]
- 27.Foekens JA, Ries C, Look MP, Gippner-Steppert C, Klijn JG, Jochum M. The prognostic value of polymorphonuclear leukocyte elastase in patients with primary breast cancer. Cancer Res. 2003 Jan 15;63(2):337–41. [PubMed] [Google Scholar]
- 28.Di Carlo E, Forni G, Lollini P, Colombo MP, Modesti A, Musiani P. The intriguing role of polymorphonuclear neutrophils in antitumor reactions. Blood. 2001 Jan 15;97(2):339–45. doi: 10.1182/blood.v97.2.339. [DOI] [PubMed] [Google Scholar]
- 29.Buonocore S, Haddou NO, Moore F, Florquin S, Paulart F, Heirman C, et al. Neutrophil-dependent tumor rejection and priming of tumoricidal CD8+ T cell response induced by dendritic cells overexpressing CD95L. J Leukoc Biol. 2008 Sep;84(3):713–20. doi: 10.1189/jlb.0108075. [DOI] [PubMed] [Google Scholar]
- 30.Schaider H, Oka M, Bogenrieder T, Nesbit M, Satyamoorthy K, Berking C, et al. Differential response of primary and metastatic melanomas to neutrophils attracted by IL-8. Int J Cancer. 2003 Jan 20;103(3):335–43. doi: 10.1002/ijc.10775. [DOI] [PubMed] [Google Scholar]
- 31.Kowanetz M, Wu X, Lee J, Tan M, Hagenbeek T, Qu X, et al. Granulocyte-colony stimulating factor promotes lung metastasis through mobilization of Ly6G+Ly6C+ granulocytes. Proc Natl Acad Sci U S A. 2010 Dec 14;107(50):21248–55. doi: 10.1073/pnas.1015855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granot Z, Henke E, Comen EA, King TA, Norton L, Benezra R. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell. 2011 Sep 13;20(3):300–14. doi: 10.1016/j.ccr.2011.08.012. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004 Jul;4(7):540–50. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 34.Monti P, Leone BE, Marchesi F, Balzano G, Zerbi A, Scaltrini F, et al. The CC chemokine MCP-1/CCL2 in pancreatic cancer progression: regulation of expression and potential mechanisms of antimalignant activity. Cancer Res. 2003 Nov 1;63(21):7451–61. [PubMed] [Google Scholar]
- 35.Gorbachev AV, Kobayashi H, Kudo D, Tannenbaum CS, Finke JH, Shu S, et al. CXC chemokine ligand 9/monokine induced by IFN-gamma production by tumor cells is critical for T cell-mediated suppression of cutaneous tumors. J Immunol. 2007 Feb 15;178(4):2278–86. doi: 10.4049/jimmunol.178.4.2278. [DOI] [PubMed] [Google Scholar]
- 36.Kondo T, Ito F, Nakazawa H, Horita S, Osaka Y, Toma H. High expression of chemokine gene as a favorable prognostic factor in renal cell carcinoma. J Urol. 2004 Jun;171(6 Pt 1):2171–5. doi: 10.1097/01.ju.0000127726.25609.87. [DOI] [PubMed] [Google Scholar]
- 37.Schadendorf D, Moller A, Algermissen B, Worm M, Sticherling M, Czarnetzki BM. IL-8 produced by human malignant melanoma cells in vitro is an essential autocrine growth factor. J Immunol. 1993 Sep 1;151(5):2667–75. [PubMed] [Google Scholar]
- 38.Singh RK, Gutman M, Radinsky R, Bucana CD, Fidler IJ. Expression of interleukin 8 correlates with the metastatic potential of human melanoma cells in nude mice. Cancer Res. 1994 Jun 15;54(12):3242–7. [PubMed] [Google Scholar]
- 39.Arenberg DA, Kunkel SL, Polverini PJ, Glass M, Burdick MD, Strieter RM. Inhibition of interleukin-8 reduces tumorigenesis of human non-small cell lung cancer in SCID mice. J Clin Invest. 1996 Jun 15;97(12):2792–802. doi: 10.1172/JCI118734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee LF, Hellendall RP, Wang Y, Haskill JS, Mukaida N, Matsushima K, et al. IL-8 reduced tumorigenicity of human ovarian cancer in vivo due to neutrophil infiltration. J Immunol. 2000 Mar 1;164(5):2769–75. doi: 10.4049/jimmunol.164.5.2769. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi H, Numasaki M, Lotze MT, Sasaki H. Interleukin-17 enhances bFGF-, HGF-and VEGF-induced growth of vascular endothelial cells. Immunol Lett. 2005 May 15;98(2):189–93. doi: 10.1016/j.imlet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Arenberg DA, Keane MP, DiGiovine B, Kunkel SL, Morris SB, Xue YY, et al. Epithelial-neutrophil activating peptide (ENA-78) is an important angiogenic factor in non-small cell lung cancer. J Clin Invest. 1998 Aug 1;102(3):465–72. doi: 10.1172/JCI3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong YF, Cheung TH, Lo KW, Yim SF, Siu NS, Chan SC, et al. Identification of molecular markers and signaling pathway in endometrial cancer in Hong Kong Chinese women by genome-wide gene expression profiling. Oncogene. 2007 Mar 22;26(13):1971–82. doi: 10.1038/sj.onc.1209986. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki H, Patel V, Wang H, Edmunds RK, Gutkind JS, Yeudall WA. Down-regulation of CXCL5 inhibits squamous carcinogenesis. Cancer Res. 2006 Apr 15;66(8):4279–84. doi: 10.1158/0008-5472.CAN-05-4398. [DOI] [PubMed] [Google Scholar]
- 45.Speetjens FM, Kuppen PJ, Sandel MH, Menon AG, Burg D, van de Velde CJ, et al. Disrupted expression of CXCL5 in colorectal cancer is associated with rapid tumor formation in rats and poor prognosis in patients. Clin Cancer Res. 2008 Apr 15;14(8):2276–84. doi: 10.1158/1078-0432.CCR-07-4045. [DOI] [PubMed] [Google Scholar]
- 46.Linehan WM, Walther MM, Zbar B. The genetic basis of cancer of the kidney. J Urol. 2003 Dec;170(6 Pt 1):2163–72. doi: 10.1097/01.ju.0000096060.92397.ed. [DOI] [PubMed] [Google Scholar]
- 47.Jiang F, Desper R, Papadimitriou CH, Schaffer AA, Kallioniemi OP, Richter J, et al. Construction of evolutionary tree models for renal cell carcinoma from comparative genomic hybridization data. Cancer Res. 2000 Nov 15;60(22):6503–9. [PubMed] [Google Scholar]
- 48.Bissig H, Richter J, Desper R, Meier V, Schraml P, Schaffer AA, et al. Evaluation of the clonal relationship between primary and metastatic renal cell carcinoma by comparative genomic hybridization. Am J Pathol. 1999 Jul;155(1):267–74. doi: 10.1016/S0002-9440(10)65120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naito S, von Eschenbach AC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986 Aug;46(8):4109–15. [PubMed] [Google Scholar]
- 50.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):5116–21. doi: 10.1073/pnas.091062498. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.