Abstract
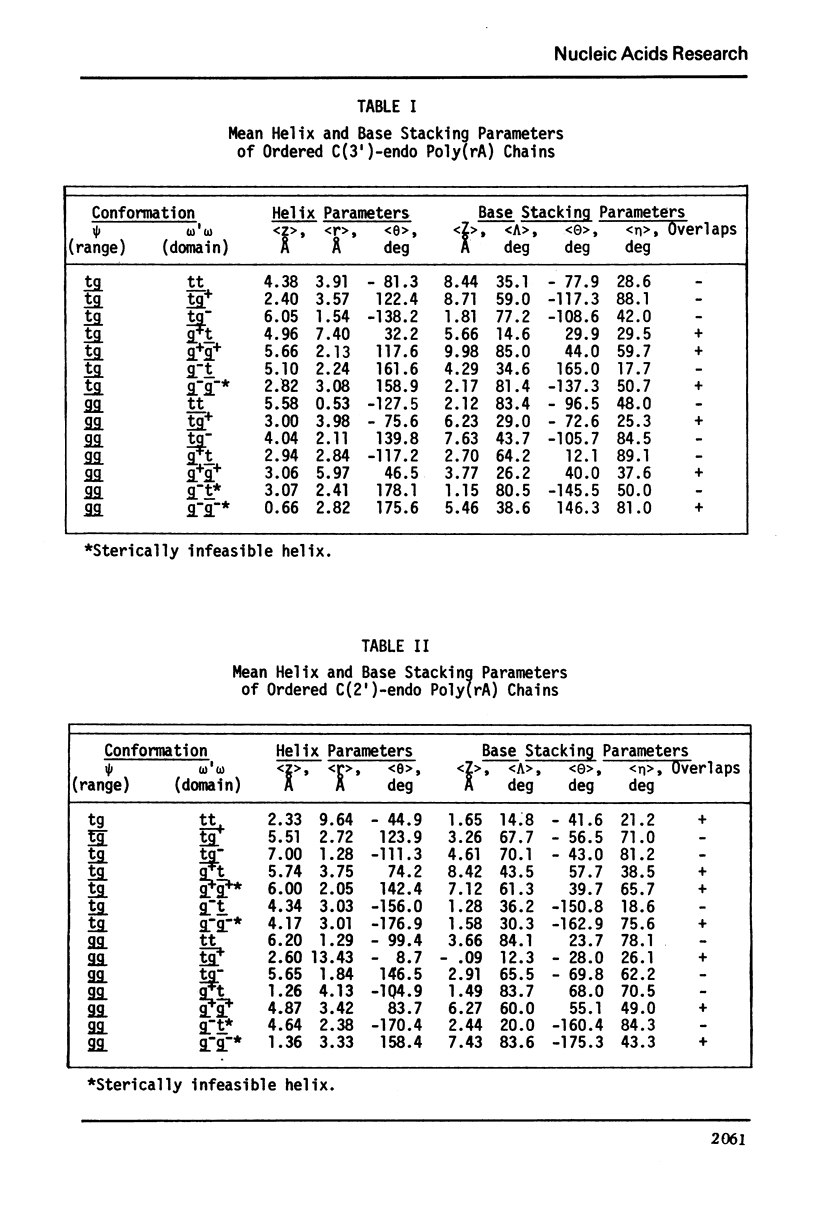
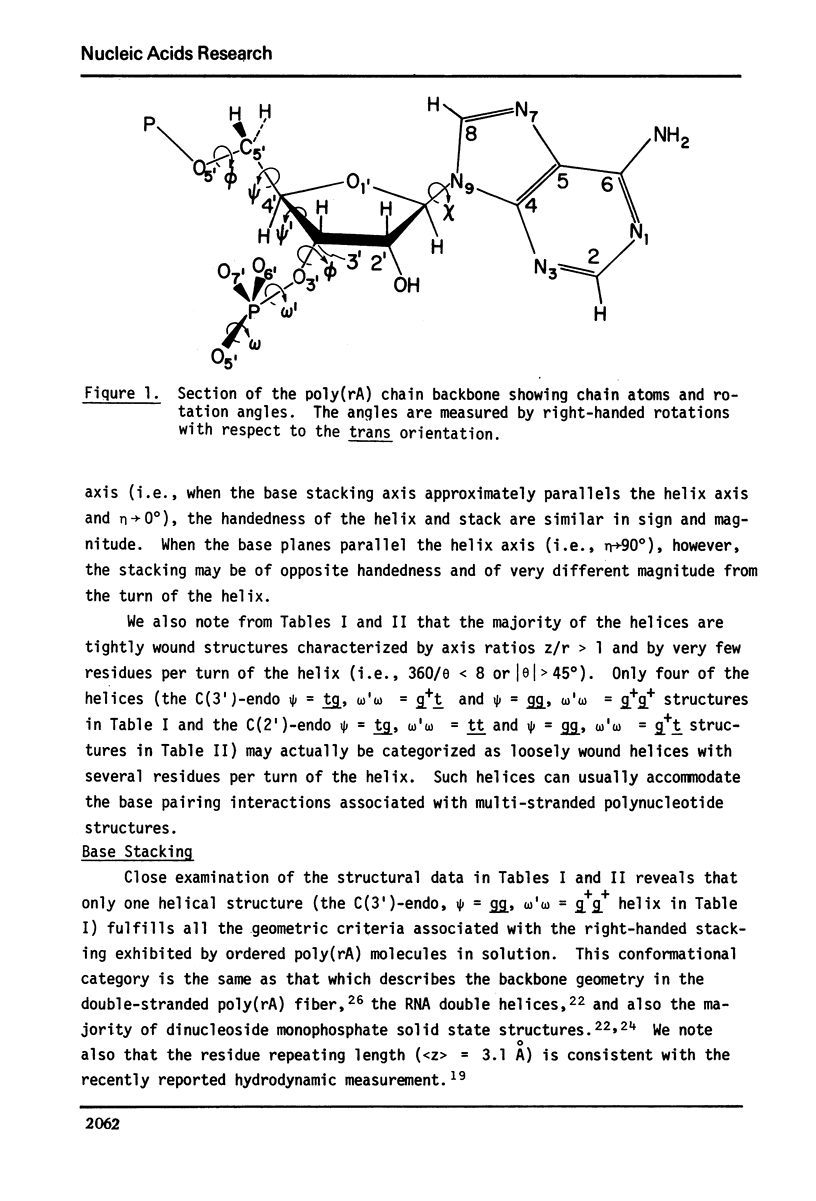
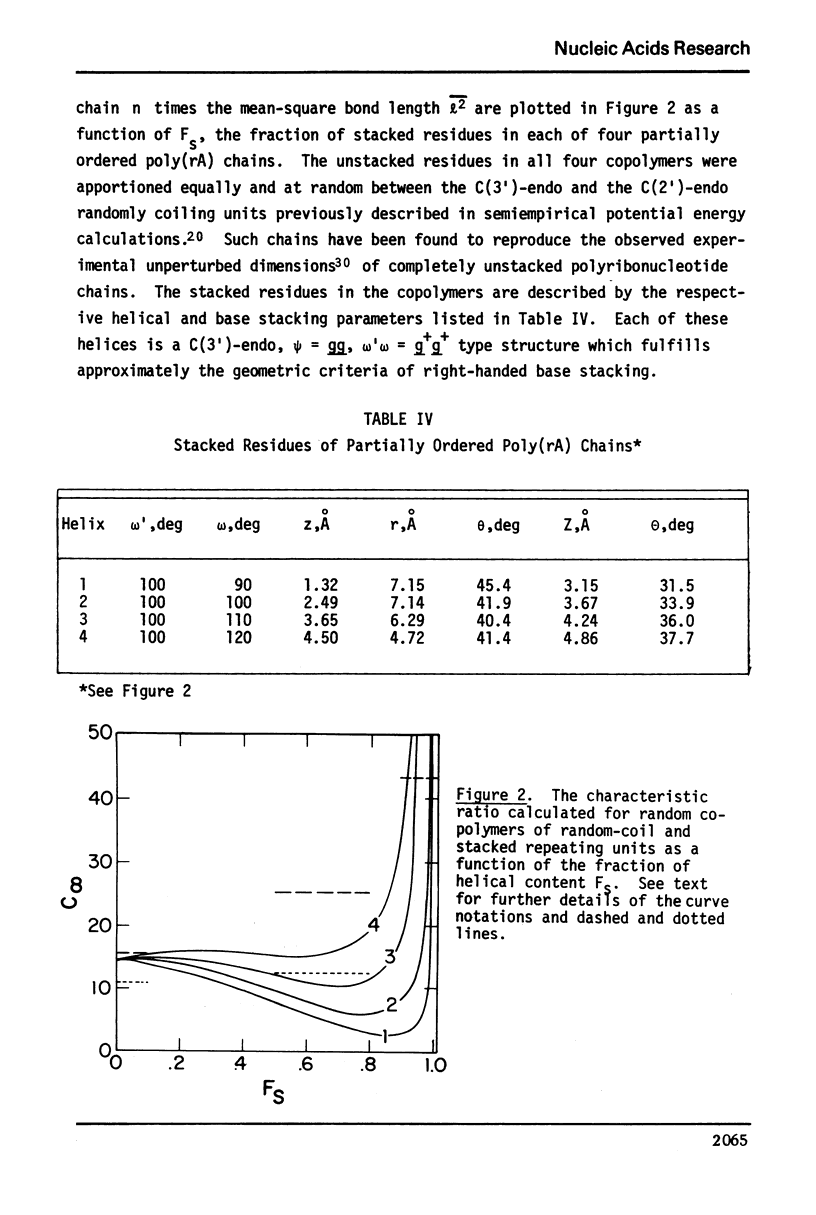
Approximate details of the spatial configuration of the ordered single-stranded poly(rA) molecule in dilute solution have been obtained in a combined theoretical analysis of base stacking and chain flexibility. Only those regularly repeating structures which fulfill the criterion of conformational flexibility (based upon all available experimental and theoretical evidence of preferred bond rotations) and which also exhibit the right-handed base stacking pattern observed in nmr investigations of poly(rA) are deemed suitable single-stranded helices. In addition, the helical geometry of the stacked structures is required to be consistent with the experimentally observed dimensions of both completely ordered and partially ordered poly(rA) chains. Only a single category of poly(rA) helices (very similar in all conformational details to the individual chains of the poly(rA) double-stranded X-ray structure) is thus obtained. Other conformationally feasible polynucleotide helices characterized simply by a parallel and overlapping base stacking arrangement are also discussed.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alderfer J. L., Tazawa I., Tazawa S., Ts'o P. Comparative studies on homopolymers of adenylic acid possessing different C-2' substituents of the furanose. Poly(deoxyriboadenylic acid), poly(riboadenylic acid),poly(2'-O-methyladenylic acid), and poly(2'-O-ethyladenylic acid). Biochemistry. 1974 Apr 9;13(8):1615–1622. doi: 10.1021/bi00705a010. [DOI] [PubMed] [Google Scholar]
- Brahms J., Michelson A. M., Van Holde K. E. Adenylate oligomers in single- and double-strand conformation. J Mol Biol. 1966 Feb;15(2):467–488. doi: 10.1016/s0022-2836(66)80122-5. [DOI] [PubMed] [Google Scholar]
