Background: A hydrophobic fusion peptide (FP) helps promote paramyxovirus F-mediated membrane fusion.
Results: Efficient membrane disordering and cell-cell fusion strongly correlate with the α-helical character of the Hendra F FP.
Conclusion: The FP adopts an α-helical structure during membrane fusion.
Significance: This study represents the first in-depth characterization of a paramyxovirus FP and offers new mechanistic insights into F-mediated fusion.
Keywords: Fusion Protein, Membrane Fusion, Virology, Virus, Virus Entry, Fusion Peptide, Paramyxovirus
Abstract
Viral fusion proteins are intriguing molecular machines that undergo drastic conformational changes to facilitate virus-cell membrane fusion. During fusion a hydrophobic region of the protein, termed the fusion peptide (FP), is inserted into the target host cell membrane, with subsequent conformational changes culminating in membrane merger. Class I fusion proteins contain FPs between 20 and 30 amino acids in length that are highly conserved within viral families but not between. To examine the sequence dependence of the Hendra virus (HeV) fusion (F) protein FP, the first eight amino acids were mutated first as double, then single, alanine mutants. Mutation of highly conserved glycine residues resulted in inefficient F protein expression and processing, whereas substitution of valine residues resulted in hypofusogenic F proteins despite wild-type surface expression levels. Synthetic peptides corresponding to a portion of the HeV F FP were shown to adopt an α-helical secondary structure in dodecylphosphocholine micelles and small unilamellar vesicles using circular dichroism spectroscopy. Interestingly, peptides containing point mutations that promote lower levels of cell-cell fusion within the context of the whole F protein were less α-helical and induced less membrane disorder in model membranes. These data represent the first extensive structure-function relationship of any paramyxovirus FP and demonstrate that the HeV F FP and potentially other paramyxovirus FPs likely require an α-helical structure for efficient membrane disordering and fusion.
Introduction
Paramyxoviruses represent a significant threat to human health and include well established pathogens such as measles, mumps, and respiratory syncytial virus in addition to the Hendra (HeV)2 and Nipah (NiV) viruses (1, 2). HeV and NiV are highly pathogenic zoonotic viruses that were identified in 1994 and 1999, respectively, as the etiological agents responsible for cases of severe respiratory disease and encephalitis in Australia and Malaysia (2–5). High mortality rates coupled with suspected human-to-human transmission has resulted in both HeV and NiV being classified as biosafety level four (BSL-4) pathogens as well as NIAID category C priority pathogens. Understanding key points within the viral lifecycle is, therefore, vital for the development of antiviral therapeutics.
Fusion between the viral and cellular membranes is essential for paramyxovirus infection, as it culminates in the deposition of the viral genome into the target cell. Generally, paramyxovirus-promoted membrane fusion requires the concerted effort of two viral surface glycoproteins (with the notable exception of Pneumovirinae subfamily members): the attachment protein (H, HN, or G), which mediates initial receptor binding, and the fusion (F) protein, which drives membrane merger through a series of extensive conformational changes (6). All paramyxovirus F proteins are trimeric type-I integral membrane proteins and, like other class I fusion proteins, contain several regions critical for fusion promotion (Fig. 1A) including a hydrophobic N-terminal fusion peptide (FP), two heptad repeat (HR) regions (HRA and HRB), and a C-terminal transmembrane domain (TMD). Following proteolytic cleavage, the metastable F protein must be triggered to undergo the vast conformational changes required for membrane fusion. Though the specific mechanism of triggering remains poorly understood, it facilitates extension of HRA toward, and FP insertion into, the target cell membrane (7–9). Fusion peptide insertion physically links both the viral and cellular membranes together, coupling the force generated by F rearrangement to membrane merger (10). Subsequent conformational changes position HRA and HRB in close proximity, culminating in the formation of a stable six-helix bundle and fusion pore formation (11). Despite the existence of pre- and post-fusion structures for some paramyxovirus F proteins (12, 13), key structural details regarding fusion intermediates remain elusive, including exactly how the FP helps drive fusion.
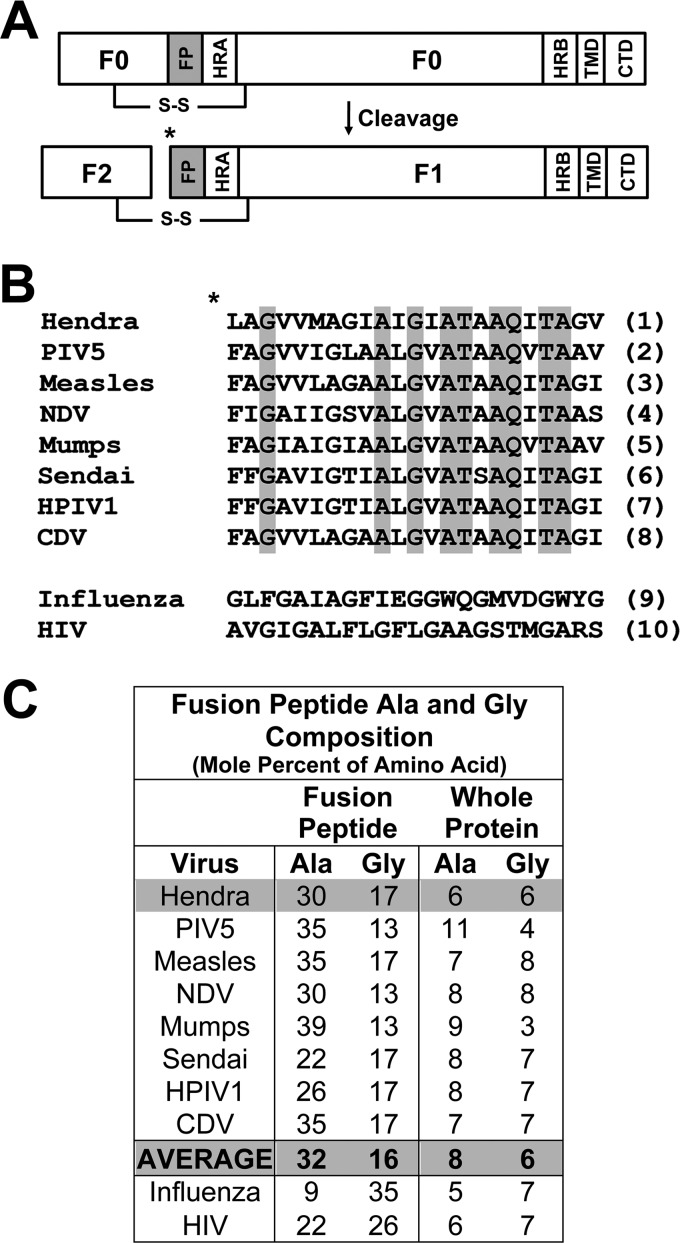
FIGURE 1.
Amino acid composition of fusion peptides from class I fusion proteins. A, shown is a diagram of a paramyxovirus F protein in the uncleaved (F0) and cleaved, fusogenically active F1+F2 forms. HR, heptad repeat; CTD, C-terminal domain (C-tail); * denotes site of proteolytic cleavage. B, shown is alignment of paramyxovirus fusion peptides with completely conserved amino acids shaded and compared with fusion peptides from influenza HA and HIV Env. References for sequences are as follows: Hendra (5), PIV5 (59), measles (60), NDV (61), Mumps (62), Sendai (63), HPIV1 (human parainfluenza virus type 1 (64)), CDV (canine distemper virus (65)), influenza (66), HIV (22). C, percent alanine and glycine composition of the fusion peptides is shown in B as compared with the whole protein.
Fusion peptides of class I fusion proteins are highly conserved within viral families, but not between viral families (Fig. 1B). On average, FPs are 20 to 30 amino acids in length and are particularly enriched in glycine and alanine residues (Fig. 1C), likely rendering these regions remarkably flexible and, in the case of isolated peptides, structurally polymorphic (14, 15). Such conformational flexibility may well be needed to promote the membrane perturbation needed for fusion (10). Studies from disparate viral systems including influenza HA (16–19), HIV Env (20–24), Sendai F (25) and Ebola GP (26) have used synthetic peptides corresponding to the FP (or fusion loop in the case of Ebola GP) to perform structure-function experiments, and to complement mutational studies within the whole protein. The influenza HA FP seems to require substantial α-helical character for membrane fusion (16, 17, 27), and is thought to adopt a kinked “boomerang” structure which is vital for membrane fusion promotion (17, 27). Despite the availability of structural data for both HIV Env (28, 29) and influenza HA (17, 18, 30) FPs, similar structural studies have not been performed for any paramyxovirus FP to date.