- Bugg C. E., Thomas J. M., Sundaralingam M., Rao S. T. Stereochemistry of nucleic acids and their constituents. X. Solid-state base-stacking patterns in nucleic acid constituents and polynucleotides. Biopolymers. 1971;10(1):175–219. doi: 10.1002/bip.360100113. [DOI] [PubMed] [Google Scholar]
- Eisenberg H., Felsenfeld G. Studies of the temperature-dependent conformation and phase separation of polyriboadenylic acid solutions at neutral pH. J Mol Biol. 1967 Nov 28;30(1):17–37. doi: 10.1016/0022-2836(67)90240-9. [DOI] [PubMed] [Google Scholar]
- Evans F. E., Lee C. H., Sarma R. H. 300 MHz NMR study on the effect of base stacking on backbone conformational flexibility in oxy- and deoxy- adenyl dinucleosides. Biochem Biophys Res Commun. 1975 Mar 3;63(1):106–114. doi: 10.1016/s0006-291x(75)80017-9. [DOI] [PubMed] [Google Scholar]
- Evans F. E., Sarma R. H. Intermolecular orientations of adenosine-5-monophosphate in aqueous solution as studied by fast fourier ytsndgotm 1H Nmr spectroscopy. Biopolymers. 1974;13(10):2117–2132. doi: 10.1002/bip.1974.360131013. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Jr, Itzkowitz M. S., Tinoco I., Jr Circular dichroism of polynucleotides: dimers as a function of conformation. Biopolymers. 1972 Jan;11(1):225–234. doi: 10.1002/bip.1972.360110117. [DOI] [PubMed] [Google Scholar]
- Kondo N. S., Fang K. N., Miller P. S., Ts'o P. O. Influence of the furanose on the conformation of adenine dinucleoside monophosphates in solution. Biochemistry. 1972 May 23;11(11):1991–2003. doi: 10.1021/bi00761a001. [DOI] [PubMed] [Google Scholar]
- Kondo N. S., Holmes H. M., Stempel L. M., Ts'o O. P. Influence of the phosphodiester linkage (3'-5', 2'-5', and 5'-5') on the conformation of dinucleoside monophosphate. Biochemistry. 1970 Sep 1;9(18):3479–3498. doi: 10.1021/bi00820a002. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Evans F. E., Sarma R. H. The gain in conformational purity and loss in flexibility as a result of 3',5' polymerization between the component mononucleotides - a 300 MHz 1H and 40.5 MHz 31 P NMR comparative study of the dynamic solution conformation of dinucleoside monophosphates and their component monomers. FEBS Lett. 1975 Mar 1;51(1):73–79. doi: 10.1016/0014-5793(75)80857-x. [DOI] [PubMed] [Google Scholar]
- Leng M., Felsenfeld G. A study of polyadenylic acid at neutral pH. J Mol Biol. 1966 Feb;15(2):455–466. doi: 10.1016/s0022-2836(66)80121-3. [DOI] [PubMed] [Google Scholar]
- MARVIN D. A., SPENCER M., WILKINS M. H., HAMILTON L. D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J Mol Biol. 1961 Oct;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- Olson W. K., Flory P. J. Spatial configuration of polynucleotide chains. II. Conformational energies and the average dimensions of polyribonucleotides. Biopolymers. 1972 Jan;11(1):25–56. doi: 10.1002/bip.1972.360110103. [DOI] [PubMed] [Google Scholar]
- Olson W. K. Syn-anti effects on the spatial configuration of polynucleotide chains. Biopolymers. 1973;12(8):1787–1814. doi: 10.1002/bip.1973.360120808. [DOI] [PubMed] [Google Scholar]
- Poland D., Vournakis J. N., Scheraga H. A. Cooperative interactions in single-strand oligomers of adenylic acid. Biopolymers. 1966;4(2):223–235. doi: 10.1002/bip.1966.360040209. [DOI] [PubMed] [Google Scholar]
- Prescott B., Gamache R., Livramento J., Thomas G. J., Jr Raman studies of nucleic acids. XII. Conformations of oligonucleotides and deuterated polynucleotides. Biopolymers. 1974;13(9):1821–1845. doi: 10.1002/bip.1974.360130914. [DOI] [PubMed] [Google Scholar]