Previous work examining paramyxovirus FPs has focused on the highly conserved glycine residues (31–34). Work with parainfluenza virus 5 (PIV5) F demonstrated that mutation of three N-terminal glycine residues (G3, G7, and G12, as named in Ref. 32) dramatically increased membrane fusion while decreasing the requirement for the homotypic attachment protein, suggesting that these residues were critical in pre-fusion F protein stability (32). Conversely, similar mutations in Newcastle disease virus (NDV) F and human parainfluenza virus 3 (HPIV3) F led to moderate increases and, in the case of the G7 mutant of NDV F, decreases in membrane fusion (32), with no change in the requirement for an attachment protein.
To examine the sequence requirements of the Hendra F FP, double and single alanine mutations were introduced into the first eight residues of the FP. Mutation of glycine residues within the FP to alanine significantly decreased F protein expression and processing, whereas the VM114/115AA double mutation completely abolished fusion. Circular dichroism spectra of synthetic fusion peptides demonstrated a strong correlation between peptide α-helicity in micelles, cell-cell fusion levels, and the degree of membrane disordering suggesting that the HeV F protein FP requires an α-helical structure for function. Together, these data demonstrate that the combination of at least two single alanine mutations within the FP can lead to dramatic fusion defects, whereas any single mutation is better tolerated. Additionally, these data point toward a role for valine residues in F-promoted fusion, whereas N-terminal glycine residues are important for efficient expression and processing of HeV F.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
Vero and BSR cells (provided by Karl-Klaus Conzelman, Pettenkofer Institut) were maintained in Dulbecco's modified Eagle's media (DMEM; Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. To select for T7 polymerase-expressing cells, 0.5 mg/ml G-418 sulfate (Invitrogen) was added to the media of the BSR cells every third passage.
Plasmids and Antibodies
Plasmids containing Hendra F and G were generously provided by Dr. Lin-Fa Wang (Australian Animal Health Laboratory). All Hendra F mutants were made in pGEM using the QuikChange site-directed mutagenesis kit (Stratagene) and subcloned into pCAGGS as described previously (35). Resultant constructs were sequenced in their entirety to confirm sequence integrity. Anti-peptide antibodies to residues 527–539 of the Hendra F cytoplasmic tail (35) were used to pull down F.
Syncytia Assay
Vero cells in 6-well plates (∼90% confluent) were transiently transfected with pCAGGS-Hendra F (0.5 μg) and pCAGGS-Hendra G (1.5 μg) using Lipofectamine and Plus Reagent (Invitrogen) per manufacturer's protocol. Syncytia formation was observed 24- to 48-h post-transfection, and photographs were taken using a Nikon digital camera mounted atop a Nikon TS100 microscope with a 10× objective.
Surface Biotinylation of Wild-type and Mutant Proteins
Sub-confluent Vero cells (80–90%) in 60-mm dishes were transiently transfected using Lipofectamine and Plus reagent (3 μg of DNA wild-type or mutant pCAGGS-Hendra F) according to the manufacturer's protocol. Eighteen to twenty-four hours post-transfection, cells were washed with PBS and starved for 45 min in DMEM deficient in cysteine and methionine. Cells were then labeled for 3 h with DMEM deficient in cysteine and methionine containing Tran35S-label (100 μCi/ml; MP Biomedicals, Irvine, CA). After the label, cells were washed 3 times with 3 ml of ice-cold PBS, and surface proteins were biotinylated by the addition of 1 mg/ml EZ-Link Sulfo-NSH-Biotin (Pierce; in PBS) rocking for 35 min at 4 °C followed by incubation at room temperature for 15 min. Cells were then washed an additional 3 times with ice-cold PBS and lysed with 500 μl of radioimmunoprecipitation assay lysis buffer (100 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% SDS, 1% Triton X-100, 1% deoxycholic acid, 1 mm phenylmethylsulfonyl fluoride (Sigma), 25 mm iodoacetaminde (Sigma), and 1 kallikrein inhibitory unit of aprotinin (Calbiochem)). The cellular lysate was centrifuged at 136,500 × g for 15 min at 4 °C. The supernatant was removed to a 1.5-ml microcentrifuge tube, and 4 μl of Hendra F peptide antibody was added and incubated for 3 h or overnight at 4 °C followed by incubation with 30 μl of Protein A-Sepharose beads (GE Healthcare) for 30 min. The beads were washed 2× with radioimmunoprecipitation assay buffer + 0.30 m NaCl, 2× with radioimmunoprecipitation assay buffer + 0.15 m NaCl, and 1× with SDS Wash II (150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 2.5 mm EDTA). Once the beads were washed, 60 μl of 10% SDS was added, and the samples were boiled for 10 min, removed to a new tube, and repeated with 40 μl of 10% SDS for a total of 100 μl. Ten microliters of the supernatant was removed to analyze total population. To the remaining supernatant, 30 μl of Streptavidin beads (Pierce) and 400 μl of biotinylation dilution buffer (20 mm Tris (pH 8), 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.2% bovine serum albumin) were then added for 1 h at 4 °C, rocking. Analysis of Hendra F was performed by 15% SDS-PAGE and visualized using the Typhoon imaging system (GE Healthcare). Band densitometry using ImageQuant 5.2 (GE Healthcare) was performed for each experiment to quantitate the amount of surface-expressed F1 present in each sample. For the DN Rab5 and TPCK (N-tosyl-l-phenylalanine chloromethyl ketone) trypsin experiments, Vero cells were transfected with 1.5 μg of either wild-type or mutant pCAGGS-Hendra F and 1.5 μg of DN-Rab5, and the experiment was performed as described above, except with the addition of 1 μg/ml of TPCK-trypsin to the label media.
Luciferase Reporter Gene Assay
Vero cells in 6-well plates were transiently transfected with 0.9 μg of wild-type pCAGGS-Hendra G, 0.3 μg of wild-type or mutant pCAGGS-Hendra F, and 0.8 μg of T7 luciferase under the control of the T7 promoter (2 μg total DNA) using Lipofectamine and Plus reagent per the manufacturer's protocol. Eighteen hours post-transfection, Vero cells were washed 1× with PBS and overlaid with BSR cells (∼1:1) stably expressing the T7 polymerase for 3 h at 37 °C. Cells were lysed and assayed for luciferase activity using the luciferase assay system from Promega (Madison, WI) according to the manufacturer's instructions. An Lmax luminometer (Molecular Devices, Sunnyvale, CA) was used to measure light emission; a 2-s delay and 8-s read time were used. Values were normalized to samples containing wild-type pCAGGS-Hendra F and G, with wild-type set at 100%, after subtraction of the pCAGGS-Hendra G alone values.
Peptide Synthesis and Purification
Five peptides were used in this study: guest, GCGKKKK; wild-type, LAGVVMAGIAIGIATA GCGKKKK; VM114/115AA; LAGVAAAGIAIGIATAGCGKKKK; V114A, LAGVAMAGIAIGIATAGCGKKKK; M115A, LAGVVAAGIAIGIATAGCGKKKK. This host guest system was first described by Han and Tamm (16). Peptides used in this study were obtained from either GenScript (Piscataway, NJ) or NEO-Peptide (Cambridge, MA) and were purified to ≥95% purity by reverse-phase HPLC, with peptide identities confirmed by mass spectrometry. The C terminus of each peptide was amidated, whereas the N terminus was unmodified. Stock peptide solutions were made by resuspending peptides in sterile, deionized water, and amino acid analysis was performed (W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University, New Haven, CT) to determine the concentrations of all stock solutions. Working stocks of 750 μm in sterile, deionized water were prepared, and all stock solutions were stored as small aliquots and kept at −20 °C.
Hemolysis Assay
Chicken red blood cells (RBCs) were purchased from Innovative Research (Novi, MI) as a 5% v/v suspension in PBS and were used within 2 weeks of purchase. For the hemolysis studies, chicken RBCs were diluted to 2% with PBS immediately before use. The final reaction volume for each sample was 300 μl, consisting of 260 μl of 2% chicken RBCs and 40 μl of either peptide, water, or a mixture of the 2 in a 1.5-ml centrifuge tube. Final peptide concentration was between 0 and 100 μm. Samples were incubated at 37 °C for 45 min and centrifuged at 3000 × g, and supernatant was removed to another tube. Absorbance of each sample was then taken at 520 nm with complete hemolysis obtained after lysing chicken RBCs with 0.5% Triton X-100. Absorbance values were then expressed as a percent of values obtained from Triton X-100 lysis.