- Pullman B., Perahia D., Saran A. Molecular orbital calculations on the conformation of nucleic acids and their constituents. 3. Backbone structure of di- and polynucleotides. Biochim Biophys Acta. 1972 Apr 26;269(1):1–14. doi: 10.1016/0005-2787(72)90068-8. [DOI] [PubMed] [Google Scholar]
- RICH A., DAVIES D. R., CRICK F. H., WATSON J. D. The molecular structure of polyadenylic acid. J Mol Biol. 1961 Feb;3:71–86. doi: 10.1016/s0022-2836(61)80009-0. [DOI] [PubMed] [Google Scholar]
- Stannard B. S., Felsenfeld G. The conformation of polyriboadenylic acid at low temperature and neutral pH. A single-stranded rodlike structure. Biopolymers. 1975 Feb;14(2):299–307. doi: 10.1002/bip.1975.360140205. [DOI] [PubMed] [Google Scholar]
- Suck D., Manor P. C., Germain G., Schwalbe C. H., Weimann G., Saenger W. X-ray study of helix, loop and base pair stacking in trinucleoside diphosphate ApApA. Nat New Biol. 1973 Dec 12;246(154):161–165. doi: 10.1038/newbio246161a0. [DOI] [PubMed] [Google Scholar]
- Thomas G. J., Jr, Hartman K. A. Raman studies of nucleic acids. 8. Estimation of RNA secondary structure from Raman scattering by phosphate-group vibrations. Biochim Biophys Acta. 1973 Jun 23;312(2):311–332. doi: 10.1016/0005-2787(73)90376-6. [DOI] [PubMed] [Google Scholar]
- Ts'o P. O., Kondo N. S., Schweizer M. P., Hollis D. P. Studies of the conformation and interaction in dinucleoside mono- and diphosphates by proton magnetic resonance. Biochemistry. 1969 Mar;8(3):997–1029. doi: 10.1021/bi00831a033. [DOI] [PubMed] [Google Scholar]
- Tsuboi M., Takahashi S., Kyogoku Y., Hayatsu H., Ukita T., Kainosho M. Phosphorus-proton spin-spin coupling and conformation of a dinucleoside phosphate. Science. 1969 Dec 19;166(3912):1504–1505. doi: 10.1126/science.166.3912.1504. [DOI] [PubMed] [Google Scholar]