Micelle Preparation and Circular Dichroism (CD) Spectroscopy
Dodecylphosphocholine (DPC) micelles were prepared 1 day before use by drying the DPC/chloroform solution under a continuous nitrogen stream. Residual chloroform was removed by placing the DPC sample under a vacuum for 30 min. Dried DPC was then resuspended in 5 mm HEPES, 10 mm MES buffer (pH 7.4) by gently shaking for 20 min and then stored at 4 °C until use the next morning. For experiments using different peptide:DPC ratios, separate micelle preparations were made for each ratio. Circular dichroism (CD) spectra were measured with a Jasco J-810 spectrapolarimeter using a quartz cuvette with a 1-mm path length. All measurements were performed at 22 °C, and each spectrum is an average of 4 scans with 3 independent spectra taken for each sample. Spectra were measured in 5 mm HEPES, 10 mm MES buffer (pH 7.4) plus or minus 10 mm DPC (Avanti Polar Lipids, Alabaster, AL) with peptide concentrations of 100 μm (peptide:DPC ratio of 1:100). Background spectra without peptide were collected and subtracted from spectra taken in the presence of peptide. Estimation of the fraction of residues in α-helical formation was performed as previously described (36). CD spectra using small unilamellar vesicles (SUVs) spectra were acquired on an Aviv model 215 spectropolarimeter in a 0.5-mm quartz cell at 22 °C. Data were collected from 200–260 nm. Experiments were collected with 100 μm peptide in 5 mm Hepes, 10 mm MES buffer with and without 5 mm POPC:POPG SUVs.
Preparation of SUVs
Appropriate amounts of stock POPC and POPG in chloroform were mixed and evaporated to a film with a nitrogen stream. The films were hydrated with 5 mm Hepes 10 mm MES buffer and sonicated with a Branson ultra-sonicator equipped with a titanium tip on ice until the vesicle solution became clear. Titanium particles were removed by centrifugation.
Preparation of Supported Bilayers
Planar single bilayers were prepared using a Langmuir trough. A monolayer of 1,2-dimyristoyl-sn-glycero-3-phosphocholine was spread on the trough. A germanium attenuated total reflectance (ATR)-IR plate was dipped into the trough and slowly removed to transfer a single 1,2-dimyristoyl-sn-glycero-3-phosphocholine monolayer. To form the second lipid leaflet, the plate was incubated with SUVs composed of POPC:POPG 95:5 in a chamber for 1–2 h. Excess vesicles were washed away with 2 ml of D2O buffer (37, 38).
ATR-Fourier Transform Infrared Spectroscopy (ATR-FTIR)
Measurements were collected using a Bruker Vector 22 Fourier transform infrared spectrometer equipped with an ATR-IR accessory. Before bilayer formation, parallel- and perpendicular-polarized attenuated total reflection measurements were obtained for the bare germanium plate in D2O buffered with 5 mm Hepes, 10 mm MES containing 150 mm NaCl. These measurements were used as references to calculate absorbance spectra of the parallel and perpendicular data collected with the single bilayer formed on the germanium plate. Dichroic ratios, RATR, were determined for the characteristic lipid methylene stretching bands at 2920 and 2850 cm−1. Lipid order parameters, SL, were calculated using the equation (38),
 |
where Ey2, Ex2, and Ez2 are 1.9691, 2.2486, and 1.8917, respectively (37, 38). |SL| can range from 0 to 1, and lower values for SL represent more disordered lipids.
RESULTS
Alanine Substitution at GV112/113 and G117 Results in Protein Misfolding and Inefficient Cathepsin L Processing
Previous studies on paramyxovirus FPs have largely focused on conserved glycine residues within this region (31–34). To examine the possible role of surrounding amino acids in fusion, scanning alanine mutagenesis was performed, resulting in four F proteins with either single or double alanine mutations (Fig. 2A). Before examining the fusogenicity of the mutant proteins, cell surface biotinylation was performed to ensure that they were properly folded, transported to the cell surface, and had undergone proteolytic processing by cathepsin L. Vero cells transfected with either wild-type or mutant F proteins were metabolically labeled, and cell surface proteins were biotinylated followed by immunoprecipitation, separation of biotinylated F proteins, and analysis by SDS-PAGE. Alanine substitution of either L110 or VM114/115 did not disrupt protein folding as both mutant proteins were expressed at or above wild-type levels (Fig. 2B). Mutation of GV112/113 to alanine significantly reduced cell surface expression (CSE) levels, as compared with wild-type, whereas the G117A mutant failed to undergo proteolytic processing, suggestive of protein misfolding. As cathepsin L-mediated cleavage of Hendra F occurs directly N-terminal to the FP, cleavage efficiency was calculated by band densitometry (F1/(F1 + F0))) to determine if mutations within the fusion peptide affect cleavage. Both L110A F and VM114/115AA F were cleaved (62.5 ± 3.4 and 65.6 ± 1.8%, respectively) at wild-type levels (67.5 ± 4.8%), demonstrating that mutation of these residues did not affect efficiency of cathepsin L processing. Cleavage efficiency of GV112/113AA F was reduced to approximately two-thirds of wild-type levels (40.2 ± 6.5%), whereas G117A F was minimally processed by cathepsin L (14.3 ± 5.7%), suggesting that glycine substitutions result in either a misfolded protein with altered cleavage site accessibility or modulate critical interactions with cathepsin L needed for cleavage.
FIGURE 2.
Cell-surface expression of Hendra F fusion peptide mutants. A, Alignment of Hendra F FP mutants is shown. CTD, C-terminal domain. B, cell-surface expression of FP mutants is shown. After transfection with empty pCAGGS vector or either wild-type or mutant pCAGGS-Hendra F, cells were starved and labeled with Tran35S-label containing Cys/Met, and surface proteins were biotinylated. After cell lysis, Hendra F was immunoprecipitated, and surface proteins were pulled down with Streptavidin beads. Protein was analyzed via 15% SDS-PAGE, and bands were visualized using the Typhoon imaging system. C, exogenous TPCK-trypsin cleavage of surface-expressed wild-type or mutant F proteins in the presence or absence of dominant negative Rab5 (DN-Rab5) is shown. Human metapneumovirus F (HMPV F) was used as a positive control for TPCK-trypsin cleavage and activity. Both B and C are representative gels from one of three independent experiments.
Unlike other paramyxovirus F proteins, Hendra F is initially surface-expressed in a non-fusogenic F0 form, which is then endocytosed and subsequently cleaved after residue Lys-109 by cathepsin L into the fusogenically active F1+F2 form (39–42). As an alternative to cathepsin L, cell surface-expressed F0 can be efficiently cleaved into F1+F2 by the addition of exogenous TPCK-trypsin, so susceptibility to TPCK-trypsin cleavage can be used to differentiate between the above hypotheses (43). To more clearly observe changes in F cleavage, uncleaved F0 was blocked at the cell surface by co-transfection with a dominant-negative form of Rab5 (DN-Rab5), a small GTPase required for endocytosis (44). Human metapneumovirus F, which can be cleaved extracellularly by trypsin (45, 46), was used as a positive control and is efficiently cleaved by addition of TPCK-trypsin (Fig. 2C). In the absence of DN-Rab5, a slight increase in wild-type F1 was observed upon the addition of TPCK-trypsin (Fig. 2C). Co-transfection of DN-Rab5 to block wild-type F at the cell surface followed by exogenous TPCK-trypsin cleavage led to a large increase in F1 (Fig. 2C). Whereas cell-surface F0 is increased in the presence of DN-Rab5 for GV112/113AA F addition of TPCK-trypsin led to a significant reduction of F0 (Fig. 2C) for GV112/113AA F with no noticeable increase in the cleaved F1 form. Furthermore, the decrease in F0 upon the addition of TPCK-trypsin observed for GV112/113AA indicates that these mutations likely expose additional TPCK-trypsin cleavage sites, suggestive of a looser overall protein fold. No increase in either F0 or F1 was observed for G117A upon TPCK-trypsin treatment suggestive of protein misfolding. These data suggest that the GV112/113AA mutations result in misfolding of the cleavage site, rendering it only partially accessible (GV112/113AA) or completely inaccessible (G117A) to TPCK-trypsin.
VM114/115AA F Does Not Promote Membrane Fusion Despite Wild-type Surface Expression Levels
Co-expression of Hendra F along with its homotypic attachment (G) protein is sufficient to induce cell-cell fusion, so syncytia formation can be used to directly assess F protein fusogenicity. 24–36 h post-transfection, Vero cells expressing wild-type Hendra F and G formed large syncytia, whereas expression of G alone was not sufficient for syncytia formation (Fig. 3A). Syncytia formation for L110A F was slightly reduced from wild-type levels, whereas GV112/113AA F and G117A showed little syncytia formation, consistent with reduced surface expression levels of the cleaved fusogenically active form. Surprisingly, the mutant VM114/115AA F could not promote syncytia formation despite being surface-expressed above wild-type levels.
FIGURE 3.
Fusogenicity of wild-type and mutant Hendra F proteins. A, syncytia assays demonstrating cell-cell fusion promotion of wild-type or mutant F proteins in the presence of the Hendra attachment (G) protein are shown. Vero cells transfected with either empty pCAGGS and wild-type pCAGGS-Hendra G or wild-type or mutant pCAGGS-Hendra F and wild-type pCAGGS-Hendra G were monitored for syncytia formation as described under “Experimental Procedures.” 36–48 h after transfection, 10 images from different fields were taken using a Nikon Coolpix995 camera mounted on a Nikon TS100 inverted phase-contrast microscope. Representative images are shown from one of three independent experiments. B, quantitative luciferase reporter gene assays show cell-cell fusion levels (black bars) alongside quantitation of F1 CSE levels from Fig. 2B. Band densitometry was performed using ImageQuant 5.2 (GE Healthcare) software. Vero cells were transfected with a T7 luciferase construct and either empty pCAGGS and wild-type pCAGGS-Hendra G or wild-type or mutant pCAGGS-Hendra F and wild-type pCAGGS-Hendra G at a 1:3 F:G ratio as described under “Experimental Procedures.” 18–24 h post-transfection Vero cells were overlaid with BSR cells stably expressing the T7 polymerase, cells were incubated for 3 h at 37 °C and lysed, and luciferase activity was measured. All data are presented as the means ± S.E. for at least three independent experiments.
For a more quantitative measure of F-mediated cell-cell fusion, a luciferase reporter gene assay was performed. Vero cells transfected with wild-type or mutant Hendra F and wild-type G along with a plasmid containing the luciferase gene under the control of the T7 promoter were overlaid with BSR cells, which stably express the T7 polymerase. Cell-cell fusion allows for expression of luciferase, thus providing a quantitative measure of cell-cell fusion. Consistent with the syncytia data, L110A F promoted fusion at slightly reduced levels (65.4 ± 9.6%), whereas cell-cell fusion promoted by GV112/113AA F and G117A F was extremely low and proportional with F1 cell-surface expression (Fig. 3B). No fusion was observed for VM114/115AA, clearly demonstrating that mutation of both of these residues together dramatically affects F-promoted membrane fusion without altering protein expression or cathepsin L processing.
Peptides Corresponding to the VM114/115AA FP Do Not Cause Efficient Hemolysis
Viral fusion peptides have been hypothesized to play a major role in driving membrane fusion through modulation of membrane curvature and lipid organization of the target cell membrane (6, 10). Thus the VM114/115AA mutation could potentially alter the ability of the FP to interact with and insert into the target cell membrane after triggering of F. To examine potential changes in membrane interactions, synthetic peptides corresponding to the first 16 amino acids of the FP (guest) for both wild-type and VM114/115AA F were designed coupled to a highly charged, flexible host peptide (Fig. 4A). Previous work studying influenza HA has extensively characterized the biophysical properties of this host-guest system and has shown that these peptides are highly water-soluble and remain monomeric in low ionic strength solutions (16, 36). Synthetic FPs have also been shown to cause RBC lysis (hemolysis) (16), and the degree of hemolysis can be used to assay changes in the extent or nature of FP-membrane interactions. Chicken RBCs were incubated with increasing concentrations of either guest, wild-type, or VM114/115AA peptide, the supernatant was clarified through centrifugation, and hemolysis was assayed by measuring the absorbance of hemoglobin at 520 nm. Maximum hemolysis was defined by incubating RBCs with 0.5% Triton X-100, and all subsequent absorbance values were expressed as a percentage of this maximum. Host peptide alone at concentrations up to 100 μm was unable to cause significant levels of hemolysis (Fig. 4B, circle), whereas wild-type peptide (Fig. 4B, square) resulted in a concentration-dependent increase in hemolysis with a maximum value of 56.1%±6.9. Incubation with VM114/115AA peptide (Fig. 4B, diamond) resulted in a 75% reduction of hemolysis (18.0 ± 0.9%) demonstrating that mutations at these positions affect either the ability of the peptide to interact with membrane or, more likely, change the nature of this membrane interaction.
FIGURE 4.
Hemolytic ability and CD spectra of synthetic fusion peptides. A, shown are synthesized peptides where the first 16 amino acids of the FP for either wild-type or mutant Hendra F are coupled to a flexible highly charged host peptide. B, hemolytic ability of synthetic fusion peptides is shown. Chicken RBCs were incubated with increasing concentrations of peptide for 45 min at 37 °C. Supernatants were then cleared by centrifugation at 11,000 × g, and absorbance was measured at 520 nm. All values are expressed as a percent of maximum hemolysis determined by RBC lysis with 0.5% Triton X-100. Data are presented as the means ± S.E. for at least three independent experiments. C, CD spectra of all five peptides in the absence of DPC in 5 mm Hepes, 10 mm MES pH 7.4 with peptide concentrations of 100 μm are shown. D, shown are spectra in the presence of 10 mm DPC micelles prepared in the above buffer as described under “Experimental Procedures.” Spectra in both C and D are representative of one of three independent experiments where each spectrum is an average of four scans.
VM114/115AA Peptide in Detergent Micelles Is Significantly Less α-Helical Than the Wild-type Peptide
Previous studies (for review, see Ref. 15) in other viral systems suggested that α-helical structure in at least a part of the FP correlates with fusion (47). The FP of influenza HA adopts a boomerang-like shape within model membranes, and point mutations that disrupt this structure abolish or reduce HA-mediated membrane fusion in the context of the whole protein (17, 18, 27). Thus, the lack of membrane fusion promotion and the significantly reduced hemolysis observed for VM114/115AA F could be a result of altered secondary structure within membranes. To examine this possibility, CD spectra were obtained for the host, wild type, and VM114/115AA peptides in the presence or absence of DPC micelles, used to mimic the hydrophobic membrane environment. DPC micelles appropriately mimicked a membranous environment, as subsequent experiments using POPC:POPG SUVs provided similar results (see Fig. 7). In aqueous solution all three peptides were disordered (Fig. 4C). The addition of 10 mm DPC (1:100 peptide:DPC ratio) caused a large structural change in both the WT and VM114/115AA peptides resulting in more ordered and partially α-helical conformations (Fig. 4D). Interestingly, the wild-type (solid black line) and VM114/115AA spectra were considerably different from one another, demonstrating that alanine substitution at positions 114/115 resulted in structural changes within a membrane-mimicking environment. To more clearly differentiate between these two spectra, the α-helical content of small peptides can be approximated using the signal at 222 nm (θ222), which is predominately due to the presence of α-helical structure (36). Wild-type peptide has an estimated α-helical content of 35.8 ± 0.1%, whereas the VM114/115AA peptide has one-third less α-helical character (24.6 ± 0.4%) (Fig. 4D and Table 1). These data suggest that the wild-type Hendra F FP requires a certain degree of α-helical structure to drive efficient membrane fusion.
FIGURE 7.
CD spectra of synthetic fusion peptides in POPG:POPG SUVs. A, shown are CD spectra of wild-type Hendra F fusion peptides in the presence or absence of SUVs composed of differing POPC:POPG ratios. B and C, CD spectra of wild-type and mutant Hendra F fusion peptides in the absence (B) or presence (C) of POPC:POPG 95:5 SUVs are shown. Spectra were obtained using a peptide concentration of 100 μm in solution or with 5 mm SUVs composed of POPC 100% or POPC:POPG 98:2, 95:5, and 92:8. All spectra were recorded at 22 ± 2 °C in a 0.5-mm quartz cuvette. Spectra were obtained from two independent sets of SUVs where each spectrum is an average of five scans.
TABLE 1.
Data summary for transiently expressed F protein and synthetic FPs
All errors are reported as S.E. ND, not determined.
| Fusion/F1 CSEa | Cleavage | Hemolysis | α-Helicity | |
|---|---|---|---|---|
| % | % | % | % | |
| WT | 100.0 ± 0.0 | 67.5 ± 4.8 | 56.1 ± 6.9 | 35.8 ± 0.1 |
| L110A | 48.8 ± 13.6 | 62.5 ± 3.4 | ND | ND |
| GV112/113AA | 32.1 ± 4.3 | 40.2 ± 6.5 | ND | ND |
| G112A | 98.5 ± 30.5 | 43.4 ± 5.8 | ND | ND |
| G112P | 6.0 ± 1.4 | 79.9 ± 2.1 | ND | ND |
| V113A | 54.9 ± 17.3 | 64.1 ± 3.4 | ND | ND |
| VM114/115AA | 3.1 ± 1.7 | 65.6 ± 1.8 | 18.0 ± 1.0 | 24.6 ± 0.4 |
| V114A | 30.7 ± 7.1 | 70.3 ± 2.3 | 36.5 ± 3.9 | 28.7 ± 0.2 |
| M115A | 88.8 ± 17.2 | 67.0 ± 1.0 | 38.0 ± 4.3 | 38.8 ± 0.5 |
| G117A | 37.4 ± 22.3 | 14.4 ± 5.7 | ND | ND |
a Cell-cell fusion values normalized to F CSE with propagated S.E.
Mutation of G112 to Alanine Is Responsible for the Decreased Surface Expression of GV112/113AA
As the lack of fusion observed for VM114/115AA could be due to mutation of either or both of the two residues, mutant F proteins containing single point mutations of residues 112–115 were generated (Fig. 5A). All single alanine mutants were surface-expressed and proteolytically processed (Fig. 5B), as expected based on the double alanine mutant data. Three single mutants, V113A, V114A, and M115A, were expressed at or above wild type, whereas G112A was expressed at approximately one-third of wild-type levels (Fig. 5B). Cleavage efficiency for V113A, V114A, and M115A was very similar to wild-type (64.1 ± 6.9, 70.3 ± 4.6, and 67.0 ± 2.1%, respectively, compared with 67.5 ± 4.8%), whereas G112A was less efficiently cleaved (43.4 ± 11.6%). Together, these results demonstrate that mutation of G112 to alanine is sufficient to dramatically reduce F surface expression levels likely by misfolding portions of the protein.
FIGURE 5.
Cell-surface expression of single alanine FP mutants. A, alignment of the four single alanine FP mutants is shown. B, cell-surface biotinylation of the four single alanine mutants performed as described in Fig. 2 and under “Experimental Procedures” is shown. These lanes are from the same gel as Fig. 2, thus allowing for a direct comparison between mutants. This is a representative gel from one of three independent experiments. CTD, C-terminal domain.
Valine Residues within the Hendra F FP Are Important for Membrane Fusion
Once surface expression levels were determined for the four single alanine mutants, syncytia and reporter gene assays were performed to determine fusogenicity of these mutant F proteins. Additionally, to further examine the sequence requirements at G112, an additional mutation (G112P) was made. A proline side chain is considerably larger than glycine and should render this region considerably less flexible. Unlike the two double mutants GV112/113AA and VM114/115AA, all single alanine mutants promoted some degree of syncytia formation (Fig. 6A). Cell-cell fusion levels for M115A were close to wild type, whereas G112A, V113A, and V114A showed reduced fusion. Quantitative reporter gene analysis for these four mutants was consistent with data from the syncytia assays, as M115A promoted cell-cell fusion at almost wild-type levels (88.0 ± 2.6%), whereas the remaining three mutants had lower cell-cell fusion levels (Fig. 6B). Comparison of cell-cell fusion levels with F1 CSE reveals that only V113A and V114A are impaired for fusion, as both G112A and M115A promote fusion at levels consistent with CSE data. Surprisingly, the G112P mutation increased CSE levels as compared with wild-type and G112A (Fig. 6C). Cathepsin L processing of G112P was also higher (79.9 ± 2.1% compared with the wild-type value of 67.5 ± 4.9). G112P was severely impaired for cell-cell fusion promotion (Fig. 6C; 15.3 ± 0.6%) as compared with wild type. These data demonstrate that the combination of at least two single alanine mutations at the N terminus of the FP can lead to dramatic fusion defects, whereas the single mutants are better tolerated. Furthermore, these data indicate that valine residues within the FP are essential for efficient membrane fusion promotion, whereas N-terminal glycine residues are important for maintaining F protein stability and proper folding.
FIGURE 6.
Fusogenicity of wild-type and single alanine mutant F proteins. A, shown is a syncytia assay for each of the four mutants as compared with wild type. The experiment was performed as described in Fig. 3A and under “Experimental Procedures.” Images are representative of one of three independent experiments. B and C, shown is a quantitative reporter gene analysis and quantitation of F1 cell-surface expression for FP mutants as compared with wild-type F performed as described in Fig. 3B and under “Experimental Procedures.” All data are presented as the means ± S.E. for at least three independent experiments.
V114A and M115A F Promote Hemolysis More Efficiently Than the V114/115AA Double Mutant
As V114A and M115A had increased levels of cell-cell fusion over the corresponding double mutant, RBC hemolysis was performed as described above using synthetic peptides corresponding to the first 16 amino acids of the V114A and M115A FPs (Fig. 4A). Both the V114A and M115A peptides were able to efficiently promote hemolysis, at ∼65–70% of wild type (Fig. 4B). As fusion levels for V114A were lower than hemolysis levels as compared with wild type, this suggests that the ability to disrupt membranes is necessary but not sufficient for efficient membrane fusion.
Percentage of α-Helical Structure in Synthetic Peptides Correlates with Membrane Fusion Levels
Circular dichroism spectra of peptides containing either the V114A and Met-115 mutations in the absence of DPC were largely disordered, similar to the VM114/115AA spectrum (Fig. 4C). In the presence of 10 mm DPC, both the V114A and M115A peptides adopted ordered secondary structures with considerable amounts of α-helicity (Fig. 4D). Surprisingly, the M115A peptide adopted a conformation distinct from wild type despite promoting almost wild-type levels of cell-cell fusion within the context of the whole protein. The V114A peptide adopted a conformation similar to VM114/115AA, suggesting that the V114A substitution was the dominant determinant of peptide structure (Fig. 4D). Comparing these spectra with those of wild-type and VM114/115AA reveals a strong correlation between estimated peptide α-helical content and fusogenicity (Table 1). The difference in α-helical content between the VM114/115AA peptide and the wild-type peptide is estimated to be around 10% (24.6 ± 0.4 and 35.8 ± 0.1%, respectively). As V114A is estimated to be around 24.6 ± 0.4% helical, these data suggest that decreases in peptide α-helical content can dramatically affect membrane fusion promotion.
Synthetic Fusion Peptide α-Helicity Directly Correlates to the Degree of Membrane Disordering
One way in which fusion peptides are thought to help drive cell-cell and virus-cell membrane fusion is through disordering of the target cell membrane. Before examining peptide-induced membrane disordering, CD spectra of each synthetic fusion peptide were determined in the presence of SUVs to ensure similar secondary structures in lipid bilayers as in detergent micelles. Vesicles composed of both POPC and POPG were prepared as described under “Experimental Procedures.” Little structural change was observed for the wild-type peptide in pure POPC SUVs; however, the addition of peptide to SUVs containing 5–8% POPG resulted in a dramatic shift to a predominately α-helical structure (Fig. 7A), presumably because the presence of the negatively charged lipids facilitated peptide binding through the tetralysine motif. All peptides were disordered in the absence of SUVs (Fig. 7B) but exhibited high degrees of α-helical structure in the presence of SUVs (Fig. 7C), similar to spectra taken in DPC. Thus, these data demonstrate that DPC is an acceptable membrane mimic and confirm that the Hendra F FP adopts an α-helical structure in membranes.
To examine if membrane disordering is impaired by the VM114/115AA or individual single mutations, ATR-FTIR experiments were performed on peptides bound to single planar bilayers. The degree of lipid acyl chain order, denoted as SL, can be determined by monitoring the dichroism of two prominent absorption bands at 2920 and 2850 cm−1 (38). Spectra were collected with perpendicular- and parallel-polarized light, and the dichroic ratios (RATR) of each peak were determined. These ratios were then used to calculate the overall order of the membrane (Equation 1 under “Experimental Procedures”). In Fig. 8, the lipid order results are presented for a single planar bilayer system before and after incubation with the synthetic Hendra F fusion peptides. After incubation, the fusion-competent wild-type FP readily destabilized the bilayer. Interestingly, the magnitude of membrane disordering correlated with the degree of peptide α-helical structure (in both DPC micelles and POPC:POPG SUVs) and with their fusion activity (Fig. 8). Thus, the fusion defects observed for the V11A and VM114/115AA mutants might be caused by an inability to drive sufficient membrane disordering due to less α-helical secondary structure.
FIGURE 8.
Peptide-induced membrane disordering obtained using ATR-FTIR. A, lipid order parameters, SL, obtained for single planar bilayers before and after incubation with 100 μm Hendra WT, M115A, and VM114/115AA peptides are shown. B, ATR dichroic ratios of lipid methylene stretching vibrations and derived acyl chain order parameters in the absence and presence of wild-type or mutant Hendra F fusion peptides are shown. For all cases the bottom layer of the bilayer was composed of 1,2-dimyristoyl-sn-glycero-3-phosphocholine, and the top was formed with POPC:POPG 95:5. Experiments were performed in D2O buffered with 5 mm HEPES and 10 mm MES with 150 mm NaCl. Data represent two independent bilayer preparations for each peptide.
Increased Effective Concentrations of VM114/115AA and Wild-type Peptides Show Different Higher-ordered Structures
As fusion peptide self-association has been hypothesized to provide a mechanism of recruitment of multiple F proteins to the site of fusion and in fusion pore formation (10, 15, 36, 48), CD spectra of both wild-type and VM114/115AA peptide were obtained under conditions that increased the effective peptide concentration. Increasing or decreasing the concentration of DPC (critical micelle concentration (CMC) of 1.1 mm), whereas keeping the peptide concentration at 100 μm will either decrease or increase the effective peptide concentration. Peptide to DPC ratios of 1:100 or lower resulted in no significant change in peptide secondary structure, suggesting that the spectra obtained at these ratios was of mostly monomeric peptide in solution (Fig. 9). As the effective peptide concentration was increased (peptide:DPC ratios from 1:100 to 1:25), wild-type peptide showed a dramatic shift to a predominately β-sheet structure (Fig. 9A). Surprisingly, at these same ratios, the VM114/115AA peptide failed to exhibit this dramatic structural shift, maintaining at least some degree of α-helical structure even at a peptide to DPC ratio of 1:25 (Fig. 9B). Combined, these data suggest that mutation of VM114/115AA to alanine affects the ability of the mutant peptides to self-interact or alters the higher order structure adopted upon FP self-interaction.
FIGURE 9.
Dependence of peptide secondary structure on effective peptide concentration. A, CD spectra of wild-type peptide at various DPC concentrations are shown. B, CD spectra of VM114/115AA peptide at various DPC concentrations are shown. Spectra in both A and B are representative of one of three independent experiments where each spectrum is an average of four scans.
DISCUSSION
Fusion peptides of enveloped virus fusion proteins serve multiple functions throughout the fusion process (10, 49). Early studies on viral FPs demonstrated that these domains can act as membrane anchors and thus have the ability to stably insert into and associate with membranes (50). Synthetic peptides corresponding to wild-type or mutant FP sequences have been used for structure determination in addition to liposome binding and fusion experiments, providing further evidence that these domains participate in membrane merger (14, 18, 25–27). Although a wealth of data exists on the FPs of HIV Env and influenza HA, little is known about the structure of paramyxovirus FPs in a membrane environment and whether particular amino acids within these domains are critical for fusion. Data presented here demonstrate that glycine residues within the FP are important for efficient expression and processing of HeV F. Furthermore, these data indicate that valine residues within the FP are key modulators of F-mediated membrane fusion, most likely by affecting the membrane-bound structure of the FP. This study also demonstrates that altering the α-helical secondary structure of the HeV F FP within a hydrophobic environment dramatically reduces membrane fusion, potentially through two mechanisms: reduced membrane disordering and altered FP-FP association.
Conserved glycine residues within viral FPs have been shown to be key modulators of membrane fusion, and our data demonstrate a role for FP glycine residues in F protein folding and stability. Failure of exogenous TPCK-trypsin cleavage in the presence of DN-Rab5 to significantly increase the amount of F1 on the surface for either GV112/113AA or G112A suggests that the G112A mutation results in misfolding around the cleavage site (Fig. 2C). Surprisingly, G112P drastically decreased cell-cell fusion despite greater-than-wild-type CSE and processing levels (Fig. 6C). Proline is strongly disruptive of α-helix and β-sheet in globular proteins; however, studies using model peptides demonstrate that proline has a helical propensity near that of alanine within a membrane-mimicking environment (51). Glycine, also a potent helix-breaker in globular proteins, promotes α-helix formation to a similar extent as alanine in a membrane environment (52–54). Given the similar helical propensities of alanine and proline in membranes, G112P might not affect the conformation of the FP once inserted into membranes but might increase the stability of the protein such that it is not triggered effectively, as proline has been shown to increase the thermostability of proteins in certain contexts (55). Mutation of a more C-terminal glycine, G117, to alanine results in inefficient protein expression and cathepsin L processing (Fig. 2B), and G117A is unable to be cleaved by exogenous TPCK-trypsin (Fig. 2C), starkly contrasting with identical mutations in HPIV3 and NDV F. Furthermore, the corresponding mutation in PIV5 F (G7A) increased membrane fusion to approximately 4 times that of wild type while reducing CSE to about 20% (33). Together these data point toward a model in which residues within the FP can modulate protein stability and/or folding. Similarly, previous work with Hendra F (56) suggested that side-chain packing beneath the fusion peptide can act in regulation of F protein triggering and the final extent of fusion. Ultimately, the phenotypic differences between glycine mutants of HeV, NDV, PIV5, and HPIV3 F underscore the essential linkage between proper folding of the prefusion metastable F protein and efficient membrane fusion promotion.
Aside from glycine residues, little is known about the role of other amino acids present within the FP in regulating membrane fusion. For all paramyxovirus FPs, the N-terminal amino acid has a large, hydrophobic side chain, typically a phenylalanine or leucine. Mutation of the first FP residue in HeV F, L110, to alanine reduced cell-cell fusion to 60–65% that of wild-type levels without altering CSE levels (Fig. 3B), indicating the slight preference for a large, hydrophobic residue in this position. Because both leucine and phenylalanine (57) at position 110 are fusion-competent, substitution with an alanine might decrease membrane interaction or alter depth or angle of membrane insertion as this would substantially reduce the hydrophobicity of the N terminus of the FP. Fitting with this idea, mutation of the N-terminal glycine (Gly-1) within the influenza HA FP to residues other than alanine significantly impaired membrane fusion (58); thus FPs from various viruses seem to have strict requirements for the N-terminal residue. The fact that the chemical properties (large and hydrophobic for the paramyxoviruses, versus small and aliphatic for influenza) of the N-terminal residue differ dramatically suggests that despite performing almost identical biological functions, viral families have evolved distinct FP structures all able to efficiently catalyze membrane fusion.
Data presented here also demonstrate that valine residues are critical in F-mediated membrane fusion. Cell-cell fusion assays show that the GV112/113AA and VM114/115AA mutants were defective for fusion, as both mutant proteins promoted membrane fusion at levels below what was expected based on CSE (Fig. 3B). Generation of the single mutants G112A, V113A, V114A, and M115A strongly indicate that the valine to alanine substitutions were largely responsible for the decreases in fusion, as both G112A and M115A drove membrane fusion at levels expected based on CSE (Fig. 6B). Although this phenotype could result from stabilization of the pre-fusion structure, the position of these mutations suggest that the reduction in fusion may be due to alterations in the ability of the FP to disrupt membranes and/or the membrane-induced secondary structure. Hemolysis data indicate that the VM114/115AA peptide had a significantly reduced ability to disrupt membranes as compared with wild type (Fig. 4B), whereas similar experiments on the two single mutants, V114A and M115A, show almost identical hemolytic tendencies (Fig. 4B). Such a disconnect between hemolytic ability and fusogenicity for the V114A and M115A peptides suggest that, although the ability of synthetic peptides to disrupt membranes often correlates with the fusogenicity of the full-length protein, membrane disruption alone is not sufficient for fusion.
Circular dichroism spectra of the synthetic FPs in DPC micelles show that substitution of valine with alanine significantly decreases α-helical content of the V114A peptide, whereas the M115A peptide has wild-type levels of α-helix (Fig. 4D). Although valine is typically thought of as a β-sheet-promoting residue in globular proteins, work by Deber and Li (52) shows that valine promotes α-helix formation to a stronger degree than alanine in the context of membrane-mimicking environments. Additionally, CD spectra of each of the peptides in POPC:POPG SUVs also demonstrate that both of the mutant peptides, V114A and VM114/115AA, adopt a less α-helical structure (Fig. 7C). This observation is supported by other studies (for review, see Ref. 14, 49)) which suggest that the α-helical structure of FPs within model membranes correlates with F-mediated membrane fusion promotion. Mechanistically, an α-helical FP structure could facilitate subsequent stages in membrane fusion including changes within the target membrane. Incubation of planar lipid bilayers with the wild-type peptide increased membrane disorder (Fig. 8), whereas incubation with the non-fusogenic VM114/115AA peptide failed to alter membrane ordering. Thus, these data provide a strong correlation between cell-cell fusion levels and membrane disordering, where only the FPs from fusogenic F proteins cause an increase in membrane disordering. Such a mechanism for FP function fits very nicely within the context of the currently accepted models for F-mediated membrane fusion, whereby the FP is thought to insert into the target cell membrane, adopt a specific secondary structure, and help to drive fusion between both the cellular and viral membranes. An increase in membrane disordering could presumably facilitate mixing between both membranes and could also help drive the stalk-to-pore transition.
In addition to changes in α-helicity modulating membrane fusion and disordering, mutations within the FP could affect potential FP-FP or FP-TMD interactions. This is an appealing hypothesis because the FPs of each F monomer are likely positioned in close proximity after extension of HRA. Furthermore, the TMD and FP are positioned on the same end of the molecule in the post-fusion structure of paramyxovirus F proteins. Thus it is possible that both FP-FP and FP-TMD interactions could regulate membrane fusion. A recent report from Degrado and co-workers (48) demonstrated that synthetic FPs of PIV5 F interact with one another in a reversible monomer-hexamer equilibrium. Additionally, they also demonstrated interactions between both isolated fusion peptides and transmembrane domains, suggesting that these two domains might associate later in the fusion process. Circular dichroism spectroscopy with the host-guest system used in this study examined the self-interaction of FPs over a range of solution ionic conditions (36). In this study, Han and Tamm (36) demonstrated that in low ionic strength solutions the synthetic FPs were monomeric. However, increasing the ionic strength of the solution led to peptide self-aggregation on the surface of model membranes and formation of an antiparallel β-sheet structure. Similar experiments, except by altering the peptide-to-lipid ratio on both VM114/115AA and wild-type peptides, demonstrated that the wild-type peptide quickly shifted to a predominately β-sheet structure as the effective peptide concentration was increased (Fig. 9). Although there was a decrease in α-helical structure for the VM114/115AA peptide as the effective peptide concentration was increased, the dramatic shift to β-sheet was absent. This lack of β-sheet formation could be interpreted in two ways; the VM114/115AA peptide self-interacts to a lesser extent than WT, or that upon interacting, these peptides adopt a different higher-ordered structure. Ultimately, additional experiments (including mutagenesis of other branched hydrophobic amino acids within the FP) will be needed to discern between these two hypotheses, but the data shown here present a model in which the Hendra F FP must adopt an α-helical structure for efficient membrane fusion promotion and target membrane disordering. Given the high sequence homology to other paramyxovirus FPs, these data also suggest that an α-helical structure is a requirement for all paramyxovirus F-mediated fusion and that mutations which alter this helicity significantly decrease fusion.
Acknowledgments
We thank Lin-fa Wang (Australian Animal Health Laboratory) for generously providing the Hendra F and G plasmids, Karl-Klaus Conzelman (Pettenkofer Institut) for providing the BSR cells, and Andrew Morris (University of Kentucky) and Michael Chika for help in preparing the DPC micelles. We also thank members of the Dutch laboratory for critically reviewing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI051517 and 2P20 RR020171 (National Center for Research Resources; to R. E. D. and T. P. C.) and R37AI30557 (to L. K. T.).
- HeV
- Hendra virus
- NiV
- Nipah virus
- F
- fusion protein
- FP
- fusion peptide
- TMD
- transmembrane domain
- HRB
- heptad repeat (HR) B
- HRA
- heptad repeat A
- DPC
- dodecylphosphocholine
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
- 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1′-rac-glycerol)
- PIV5
- parainfluenza virus 5
- SUV
- small unilamellar vesicle
- ATR-FTIR
- attenuated total reflectance-fourier transform infrared spectroscopy
- TPCK-trypsin
- L-(tosylamido-2-phenyl) ethyl chloromethyl ketone-trypsin
- CSE
- cell surface expression
- NDV
- Newcastle disease virus
- HPIV3
- human parainfluenza virus 3
- DN
- dominant negative
- CTD
- C-terminal domain.
REFERENCES
- 1. Lamb R. A., Parks G. D. (2007) in Fields Virology (Knipe D. M., Howley P. M., eds.) pp 1449–1496, Fifth Ed., Lippincott, Williams and Wilkins, Philadelphia [Google Scholar]
- 2. Eaton B. T., Broder C. C., Middleton D., Wang L. F. (2006) Hendra and Nipah viruses. Different and dangerous. Nat. Rev. Microbiol. 4, 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Sullivan J. D., Allworth A. M., Paterson D. L., Snow T. M., Boots R., Gleeson L. J., Gould A. R., Hyatt A. D., Bradfield J. (1997) Fatal encephalitis due to novel Paramyxovirus transmitted from horses. Lancet 349, 93–95 [DOI] [PubMed] [Google Scholar]
- 4. Harcourt B. H., Tamin A., Ksiazek T. G., Rollin P. E., Anderson L. J., Bellini W. J., Rota P. A. (2000) Molecular characterization of Nipah virus, a newly emergent paramxyovirus. Virology 271, 334–349 [DOI] [PubMed] [Google Scholar]
- 5. Murray K., Selleck P., Hooper P., Hyatt A., Gould A., Gleeson L., Westbury H., Hiley L., Selvey L., Rodwell B. (1995) A morbillivirus that caused fatal disease in horses and humans. Science 268, 94–97 [DOI] [PubMed] [Google Scholar]
- 6. White J. M., Delos S. E., Brecher M., Schornberg K. (2008) Structures and mechanisms of viral membrane fusion proteins. Multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43, 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asano K., Asano A. (1985) Why is a specific amino acid sequence of F glycoprotein required for the membrane fusion reaction between envelope of HVJ (Sendai virus) and target cell membranes? Biochem. Int. 10, 115–122 [PubMed] [Google Scholar]
- 8. Novick S. L., Hoekstra D. (1988) Membrane penetration of Sendai virus glycoproteins during the early stage of fusion with liposomes as determined by hydrophobic affinity labeling. Proc. Natl. Acad. Sci. U.S.A. 85, 7433–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim Y. H., Donald J. E., Grigoryan G., Leser G. P., Fadeev A. Y., Lamb R. A., DeGrado W. F. (2011) Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Natl. Acad. Sci. U.S.A. 108, 20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tamm L. K., Crane J., Kiessling V. (2003) Membrane fusion. A structural perspective on the interplay of lipids and proteins. Curr. Opin. Struct. Biol. 13, 453–466 [DOI] [PubMed] [Google Scholar]
- 11. Baker K. A., Dutch R. E., Lamb R. A., Jardetzky T. S. (1999) Structural basis for Paramyxovirus-mediated membrane fusion. Mol. Cell 3, 309–319 [DOI] [PubMed] [Google Scholar]
- 12. Yin H. S., Wen X., Paterson R. G., Lamb R. A., Jardetzky T. S. (2006) Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin H. S., Paterson R. G., Wen X., Lamb R. A., Jardetzky T. S. (2005) Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U.S.A. 102, 9288–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamm L. K., Han X. (2000) Viral fusion peptides. A tool set to disrupt and connect biological membranes. Biosci. Rep. 20, 501–518 [DOI] [PubMed] [Google Scholar]
- 15. Martin I., Ruysschaert J. M. (2000) Common properties of fusion peptides from diverse systems. Biosci. Rep. 20, 483–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han X., Tamm L. K. (2000) A host-guest system to study structure-function relationships of membrane fusion peptides. Proc. Natl. Acad. Sci. U.S.A. 97, 13097–13102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han X., Bushweller J. H., Cafiso D. S., Tamm L. K. (2001) Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 8, 715–720 [DOI] [PubMed] [Google Scholar]
- 18. Li Y., Han X., Lai A. L., Bushweller J. H., Cafiso D. S., Tamm L. K. (2005) Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. J. Virol. 79, 12065–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Durrer P., Galli C., Hoenke S., Corti C., Glück R., Vorherr T., Brunner J. (1996) H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J. Biol. Chem. 271, 13417–13421 [DOI] [PubMed] [Google Scholar]
- 20. Peisajovich S. G., Epand R. F., Pritsker M., Shai Y., Epand R. M. (2000) The polar region consecutive to the HIV fusion peptide participates in membrane fusion. Biochemistry 39, 1826–1833 [DOI] [PubMed] [Google Scholar]
- 21. Martin I., Schaal H., Scheid A., Ruysschaert J. (1996) Lipid membrane fusion induced by the human immunodeficiency virus type 1 gp41 N-terminal extremity is determined by its orientation in the lipid bilayer. J. Virol. 70, 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nieva J. L., Nir S., Muga A., Goñi F. M., Wilschut J. (1994) Interaction of the HIV-1 fusion peptide with phospholipid vesicles. Different structural requirements for fusion and leakage. Biochemistry 33, 3201–3209 [DOI] [PubMed] [Google Scholar]
- 23. Rafalski M., Lear J. D., DeGrado W. F. (1990) Phospholipid interactions of synthetic peptides representing the N terminus of HIV gp41. Biochemistry 29, 7917–7922 [DOI] [PubMed] [Google Scholar]
- 24. Sáez-Cirión A., Nieva J. L. (2002) Conformational transitions of membrane-bound HIV-1 fusion peptide. Biochim. Biophys. Acta 1564, 57–65 [DOI] [PubMed] [Google Scholar]
- 25. Rapaport D., Shai Y. (1994) Interaction of fluorescently labeled analogues of the amino-terminal fusion peptide of Sendai virus with phospholipid membranes. J. Biol. Chem. 269, 15124–15131 [PubMed] [Google Scholar]
- 26. Gregory S. M., Harada E., Liang B., Delos S. E., White J. M., Tamm L. K. (2011) Structure and function of the complete internal fusion loop from Ebola virus glycoprotein 2. Proc. Natl. Acad. Sci. U.S.A. 108, 11211–11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lai A. L., Park H., White J. M., Tamm L. K. (2006) Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J. Biol. Chem. 281, 5760–5770 [DOI] [PubMed] [Google Scholar]
- 28. Li Y., Tamm L. K. (2007) Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys. J. 93, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang J., Gabrys C. M., Weliky D. P. (2001) Solid-state nuclear magnetic resonance evidence for an extended β strand conformation of the membrane-bound HIV-1 fusion peptide. Biochemistry 40, 8126–8137 [DOI] [PubMed] [Google Scholar]
- 30. Lorieau J. L., Louis J. M., Bax A. (2010) The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc. Natl. Acad. Sci. U.S.A. 107, 11341–11346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sergel T. A., McGinnes L. W., Morrison T. G. (2001) Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J. Virol. 75, 7934–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Russell C. J., Jardetzky T. S., Lamb R. A. (2004) Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J. Virol. 78, 13727–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horvath C. M., Lamb R. A. (1992) Studies on the fusion peptide of a paramyxovirus fusion glycoprotein. Roles of conserved residues in cell fusion. J. Virol. 66, 2443–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sergel-Germano T., McQuain C., Morrison T. (1994) Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68, 7654–7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pager C. T., Wurth M. A., Dutch R. E. (2004) Subcellular localization and calcium and pH requirements for proteolytic processing of the Hendra virus fusion protein. J. Virol. 78, 9154–9163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han X., Tamm L. K. (2000) pH-dependent self-association of influenza hemagglutinin fusion peptides in lipid bilayers. J. Mol. Biol. 304, 953–965 [DOI] [PubMed] [Google Scholar]
- 37. Frey S., Tamm L. K. (1991) Orientation of melittin in phospholipid bilayers. A polarized attenuated total reflection infrared study. Biophys. J. 60, 922–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tamm L. K., Tatulian S. A. (1997) Infrared spectroscopy of proteins and peptides in lipid bilayers. Q. Rev. Biophys. 30, 365–429 [DOI] [PubMed] [Google Scholar]
- 39. Pager C. T., Craft W. W., Jr., Patch J., Dutch R. E. (2006) A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346, 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pager C. T., Dutch R. E. (2005) Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79, 12714–12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meulendyke K. A., Wurth M. A., McCann R. O., Dutch R. E. (2005) Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79, 12643–12649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diederich S., Moll M., Klenk H. D., Maisner A. (2005) The nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 280, 29899–29903 [DOI] [PubMed] [Google Scholar]
- 43. Carter J. R., Pager C. T., Fowler S. D., Dutch R. E. (2005) Role of N-linked glycosylation of the Hendra virus fusion protein. J. Virol. 79, 7922–7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stenmark H., Parton R. G., Steele-Mortimer O., Lütcke A., Gruenberg J., Zerial M. (1994) Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van den Hoogen B. G., de Jong J. C., Groen J., Kuiken T., de Groot R., Fouchier R. A., Osterhaus A. D. (2001) A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7, 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schowalter R. M., Smith S. E., Dutch R. E. (2006) Characterization of human metapneumovirus F protein-promoted membrane fusion. Critical roles for proteolytic processing and low pH. J. Virol. 80, 10931–10941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y., Han X., Tamm L. K. (2003) Thermodynamics of fusion peptide-membrane interactions. Biochemistry 42, 7245–7251 [DOI] [PubMed] [Google Scholar]
- 48. Donald J. E., Zhang Y., Fiorin G., Carnevale V., Slochower D. R., Gai F., Klein M. L., DeGrado W. F. (2011) Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc. Natl. Acad. Sci. U.S.A. 108, 3958–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Epand R. M. (2003) Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta 1614, 116–121 [DOI] [PubMed] [Google Scholar]
- 50. Paterson R. G., Lamb R. A. (1987) Ability of the hydrophobic fusion-related external domain of a paramyxovirus F protein to act as a membrane anchor. Cell 48, 441–452 [DOI] [PubMed] [Google Scholar]
- 51. Li S. C., Goto N. K., Williams K. A., Deber C. M. (1996) α-Helical, but not β-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. U.S.A. 93, 6676–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li S. C., Deber C. M. (1994) A measure of helical propensity for amino acids in membrane environments. Nat. Struct. Biol. 1, 368–373 [DOI] [PubMed] [Google Scholar]
- 53. Li S. C., Deber C. M. (1992) Influence of glycine residues on peptide conformation in membrane environments. Int. J. Pept. Protein Res. 40, 243–248 [DOI] [PubMed] [Google Scholar]
- 54. Li S. C., Deber C. M. (1992) Glycine and β-branched residues support and modulate peptide helicity in membrane environments. FEBS Lett. 311, 217–220 [DOI] [PubMed] [Google Scholar]
- 55. Matthews B. W., Nicholson H., Becktel W. J. (1987) Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc. Natl. Acad. Sci. U.S.A. 84, 6663–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith E. C., Dutch R. E. (2010) Side chain packing below the fusion peptide strongly modulates triggering of the Hendra F protein. J. Virol. 84, 10928–10932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moll M., Diederich S., Klenk H. D., Czub M., Maisner A. (2004) Ubiquitous activation of the Nipah virus fusion protein does not require a basic amino acid at the cleavage site. J. Virol. 78, 9705–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qiao H., Armstrong R. T., Melikyan G. B., Cohen F. S., White J. M. (1999) A specific point mutation at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell 10, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paterson R. G., Harris T. J., Lamb R. A. (1984) Fusion protein of the paramyxovirus simian virus 5. Nucleotide sequence of mRNA predicts a highly hydrophobic glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 81, 6706–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Richardson C., Hull D., Greer P., Hasel K., Berkovich A., Englund G., Bellini W., Rima B., Lazzarini R. (1986) The nucleotide sequence of the mRNA encoding the fusion protein of measles virus (Edmonston strain). A comparison of fusion proteins from several different paramyxoviruses. Virology 155, 508–523 [DOI] [PubMed] [Google Scholar]
- 61. McGinnes L. W., Morrison T. G. (1986) Nucleotide sequence of the gene encoding the Newcastle disease virus fusion protein and comparisons of paramyxovirus fusion protein sequences. Virus Res. 5, 343–356 [DOI] [PubMed] [Google Scholar]
- 62. Waxham M. N., Server A. C., Goodman H. M., Wolinsky J. S. (1987) Cloning and sequencing of the mumps virus fusion protein gene. Virology 159, 381–388 [DOI] [PubMed] [Google Scholar]
- 63. Hsu M., Choppin P. W. (1984) Analysis of Sendai virus mRNAs with cDNA clones of viral genes and sequences of biologically important regions of the fusion protein. Proc. Natl. Acad. Sci. U.S.A. 81, 7732–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Merson J. R., Hull R. A., Estes M. K., Kasel J. A. (1988) Molecular cloning and sequence determination of the fusion protein gene of human parainfluenza virus type 1. Virology 167, 97–105 [DOI] [PubMed] [Google Scholar]
- 65. Varsanyi T. M., Jörnvall H., Orvell C., Norrby E. (1987) F1 polypeptides of two canine distemper virus strains. Variation in the conserved N-terminal hydrophobic region. Virology 157, 241–244 [DOI] [PubMed] [Google Scholar]
- 66. Schäfer J. R., Kawaoka Y., Bean W. J., Süss J., Senne D., Webster R. G. (1993) Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology 194, 781–788 [DOI] [PubMed] [Google Scholar]